Abstract
The processes involved in sexual reproduction have been diversified during plant evolution. Whereas charales, bryophytes, pteridophytes, and some gymnosperms utilize motile sperm as male gametes, in other gymnosperms and angiosperms the immotile sperm cells are delivered to the egg cells through elongated pollen tubes. During formation of the motile sperms, cells undergo a dynamic morphological transformation including drastic changes in shape and the generation of locomotor architecture. The molecular mechanism involved in this process remains mostly unknown. Membrane trafficking fulfills the exchange of various proteins and lipids among single membrane-bound organelles in eukaryotic cells, contributing to various biological functions. RAB GTPases and SNARE proteins are evolutionarily conserved key machineries of membrane trafficking mechanisms, which regulate tethering and fusion of the transport vesicles to target membranes. Our observation of fluorescently tagged plasma membrane-resident SNARE proteins demonstrated that these proteins relocalize to spherical structures during the late stages in spermiogenesis. Similar changes in subcellular localization were also observed for other fluorescently tagged SNARE proteins and a RAB GTPase, which acts on other organelles including the Golgi apparatus and endosomes. Notably, a vacuolar SNARE, MpVAMP71, was localized on the membrane of the spherical structures. Electron microscopic analysis revealed that there are many degradation-related structures such as multi-vesicular bodies, autophagosomes, and autophagic bodies containing organelles. Our results indicate that the cell-autonomous degradation pathway plays a crucial role in the removal of membrane components and the cytoplasm during spermiogenesis of Marchantia polymorpha. This process differs substantially from mammalian spermatogenesis in which phagocytic removal of excess cytoplasm involves neighboring cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sexual reproduction is one of the most important events for a majority of multicellular organisms. Reproductive cells, which are generally classified into male and female gametes, are generated through a tightly regulated differentiation and maturation process. Male and female gametes fuse to generate zygotes, leading to the development of their progenies. The morphology and motility of male gametes have changed substantially during land plant evolution. Charales, bryophytes, lycophytes, pteridophytes and some gymnosperms generate motile sperm as male gametes, which are equipped with two or more motile flagella and move to the egg cells in water in order to accomplish fertilization. In contrast, most seed plants generate non-motile gametes without flagellum instead of sperm. The non-motile gametes are transported to the egg cells via pollen tubes during the process of fertilization.
The architecture of plant sperm has been investigated by electron microscopic studies (Carothers and Kreitner 1968; Graham and McBride 1979; Li et al. 1989; Renzaglia and Duckett 1987; Renzaglia and Garbary 2001; Ueda 1979; Vaughn and Renzaglia 2006). A common feature in plant sperm is the helically shaped cell body, which confers the name “Streptophyta” to the charophytes and land plants. The spiral morphology is rendered by the spline, which is a band of microtubules extending from the multilayered structure (MLS) located at the apical region of the sperm cell. The MLS is characteristic to sperm cells in charophytes and land plants, above which basal bodies associated with the flagella are located (Carothers and Kreitner 1968; Graham and McBride 1979; Li et al. 1989; Renzaglia and Duckett 1987). The number of flagella is divergent among land plants; bryophytes generate biflagellate sperm, while the sperm of pteridophytes and lycophytes harbor larger numbers of flagella (Renzaglia and Garbary 2001).
The liverwort, Marchantia polymorpha, is an emerging model organism representing basal land plants, whose life cycle is predominantly occupied by haploid gametophytic generations (Ishizaki et al. 2016). M. polymorpha propagates through both sexual and asexual reproduction. Sexual reproduction involves the sperm having two motile flagella as the male gamete. M. polymorpha is a dioecious species; thus, when the male plant enters the sexual reproductive phase, the reproductive organ, the antheridiophore, develops near the meristematic region of the thallus. Antheridia, which are composed of outer jacket cells and inner reproductive cells, are buried in the antheridial receptacles. Within the antheridium, spermatogenous cells divide transversely and vertically to increase the number of cells that form spermatid mother cells. The spermatid mother cell then divides diagonally to generate the spermatids, which then undergo transformation into the individual sperm (Shimamura 2016). The transformation sequence is accompanied by complete morphological alteration in a process called spermiogenesis.
Spermiogenesis in liverworts comprises various dynamic cellular events including the following: reduction of the cytoplasm, condensation of the nucleus, replacement of organelles, and formation of the locomotory apparatus. The completion of spermiogenesis culminates in the formation of mature sperm cells consisting of the helical cell body and two flagella, which are released into the water and move towards the female gametes produced in the female reproductive organ, the archegoniophore. The process of sperm development in liverworts has been of great interest to cell and developmental biologists (Higo et al. 2016; Renzaglia and Garbary 2001). However, details of cell and organelle dynamics during plant spermatogenesis remain largely unknown. This is in part because of a lack of appropriate tools with which to visualize cellular dynamics and structures in cells undergoing spermatogenesis.
Membrane trafficking enables the exchange of proteins and lipids among single membrane-bounded organelles and the plasma membrane (Uemura 2016). It also plays a critical role in the proper functioning of organelles. The molecular framework of membrane trafficking is highly conserved in eukaryotic lineages, and generally consists of several sequential processes: (1) sorting cargo molecules and the formation of transport vesicles/tubules from donor membranes, which are generally mediated by coat protein complexes; (2) dissociation of coat protein complexes and the transport of transport vesicles; (3) tethering transport vesicles to target membranes; and (4) the fusion of vesicles to target membranes (Fujimoto and Ueda 2012). These steps are mediated by evolutionarily conserved machinery components such as RAB GTPases and soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs), which are conserved machineries mediating the tethering and fusion of transport vesicles to target membranes, respectively. The RAB GTPase family is a subfamily of the Ras superfamily and functions as molecular switches by cycling between GTP-bound active and GDP-bound inactive forms. The RAB GTPases are activated by guanine nucleotide exchange factors to interact with specific effector molecules comprising tethering factors that are responsible for the docking of transport vesicles to specific target membranes (Saito and Ueda 2009; Zhen and Stenmark 2015). SNARE proteins mediate membrane fusion between transport vesicles and target membranes by assembling into tight complexes in specific combinations between vesicle-localizing members and target membrane-residing partners (Saito and Ueda 2009). Subcellular localization of RAB GTPases and SNAREs is tightly regulated to ensure specificity of membrane fusion in the membrane trafficking system. Therefore, these molecules are also regarded as determinants of organelle identity and are utilized as specific organelle markers in cell biology studies.
Recently, we conducted a comprehensive analysis of SNARE proteins and RAB GTPases encoded in the M. polymorpha genome and localized almost all members of these proteins in this organism (Kanazawa et al. 2016). In this study, we investigated organelle dynamics during spermatogenesis in M. polymorpha using SNARE and RAB proteins as organelle markers. Our observations indicated that the protein content of the plasma membrane is completely reorganized during spermiogenesis in M. polymorpha, and it is associated with cell-autonomous degradative processes of plasma membrane components and other organelles, distinctive from spermiogenesis in mammalian systems.
Materials and methods
Plant materials and growth conditions
Male accession of M. polymorpha, Takaragaike-1 (Tak-1) (Ishizaki et al. 2008), was grown on 1/2 × Gamborg’s B5 medium containing 1.4% agar at 22 °C under continuous white light. To induce sexual reproduction, thalli grown on the 1/2 × Gamborg’s B5 medium were transferred to rock wool under continuous light supplemented with far-red light as described previously (Chiyoda et al. 2008).
Constructs and transformation
The constructs for the expression of monomeric Citrine (mCitrine)-tagged MpSYP12A (mCitrine-MpSYP12A), mCitrine-MpSYP13A, and mCitrine-MpSYP2 under the regulation of native promoters were described previously (Kanazawa et al. 2016). To construct mCitrine-MpRAB5 and mCitrine-MpVAMP71, the genomic sequences comprising the 5′ sequences (3.0 kb for MpRAB5 and 6.0 kb for MpVAMP71), protein-coding regions including introns (4.1 and 1.5 kb), and 3′ flanking sequences (1.0 and 1.6 kb) were amplified by PCR and subcloned into the pENTRTM/D-TOPO vector (Invitrogen). The cDNA for mCitrine was then inserted in front of the start codon using the In-Fusion HD Cloning System (Clontech) according to the manufacturer’s instructions. The chimeric genes were introduced into pMpGWB101 or pMpGWB301 (Ishizaki et al. 2015) using the Gateway LR Clonase™ II Enzyme Mix (Invitrogen) according to the manufacturer’s instructions. We constructed the pMpGWB101 containing a promoter sequence with 4.2 kb of MpSYP2, and the cDNA for mCitrine ( pro MpSYP2:mCitrine-Gateway). The pro MpSYP2:mCitrine sequence was then amplified from the mCitrine-MpSYP2 plasmid previously described (Kanazawa et al. 2016) and inserted into the HindIII site of pMpGWB101. The cDNA for MpGOS11 was then introduced into pro MpSYP2:mCitrine-Gateway to obtain pro MpSYP2:mCitrine-MpGOS11. The primer sequences used in this study are listed in Supplementary Table 1.
Transformation was performed according to the method described previously (Kubota et al. 2013). Selection of transformants was performed with 10 mg l−1 hygromaycin B and 100 mg l−1 cefotaxime for pMpGWB101-based binary vectors and 0.5 µM chlorsulfuron and 100 mg l−1 cefotaxime for pMpGWB301-based binary vectors (Ishizaki et al. 2015).
Fluorescence microscopy
The antheridial receptacles between stages 3 and 5 (Higo et al. 2016) were sliced manually with a razor blade, placed on a glass slide and then covered with a cover slip. Antheridia were observed under the confocal microscope (LSM780, Carl Zeiss) with an oil immersion lens (×63). Spectral unmixing and processing of the obtained images were performed using ZEN2012 software (Carl Zeiss). The samples were excited at 488 nm (Argon 488) and the emission fluorescence was between 482 and 650 nm, under which the images were collected. The obtained images were processed digitally with ImageJ (National Institute of Health) and Photoshop software (Adobe Systems).
Electron microscopy
Tak-1 antheridia were collected at different stages and fixed with 2% paraformaldehyde and 2% glutaraldehyde in 0.05 M cacodylate buffer pH 7.4 at 4 °C overnight. The fixed samples were washed 3 times with 0.05 M cacodylate buffer for 30 min each and were then post-fixed with 2% osmium tetroxide in 0.05 M cacodylate buffer at 4 °C for 3 h. The samples were dehydrated in graded ethanol solutions (50 and 70% ethanol for 30 min each at 4 °C, 90% for 30 min at room temperature, 4 times with 100% for 30 min each at room temperature, and 100% overnight at room temperature). The samples were infiltrated with propylene oxide (PO) two times for 30 min each, and then placed into a 70:30 mixture of PO and resin (Quetol-651; Nisshin EM Co.) for 1 h. The caps of tubes were opened overnight to volatilize PO. The samples were transferred to fresh 100% resin and polymerized at 60 °C for 48 h. Ultra-thin sample sections were mounted on copper grids, stained with 2% uranyl acetate and lead stain solution (Sigma–Aldrich), and observed under a transmission electron microscope (JEM-1400Plus; JEOL Ltd) at an acceleration voltage of 80 kV. Digital images (2048 × 2048 pixels) were obtained using a CCD camera (VELETA; Olympus Soft Imaging Solutions GmbH).
Results
Dynamic relocalization of MpSYP12A during spermiogenesis
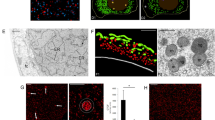
To analyze morphological changes in cells undergoing spermatogenesis, we visualized cell shapes using a fluorescent marker targeted to the plasma membrane. To achieve this we employed MpSYP12A (mCitrine-MpSYP12A), which is a member of the SYP1 group mediating membrane fusion at the plasma membrane (Kanazawa et al. 2016; Sanderfoot 2007), tagged with mCitrine. mCitrine is a monomeric version of yellow-fluorescent Citrine, which is resistant to low pH (Griesbeck et al. 2001; Shaner et al. 2005). In thallus cells, mCitrine-MpSYP12A, driven by its own promoter, was localized to the plasma membrane as we reported previously (Kanazawa et al. 2016; Fig. 1a). However, we did not detect any fluorescence from mCitrine-MpSYP12A on the plasma membrane of sperm (Fig. 1b). We then examined if we could detect mCitrine-MpSYP12A in actively dividing spermatogenous cells and found that mCitrine-MpSYP12A was expressed at this stage and was localized to the plasma membrane (Fig. 1c). These results suggested that mCitrine-MpSYP12A is degraded during sperm formation in M. polymorpha.
Subcellular localization of mCitrine-MpSYP12A and mCitrine-MpSYP13A. a mCitrine-MpSYP12A localized on the plasma membrane in thallus cells. b Differential interface contrast (DIC) and fluorescent (FL) images of a sperm cells collected from the transgenic line presented in a. Fluorescence from mCitrine-MpSYP12A was not detected in the spermatids. Green dots in the FL image are image noise. c–f Changes in subcellular localization of mCitrine-MpSYP12A during spermatogenesis. Images were aligned in a young-to-mature order from left to right. g, h Subcellular localization of mCitrine-MpSYP13A at the earlier (g) and later (h) stages of spermatogenesis. Scale bars correspond to 10 μm
To determine the stage at which mCitrine-MpSYP12A disappears from the plasma membrane, we observed different stages of spermatogenesis. Since antheridia are aligned from young to mature starting from the edge of the antheridiophore (Shimamura 2016), we observed antheridia at different developmental stages in a single antheridiophore. Plasma membrane localization of mCitrine-MpSYP12A was also observed in spermatids just after the last cell division occurred in a diagonal direction. The localization on punctate structures in the cytoplasm was also detected (Fig. 1d). When the cytoplasm of spermatids began to shrink, the plasma membrane localization of MpSYP12A was gradually decreased. Instead, large spherical structures containing mCitrine fluorescence became evident in the cytoplasm (Fig. 1e). In the later stage, MpSYP12A completely disappeared from the plasma membrane and instead accumulated in the spherical structures (Fig. 1f).
To examine whether this relocalization from the plasma membrane to the spherical structures was an event specific to MpSYP12A, we examined the subcellular localization of another SYP1 member, MpSYP13A. While mCitrine-MpSYP13A was localized almost exclusively in the plasma membrane during the proliferative phase (Fig. 1g), it accumulated only in the spherical structures at the later stages (Fig. 1h). These results indicated that the protein content of the plasma membrane was completely reorganized during spermiogenesis in M. polymorpha.
Other organelle proteins also accumulate in the spherical structures
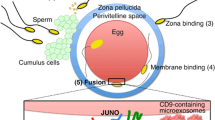
To observe the behavior of other organelles during spermatogenesis, we generated transgenic M. polymorpha expressing a fluorescently tagged Golgi-resident SNARE protein, MpGOS11, under the regulation of the MpSYP2 promoter (mCitrine-MpGOS11) (Kanazawa et al. 2016), or mCitrine-tagged endosomal RAB GTPase, MpRAB5, driven by its own promoter (mCitrine-MpRAB5). mCitrine-MpGOS11 was localized to the disc- or ring-shaped structures during the early stages of antheridia development (Fig. 2a). In the later stages of antheridia development, during spermiogenesis, mCitrine-MpGOS11 accumulated in the larger spherical structures similar to the observations with MpSYP12A and MpSYP13A (Fig. 2b). Similar translocation from punctate structures in the cytoplasm to larger spherical structures during spermiogenesis was also observed for mCitrine-MpRAB5 (Fig. 2c, d). These data suggested that the organelles and cytoplasmic components including the Golgi apparatus and endosomes were transported to the spherical structures during spermatogenesis.
Subcellular localization of membrane trafficking components during spermatogenesis. a, b Localization of mCitrine-MpGOS11 in the early (a) and late (b) stages of spermatogenesis. c, d Localization of mCitrine-MpRAB5 in the early (c) and late (d) stages of spermatogenesis. e–g Localization of mCitrine-MpVAMP71 in the early (e) and late (f, g) stages of spermatogenesis. mCitrine-MpVAMP71 remains on the membrane of the spherical structures (g). h–j Localization of mCitrine-MpSYP2 in the early (h) and late (i, j) stages of spermatogenesis. mCitrine-MpSYP2 changed its localization to the luminal space in the spherical structures during spermiogenesis (j). Scale bars correspond to 10 μm
We predicted that these spherical structures represent vacuoles because of their sizes and spherical shapes, therefore we next observed the transgenic plants expressing fluorescently tagged vacuolar SNARE proteins, MpVAMP71 (mCtirine-MpVAMP71) or MpSYP2 (mCitrine-MpSYP2). Both of these proteins were expressed under the regulation of their own promoters. Both of these SNARE proteins were localized at the vacuolar membrane during the early stages of antheridia development (Fig. 2e, h) and were localized to the membrane on the larger spherical structures in later stages (Fig. 2f, i). These results showed the vacuolar nature of the spherical structures. Intriguingly, the localization of mCitrine-MpSYP2 fluorescence changed from being limited to the membrane of the spherical structures to including the luminal spaces as spermiogenesis progressed (Fig. 2j), whereas mCitrine-MpVAMP71 remained on the membrane of the spherical structures (Fig. 2g). The distinct dynamics of the two vacuolar SNARE proteins may indicate that these two SNAREs play distinct roles during spermiogenesis.
Degradative organelles observed in spermatids undergoing spermiogenesis
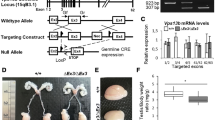
To obtain more information on the spherical structures, we performed electron microscopic observations of the developing antheridia. Antheridia from antheridiophores were divided into two groups according to their developmental stages, those in the young stage and those in the maturing stage. The samples in the young stage contained antheridia that had dividing spermatogenous cells and spermatids before shrinkage of the cytoplasm but contained no spermatids undergoing spermiogenesis (Fig. 3a). Antheridia in the maturing stage contained spermatids undergoing spermiogenesis and were associated with shrinking cytoplasm (Fig. 3b). Spermatids in the maturing stage contained large vacuole-like compartments with sizes comparable to those of the spherical structures observed in fluorescence microscopy (Fig. 3b). The multi-vesicular bodies were also observed in the spermatids (Fig. 3c), suggesting that the endocytic degradation pathway was active during spermiogenesis. Notably, autophagy-related structures such as autophagosomes (Fig. 3d) and autophagic bodies (Fig. 3e) were observed frequently in antheridia cells in the maturing stage. We also observed vacuoles containing the Golgi apparatus in the luminal space (Fig. 3f, g), suggesting that active degradation of cytoplasmic components including whole organelles occurred during spermiogenesis. The enhanced autophagic degradation during spermiogenesis was also supported by the result of quantification. A significantly larger number of autophagosomes was observed in cells in the maturing stage than in the young stage (Fig. 3h). These results indicate that autophagy became highly active and degraded cytoplasmic components during spermiogenesis in M. polymorpha. We also noted that autophagic bodies and residuals of the cytosol were present between the plasma membrane of shrinking cell bodies and the cell wall (arrowheads in Fig. 3d). These might represent remnants of cytoplasm that are internalized into the spherical structures by the autophagic processes and released during fusion between the membrane of spherical structures and the plasma membrane of spermatids.
Electron micrographs of antheridial cells in M. polymorpha. a Spermatogenous cells during proliferation phase. b Spermatids in which the cytoplasm is shrinking. Spherical structures (arrowheads) and autophagosomes (arrows) were frequently observed. c A multivesicular body in the spermatid undergoing spermiogenesis. d Autophagosomes were observed frequently in spermatids undergoing spermiogenesis. Arrowheads indicate an autophagic body-like structure (right) and remnants of the cytoplasm (left) in the paramural space. The cell wall (CW) and plasma membrane (PM) are indicated by arrows. e An autophagic body in the luminal space of a spherical structure. f A spherical structure containing the Golgi apparatus in the luminal space. g High magnification images of the Golgi apparatus shown in (f). Scale bars are 5 μm for a and b, 500 nm for c–f, and 250 nm for g. h The average number of autophagic structures in a unit area (24.9 μm × 24.9 μm). Samples were classified into two groups; young cells (young) in which the cytoplasm had not shrunk (n = 11 pictures) and maturing cells (maturing) in which the cytoplasm was shrinking (n = 8 pictures). *P < 0.0001, Student’s t test
Discussion
Reorganization of the plasma membrane during spermiogenesis of M. polymorpha
Formation of sperm cells in M. polymorpha involves complete cellular reorganization, which includes the condensation of the nucleus, reduction of cytoplasm volume, removal of unnecessary organelles, redistribution of the remaining organelles, and formation of a locomotory apparatus such as the MLS and flagella (Renzaglia and Garbary 2001). In addition to these processes, the plasma membrane of sperm cells is also reorganized during spermiogenesis to create a sperm-specific protein constitution in the plasma membrane. In this study, we initially aimed to monitor the morphological changes in cells during spermatogenesis in M. polymorpha using plasma membrane SNARE proteins as markers. Unexpectedly, we found that two plasma membrane SNARE proteins, MpSYP12A and MpSYP13A, were eliminated from the plasma membrane and was transferred to the spherical structures in the spermatids undergoing spermiogenesis. Using electron microscopy, we also detected multi-vesicular bodies in spermatids reducing their cell volume (Fig. 3c). We attempted to trace the endocytic pathway in spermatids using FM4-64 to examine whether the spherical structures indeed function in the endocytic pathway, which has not been successful because of strong autofluorecence with an overlapping spectrum with the emission from FM4-64 in antheridial receptacles of M. polymorpha. However, given that the MpSYP1 members are anchored to the plasma membrane via their carboxyl-terminal transmembrane domains, these data most likely indicated that endocytic transport is responsible for the translocation of these SNARE proteins in spermatids. Our observations also indicated that the plasma membrane of mature sperms comprises SYP1 members below the detection limit of our microscopic analysis. The low level of SNARE content possibly indicate that SNARE-mediated membrane fusion activity is quite low on the plasma membrane in sperm, if any, which might indicate that plasma membrane components required for sperm functions have already been transported before the maturation of sperm. Further analysis of sperm plasma membrane proteins, along with their identification and localization, should be conducted to gain a better understanding of the localization mechanism of components found at the sperm surface.
Autophagy is highly active during spermiogenesis
We found many autophagosomes and autophagic bodies in spermatids that were undergoing spermiogenesis. This suggested a pivotal role for autophagy in spermiogenesis in M. polymorpha. Autophagy is an evolutionarily conserved system that results in the degradation of macromolecules and organelles, and it plays important roles in various cellular functions including the recycling of nutrients and removal of damaged organelles (Nakatogawa et al. 2009). In the animal system, autophagy is required for normal developmental and differentiation processes (Mizushima and Komatsu 2011). Similarly, in plants, it has been suggested that autophagy is involved in many biological functions such as drought and salt stress resistance, senescence, and pathogen infection (Doelling et al. 2002; Hanaoka et al. 2002; Lai et al. 2011; Liu et al. 2014; Yoshimoto 2012). However autophagy-defective mutants of Arabidopsis have not been reported to have defective reproductive processes. Recently, it was reported that autophagy is required in the tapetal tissue for normal pollen development in rice (Hanamata et al. 2014; Kurusu et al. 2014). Although the mechanism of spermatogenesis in M. polymorpha has diverged from that of pollen development in rice, our result suggested that autophagy also plays an important role in spermiogenesis in M. polymorpha. Given that rapid removal of the cytoplasm occurs during sperm maturation, autophagy could be responsible for reduction of the volume of sperm cells. Further studies using autophagy-defective mutants and localization of machinery components of autophagy during spermatogenesis should be performed to clarify the possible contributions of autophagy to sperm formation in M. polymorpha.
Degradation pathways are coordinated during spermiogenesis
Our observations suggested that degradative pathways are highly activated during spermiogenesis. Large spherical structures are formed and organelle markers and organelles such as the Golgi apparatus are translocated from the cytoplasm to these structures. Autophagosomes were also frequently observed. During mammalian spermiogenesis, reduction of the cytoplasm is accomplished by the phagocytic processes by neighboring cells called Sertori cells (O’Donnell et al. 2011). In contrast, reduction of the cytoplasm has to be accomplished in a cell-autonomous manner in land plants because individual spermatids are surrounded by the cell walls, which prevent spermatids from directly connecting with neighboring cells except for the connection via the plasmodesmata. Our results strongly suggest that the active degradation pathways are responsible for the reduction of the cytoplasmic volume during spermiogenesis in M. polymorpha.
One of the questions that has yet to be answered is how the spherical structures are removed from the sperm cell body. One possible mechanism is direct fusion between the membrane of the spherical structures and the plasma membrane to dispose of contents of the structures to the paramural space. It is reported that the vacuolar membrane fuses with the plasma membrane in a proteasome-dependent manner in the plant cell and undergoes programmed cell death triggered by bacterial infection (Hatsugai et al. 2009). Secretion via the lysosomal compartments is also reported in the animal system (van der Sluijs et al. 2013). A similar mechanism might be active in spermiogenesis in M. polymorpha, consistent with previous assumptions (Renzaglia and Garbary 2001; Ueda 1979) and our observation that remnants of the cytosol and autophagic bodies were visible in the paramural space. It is also possible that the cytoplasm is removed by a mechanism similar to viral budding or exosome secretion in animal cells, which is mediated by subunits of the endosomal sorting complex required for transport (ESCRT) (Henne et al. 2011). An interesting future project would be to elucidate the exact mechanism of the cell-autonomous removal of the cytoplasm during spermiogenesis in plants.
References
Carothers ZB, Kreitner GL (1968) Studies of spermatogenesis in the Hepaticae. II. Blepharoplast structure in the spermatid of Marchantia. J Cell Biol 36:603–616
Chiyoda S, Ishizaki K, Kataoka H, Yamato KT, Kohchi T (2008) Direct transformation of the liverwort Marchantia polymorpha L. by particle bombardment using immature thalli developing from spores. Plant Cell Rep 27:1467–1473. doi:10.1007/s00299-008-0570-5
Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD (2002) The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 277:33105–33114. doi:10.1074/jbc.M204630200
Fujimoto M, Ueda T (2012) Conserved and plant-unique mechanisms regulating plant post-Golgi traffic. Front Plant Sci 3:197. doi:10.3389/fpls.2012.00197
Graham LE, McBride GE (1979) The occurrence and phylogenetic significance of a multilayered structure in Coleochaete spermatozoids. Am J Botany 66:887–894
Griesbeck O, Baird GS, Campbell RE, Zacharias DA, Tsien RY (2001) Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J Biol Chem 276:29188–29194. doi:10.1074/jbc.M102815200
Hanamata S, Kurusu T, Kuchitsu K (2014) Roles of autophagy in male reproductive development in plants. Front Plant Sci 5:457. doi:10.3389/fpls.2014.00457
Hanaoka H et al (2002) Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 129:1181–1193. doi:10.1104/pp.011024
Hatsugai N et al. (2009) A novel membrane fusion-mediated plant immunity against bacterial pathogens. Genes Dev 23:2496–2506. doi:10.1101/gad.1825209
Henne WM, Buchkovich NJ, Emr SD (2011) The ESCRT pathway. Dev Cell 21:77–91. doi:10.1016/j.devcel.2011.05.015
Higo A et al. (2016) Transcriptional framework of male gametogenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol 57:325–338. doi:10.1093/pcp/pcw005
Ishizaki K, Chiyoda S, Yamato KT, Kohchi T (2008) Agrobacterium-mediated transformation of the haploid liverwort Marchantia polymorpha L., an emerging model for plant biology. Plant Cell Physiol 49:1084–1091. doi:10.1093/pcp/pcn085
Ishizaki K et al (2015) Development of gateway binary vector series with four different selection markers for the liverwort Marchantia polymorpha. PloS One 10:e0138876. doi:10.1371/journal.pone.0138876
Ishizaki K, Nishihama R, Yamato KT, Kohchi T (2016) Molecular genetic tools and techniques for Marchantia polymorpha research. Plant Cell Physiol 57:262–270. doi:10.1093/pcp/pcv097
Kanazawa T et al. (2016) SNARE molecules in Marchantia polymorpha: unique and conserved features of the membrane fusion machinery. Plant Cell Physiol 57:307–324. doi:10.1093/pcp/pcv076
Kubota A, Ishizaki K, Hosaka M, Kohchi T (2013) Efficient agrobacterium-mediated transformation of the liverwort Marchantia polymorpha using regenerating thalli. Biosci Biotechnol Biochem 77:167–172. doi:10.1271/bbb.120700
Kurusu T et al (2014) OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy 10:878–888. doi:10.4161/auto.28279
Lai Z, Wang F, Zheng Z, Fan B, Chen Z (2011) A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J 66:953–968. doi:10.1111/j.1365-313X.2011.04553.x
Li Y, Wang FH, Knox RB (1989) Ultrastructural analysis of the flagellar apparatus in sperm cells of ginkgo-biloba. Protoplasma 149:57–63. doi:10.1007/Bf01623983
Liu Y, Xiong Y, Bassham DC (2014) Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 5:954–963. doi:10.4161/auto.5.7.9290
Mizushima N, Komatsu M (2011) Autophagy: renovation of cells and tissues. Cell 147:728–741. doi:10.1016/j.cell.2011.10.026
Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y (2009) Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 10:458–467. doi:10.1038/nrm2708
O’Donnell L, Nicholls PK, O’Bryan MK, McLachlan RI, Stanton PG (2011) Spermiation: The process of sperm release. Spermatogenesis 1:14–35. doi:10.4161/spmg.1.1.14525
Renzaglia KS, Duckett JG (1987) Spermatogenesis in Blasia pusilla: from young antheridium through mature spermatozoid. Bryologist 90:419. doi:10.2307/3243109
Renzaglia KS, Garbary DJ (2001) Motile gametes of land plants: diversity, development, and evolution. Crit Rev Plant Sci 20:107–213. doi:10.1080/20013591099209
Saito C, Ueda T (2009) Chap. 4: functions of RAB and SNARE proteins in plant life. Int Rev Cell Mol Biol 274:183–233. doi:10.1016/S1937-6448(08)02004-2
Sanderfoot A (2007) Increases in the number of SNARE genes parallels the rise of multicellularity among the green plants. Plant Physiol 144:6–17. doi:10.1104/pp.106.092973
Shaner NC, Steinbach PA, Tsien RY (2005) A guide to choosing fluorescent proteins. Nat Methods 2:905–909. doi:10.1038/nmeth819
Shimamura M (2016) Marchantia polymorpha: taxonomy, phylogeny and morphology of a model system. Plant Cell Physiol 57:230–256. doi:10.1093/pcp/pcv192
Ueda K (1979) Denshikenbikyou de Mita Syokubutsu no Kouzou. Baishukan, Tokyo, pp 220–237 (in Japanese)
Uemura T (2016) Physiological roles of plant post-golgi transport pathways in membrane trafficking. Plant Cell Physiol doi:10.1093/pcp/pcw149
van der Sluijs P, Zibouche M, van Kerkhof P (2013) Late steps in secretory lysosome exocytosis in cytotoxic lymphocytes. Front Immunol 4:359. doi:10.3389/fimmu.2013.00359
Vaughn KC, Renzaglia KS (2006) Structural and immunocytochemical characterization of the Ginkgo biloba L. sperm motility apparatus. Protoplasma 227:165–173. doi:10.1007/s00709-005-0141-3
Yoshimoto K (2012) Beginning to understand autophagy, an intracellular self-degradation system in plants. Plant Cell Physiol 53:1355–1365. doi:10.1093/pcp/pcs099
Zhen Y, Stenmark H (2015) Cellular functions of Rab GTPases at a glance. J Cell Sci 128:3171–3176. doi:10.1242/jcs.166074
Acknowledgements
We thank Dr. Atsuko Era for sharing unpublished results. This work was supported by Grants-in-Aid for Scientific Research of the Ministry of Education, Culture, Sports, Science, and Technology of Japan and by a Grant-in-Aid for JSPS Fellows (N.M. and T.K.). This research was also supported by JST, PRESTO.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Minamino, N., Kanazawa, T., Nishihama, R. et al. Dynamic reorganization of the endomembrane system during spermatogenesis in Marchantia polymorpha . J Plant Res 130, 433–441 (2017). https://doi.org/10.1007/s10265-017-0909-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-017-0909-5