Abstract
The role of gut microbiota on immune regulation and the development of autoimmune diseases such as rheumatoid arthritis (RA) is an emerging research topic. Multiple studies have demonstrated alterations on gut microbiota composition and/or function (referred to as dysbiosis) both in early and established RA patients. Still, research delineating the molecular mechanisms by which gut microorganisms induce the loss of immune tolerance or contribute to disease progression is scarce. Available data indicate that gut microbiota alterations are involved in RA autoimmune response by several mechanisms including the post-translational modification of host proteins, molecular mimicry between bacterial and host epitopes, activation of immune system and polarization toward inflammatory phenotypes, as well as induction of intestinal permeability. Therefore, in this review we analyze recent clinical and molecular evidence linking gut microbiota with the etiopathogenesis of RA. The potential of the gut microbiota as a diagnostic or severity biomarker is discussed, as well as the opportunity areas for the development of complementary therapeutic strategies based on the modulation of gut microbiota in the rheumatic patient.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rheumatoid arthritis (RA) is a systemic autoimmune disease characterized by chronic pain, joint inflammation, autoantibodies production and progressive disability [1]. This condition is associated with multiple comorbidities including cardiovascular disease (CVD), depression, infections, osteoporosis, cancer, among others [2]. The etiology of RA remains incompletely understood, but there is consensus that a complex interaction between genetic and environmental factors is involved on its development [3]. Genetic factors are estimated to contribute with up to 50% of RA risk. Among these, the allelic variants in the HLA-DRB1 gene that codes for the DRB1 chain of the human leukocyte antigen (HLA) molecule, with a common amino acid sequence known as the shared epitope (SE), are the strongest risk factor [1, 4], followed by multiple low-magnitude risk variants mostly on immunoregulatory genes located outside the HLA-region, including PTPN22, PADI4, STAT4, TNFAIP3, CTLA4, among others [4]. The remaining RA risk is attributed to environmental and lifestyle-related factors such as smoking, silica exposure, diet, infections and the oral, lung and intestinal mucosa-associated microbiota [3, 5].
Microbe infections have been associated with RA for more than a century, although the etiology of this condition could never be confirmed by the single presence of an infectious agent [6]. Among the microorganisms commonly associated with RA are bacteria such as Porphyromonas gingivalis, Proteus mirabilis, Streptococcus pyogenes and Mycobacterium, as well as Epstein-Barr virus and cytomegalovirus infection [7, 8]. In the last years, there has been a renewed interest in the study of microbes as implicated agents on the etiopathogenesis of autoimmune diseases, partly, due to the development and widespread use of techniques for massive DNA sequencing that allow the characterization of trillions of microorganisms, that is the microbiota, that resides within the human body in conditions of health and disease.
The mucosa-associated microbiota of the oral cavity, the lungs and the gastrointestinal tract is in close relationship with the immune system and mounting evidence suggests that the high load of microbial antigens to which these mucosal sites are constantly exposed to may lead to the loss of immune tolerance and the development of RA in genetically predisposed individuals [3, 5]. Intestinal dysbiosis, that is alterations on the composition and/or function of the gut microbiota, has been extensively demonstrated in early [9,10,11] and established RA patients [12, 13]. Still, data delineating the molecular mechanisms by which microorganisms can induce autoimmunity are scarce and constitute an emerging research topic. Therefore, in this article we provide an overview of the current clinical and molecular evidence linking the gut microbiota with RA etiopathogenesis. The potential of gut microbiota as a diagnostic or severity biomarker is presented, and finally, we discuss the opportunity areas for the development of complementary therapeutic approaches based on the modulation of gut microbiota in the rheumatic patient.
Search methodology
For this narrative review, original and review articles published until March 2020 were searched in the NCBI PubMed database using the terms rheumatoid arthritis OR arthritis AND one of the following terms: gut microbiota OR microbiome OR probiotics OR intestinal flora OR a specific bacterium. Studies of gut microbiota characterization by metagenomics in RA patients were systematically analyzed and are resumed in Table 1. The reference lists of some articles were considered for the identification of papers of interest. Additional information presented in the review is based on meta-analyses or systematic reviews; in the absence of such studies we searched for experimental studies, clinical trials or epidemiological studies.
Gut microbiota and immune response: ¿from the gut to the joints?
The gastrointestinal tract is an essential constituent of our immune system, as it hosts the greatest density and diversity of microorganisms of our body. Its major functions include the nutrient transport from the luminal space into the circulation and providing a selective barrier that prevents translocation of pathogenic bacteria toward internal tissues [14]. The intestinal barrier is composed by a monolayer of cells with predominantly absorptive functions, distributed from the small intestine (enterocytes) to the large intestine (colonocytes), as well as specialized subsets of cells, including Goblet cells, M cells, zymogenic cells, enteroendocrine cells and Paneth cells [15]. At the submucosa of digestive tract, there is the gut-associated lymphoid tissue (GALT), which is formed by organized cell structures (such as the Peyer patches, intestinal lymphoid follicles and the mesenteric lymph nodes), as well as individual cells interspersed trough the gut, including intestinal intraepithelial lymphocytes (IELs), macrophages, dendritic cells (DCs), natural killer (NK) cells, polymorphonuclear (PMN) leukocytes and innate lymphoid cells (ILCs) [15, 16]. The GALT is a key modulator of the immune response; capable of mounting effector responses against intestinal pathogens, while keeping immune tolerance to antigens from commensal bacteria, food and self-proteins [14, 17].
The gut microbiota encompasses trillions of bacteria, archaea, fungi, protozoa and viruses that establish a predominantly symbiotic relationship with the host [15]. It varies among individuals, as it is affected by several factors including genetics, nutrition, medication, age, environmental exposures, among others [18, 19]. Despite this, it has been documented that an adult ‘normal’ gut microbiota is predominantly colonized by bacteria belonging to the Firmicutes and Bacteroidetes phyla, while bacteria from the Proteobacteria, Actinobacteria, Verrucomicrobia and Fusobacteria phyla are found in lower proportion [18]. The genes of the microbial community (known as the microbiome) encode more than eight million bacterial proteins that work as an extension of our own genome [20], by performing hundreds of metabolic, neuro-endocrine and immunologic functions.
The gut microbiota is essential for the appropriate development of the immune system. Axenic mice (germ-free) or gnotobiotic mice (colonized with defined bacterial species) exhibit marked immune deficiencies such as lower number of intestinal Peyer patches and germinal centers, reduced numbers of intestinal CD4+ T lymphocytes and IgA secretory plasmatic cells [21,22,23]. Importantly, these altered immune responses do not only affect the GALT, but also systemic responses as evidenced by quantitative and functional impairments on circulating T helper 17 (Th17) cells and regulatory T cell subsets (Tregs) [22, 24, 25]. The gut-associated microbiota regulates the GALT directly, trough signaling of pattern recognition receptors (PRRs) expressed on immune cells that recognize conserved motifs on bacteria (known as pathogen-associated molecular pattern, PAMPs); and indirectly, through secretion of hundreds of metabolites that act as ligands on host receptors thereby modulating intestinal- and extra-intestinal cell physiology [22, 26]. As an example, short chain fatty acids (SCFA) are metabolites produced by the microbial fermentation of dietary fiber that exert potent immunomodulatory roles through its binding and activation of membrane G-protein-coupled receptors (e.g., GPR41, GPR43 and Olfr78), as well as the activation of histone acetyltransferases and deacetylases or transcription factors that regulate lymphocyte gene expression [26, 27]. SCFAs modulate cytokine production, the development of Treg cells, stimulate production of antimicrobial peptides, promote intestinal integrity, and in general, are recognized to induce tolerogenic and anti-inflammatory immune responses, but under certain circumstances, they can induce proinflammatory responses [27,28,29]. As an example, butyrate was shown to induce IL-23 secretion by dendritic cells [30] thereby inducing Th17 cell differentiation, a subtype involved on RA pathogenesis.
Immune system interaction with the gut microbiota is bidirectional and highly dynamic (reviewed in [17]); for instance, mice lacking MyD88 (a protein involved on the innate immune system sensing of microorganisms) exhibit alterations on the composition and function of their gut microbiota [31], evidencing that quantitative and functional changes on the innate or adaptive immune system may favor gut dysbiosis. As a result, it remains a challenge to determine if the intestinal dysbiosis frequently detected in RA patients is indeed a causal factor linked to the development of the disease, or rather a consequence of the plethora of immune alterations associated with RA.
Gut microbiota in the etiopathogenesis of RA
Epidemiologic evidence
The relationship between individual infectious agents and the development of RA was noted more than a century ago [6, 32] but until recent years, dysbiosis of mucosal sites is acknowledged as an important element on the model of RA etiopathogenesis [5, 22, 33]. The hygiene hypothesis (a term coined in the early 2000s) was significant in this matter, since it provided epidemiologic evidence of an inverse relationship between the frequency of infections and the frequency of allergic and autoimmune diseases at the population level [34, 35].
At present, there is robust epidemiologic and molecular evidence confirming the role of gut microbiota in the development and regulation of the immune system, and, it has even been suggested that the rise in incidence of immune-mediated diseases observed in the last decades may be due, in part, to microbial deprivation prevalent in the western world [36]. The gastrointestinal tract is essential for the maintenance of homeostatic immune responses and autoimmunity prevention by the generation of key T cell populations such as inducible regulatory T cells (iTregs) and IL17-producing T cells (Th17) [17]. Such precise balance appears to be mostly mediated by the synergistic effects of multiple bacterial strains rather than by single species [32].
Some factors associated with RA etiology such as polymorphisms in HLA gene, hormones, age, smoking and stress, all have demonstrated effects on gut microbiota composition [18, 37,38,39,40]. Furthermore, some drugs used for RA treatment have antimicrobial properties [41] and it is possible that its therapeutic effects may be partially mediated by modulation of the microbiota. Diet and obesity are associated with substantial modifications of the gastrointestinal bacteria, and, remarkably, both factors are associated with RA risk [19, 42].
Clinical evidence: gut dysbiosis in RA patients
Intestinal dysbiosis, that is, alterations in the composition and/or function of the gut microbiota, is a phenomenon associated with multiple intestinal and extra-intestinal diseases [18, 22]. A surge of metagenomic studies of gut microbiota characterization in RA have demonstrated that most patients exhibit microbial alterations, such as loss of diversity, altered functional profile or proliferation of specific bacteria mainly at the genus and species level (see Table 1). It should be noted that at present, a signature RA-associated microbial profile has been difficult to prove, probably by differences in the studies such as: (a) the techniques for gut microbiota analysis (e.g., qPCR, 16S rRNA gene sequencing or shotgun sequencing); (b) limitations on the sample size; (c) cohort characteristics (e.g., disease duration, genetic background, disease activity or therapeutic schemes) and (d) environmental exposures and lifestyle-related factors (e.g., geographic location, diet, smoking, physical activity level, among others).
A frequent finding in early RA patients is a significant increase of Prevotella genus [43] and P. copri species [9, 10, 44] in comparison to healthy controls (Table 1). This bacterium was also found augmented in preclinical stages of RA (i.e., with systemic autoimmunity and/or possible symptoms of RA) [11] but still, no longitudinal studies have been performed to evaluate the contribution of P. copri or Prevotella spp. to autoantibody production or to assess its role on the transition from preclinical to clinical RA stages. In addition, P. copri expansion was not replicated in two large metagenomic studies performed in Chinese RA patients [13, 45] which suggests these findings cannot be generalized.
Another common finding in RA is the proliferation of Bacillus and Lactobacillus. In early RA, an increase of Lactobacillus sp. was reported in fecal samples [46], while in untreated established RA patients, a significant expansion of Bacilli class and Lactobacillales order was found [47]. Similarly, a study by Zhang et al. [13] detected an increase of Lactobacillus salivarius in both oral an intestinal RA samples and its abundance was related to disease severity. Paradoxically, collagen induced arthritis (CIA) severity in mice was attenuated by oral administration of L. salivarius by decreasing bone erosions and synovial infiltration and augmenting both Treg cells and circulating IL-10 levels [48], whereas an enrichment of Lactobacillus genus was reported in CIA susceptible DBA/1 mice before arthritis development [49]. Therefore, the role of these bacteria in RA should be cautiously interpreted since this commensal genus is widely distributed on mucosal epithelium and some of its species are frequently administered as probiotics. In fact, L. casei and L. acidophilus supplementation in RA patients has shown to decrease inflammatory markers and disease activity [50, 51], while other species such as L. rhamnosus and L. reuteri did not had significant effects on clinical improvement in active RA patients [52, 53], this highlights the previous observation that physiological effects of probiotics are specific to each bacterial species [47, 54].
In summary, although studies performed to date are somewhat heterogeneous, they all demonstrate alterations in gut microbiota of RA patients both in early and established disease stages. Gut dysbiosis appears to play a key role on the initial stages of RA development [55] although longitudinal studies, especially in preclinical phases of the disease are still necessary to identify early changes of gut microbiota and to explore the mechanisms by which these contribute to RA development.
Molecular evidences
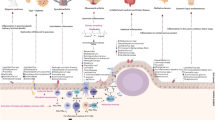
Several mechanisms by which infections or bacterial imbalances at mucosal sites can favor autoimmunity or RA progression have been proposed; in regards to gut dysbiosis these include: (a) autoantigen modification; (b) molecular mimicry between microbial and host epitopes; (c) immune system activation and polarization toward proinflammatory phenotypes; (d) induction of intestinal permeability (Fig. 1).
Molecular mechanisms by which altered-gut microbiota contributes to rheumatoid arthritis (RA) etiopathogenesis. The gut microbiota of RA patients is characterized by reduced diversity and proliferation of particular bacteria. These alterations may impact systemic immune responses and RA by several mechanisms such as: a Post-translational modification (PTM) of host proteins; oral bacteria such as Aggregatibacter actinomycetemcomitans (A.a) and Porphyromonas gingivalis augment protein citrullination by leucotoxin-A (Ltx-A) production and Porphyromonas-PAD (PPAD) enzyme activity, respectively. More than 200 citrullinated peptides are detected in colon and it is possible that some of these and other PTMs in host proteome are mediated by intestinal bacteria. Arrow indicates the dynamic relationship between gut and oral microbiomes. b Molecular mimicry; the intestinal genus Prevotella spp. has antigens that are structurally similar to an RA-citrullinated autoantigen, N-acetylglucosamine-6-sulfatase (GNS), which activates T and B cell responses in about 50% of RA patients. c Inflammatory responses; in experimental arthritis, P. copri, segmented filamentous bacteria (SFB) and Lactobacillus species have demonstrated effects on CD4+ T cells, specifically by increasing the numbers of IL-17+ Th17 cells and activating Th1 cell responses. d Intestinal permeability; in RA murine models, C. aerofaciens treatment was shown to promote intestinal permeability by increasing inflammatory mediators and chemokines (IL-17A, CXCL1, CXCL5) and reducing the expression of tight junction proteins (ZO-1 and occludin), whereas P. copri colonization increased CD4+ (IFNγ+) T cell numbers and induced colitis in mice
Post-translational modification (PTM) of self-proteins, in particular citrullination (the conversion of arginine to citrulline on a peptide chain) is a key process on RA pathogenesis. Production of antibodies against citrullinated peptides or proteins (ACPAs) can occur in preclinical stages (before clinical signs or symptoms) in a subset of patients [1, 5]. This modification is mediated by peptidylarginine deiminases (PADs) enzymes during physiological processes such as cell differentiation, apoptosis, gene expression and the formation of neutrophil extracellular traps (NETs) [56]. However, under certain circumstances, increased citrullination or “hypercitrullination” of proteins may occur and promote ACPAs production by generating multiple neo-epitopes for which T and B cell tolerance was not guaranteed [5, 56, 57]. Protein hypercitrullination may occur by overactivation of PAD enzymes, after cell membrane damage, necrosis or NETosis [56] and interestingly, those processes are boosted by environmental factors associated with RA, including smoking and specific mucosa-associated bacteria.
In the oral cavity, two citrullination-promoting bacteria were identified; Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans. The first one expresses a prokaryotic enzyme with similar catalytic function to human PADs, named Porphyromonas PAD or PPAD, that is able to citrullinate microbial and host antigens including fibrinogen and α-enolase (both frequently targeted by ACPAs) [58]. It has been hypothesized that protein citrullination by PPAD might evoke an immune response that is cross-reactive with self-peptides, leading to loss of immune tolerance and ACPA production [59]. However, this molecular mimicry mechanism for P. gingivalis is still subject to debate, as human PADs citrullinate arginine residues within polypeptide chains, while bacterial PPAD preferentially modifies C-terminal arginines [58, 60]. On the other hand, the Gram-negative A. actinomycetemcomitans, generates hypercitrullination of host proteins by production of leucotoxin A (LtxA), a bacterial toxin that forms pores on the neutrophil membrane and induces a calcium-dependent overactivation of intracellular PADs [61]. Therefore, generation of citrullinated antigens by oral bacteria could favor the production of ACPA antibodies [5, 61, 62]. Furthermore, as both microorganisms are linked to periodontal disease, such findings partly explain the epidemiological associations between RA and periodontitis [58].
It is conceivable that citrullination of neo-epitopes by P. gingivalis and A. actinomycetemcomitans is not only restricted to oral mucosa, but also may occur in distant tissues by translocation of either viable bacteria or its cellular components. Indeed, the entry of oral bacteria to systemic circulation, liver or spleen, has been demonstrated during common dental procedures [63]. Another study revealed that P. gingivalis can survive intracellularly in macrophages and DCs; therefore, immune phagocytic cells may act as “trojan horse” spreading the bacteria away from the initial infection site [64,65,66]. Moreover, detection of DNA from P. gingivalis and P. intermedia was reported in synovial fluid of RA patients with periodontitis [67, 68]. The mechanisms mediating the entry of bacterial DNA to RA joints are still unclear; however, future studies should demonstrate if bacteria-induced citrullination of proteins can occur at sites other than periodontium. Another remarkable finding that links P. gingivalis to RA pathogenesis is that, when orally administered to murine models of arthritis, it increases intestinal permeability and induces significant changes on gut microbiota composition and diversity [69, 70]. These findings highlight the close relationship between oral and gut microbiota and its effects on intestinal and systemic immune responses.
In a recent study, more than 200 citrullinated peptides were identified in colon biopsies from RA patients and healthy subjects [71]. Although this imply that protein citrullination is a general phenomenon in the gastrointestinal tract, it is possible that citrullination in the gut favor the loss of tolerance and RA development in genetically predisposed individuals; while in those who already have RA, it may promote disease severity by amplifying ACPAs production at the intestine. In this context, recent hypotheses highlight that ACPA antibodies are generated in mucosal sites in early stages of the disease [3, 5] based on several findings such as higher frequency of circulating plasmablasts and IgA ACPAs in subjects at risk of developing RA (positive to RF or ACPAs autoantibodies, in the absence of clinical symptoms) in comparison to controls [72]. ACPAs can promote RA pathogenesis by several mechanisms including immune complexes formation and induction of inflammatory responses on innate immune cells after its recognizing by Fc receptors [73], as well as induction of bone erosion by promoting osteoclastogenesis [74].
Moreover, the gut-associated microbiota likely generates other PTMs of the host proteome that can lead to production of other autoantibody systems in RA; for instance, anti-acetylated protein antibodies (anti-APAs) were recently described in approximately 35% of early RA patients and displayed a specificity from 65 to 85% [75]. Acetylation is a reversible enzymatic process that consists on the addition of acetyl groups to free amines of lysine residues. Bioinformatic studies show that most bacteria carry genes predicted to encode lysine acetyltransferases and lysine deacetylases [76]. The microbiome was shown to affect host acetylation patterns in mice [77] and strikingly, certain dietary components can modulate lysine acetylation levels in vivo [78]. Although the precise mechanisms that lead to production of anti-APAs in RA and its biologic and clinical relevance remains to be elucidated, these studies provide an interesting link between diet, gut microbiota and protein acetylation as a target for antibody response in RA.
One of the mechanisms by which the RA-associated bacteria P. copri may contribute to RA etiopathogenesis is by induction of proinflammatory immune responses [10, 79]. Germ-free SKG mice (a genetic model of autoimmune arthritis) inoculated with fecal samples from RA patients enriched with P. copri, showed increased numbers of intestinal Th17 cells and developed severe arthritis after exposure to the fungal particle zymosan [10]. While in early RA patients, P. copri-derived peptides bound to HLA-DR molecules induced Th1 type inflammatory responses in PBMCs. Furthermore, IgA and IgG anti-P.copri antibodies were detected in early and established RA patients, and its levels were correlated with Th17 cytokine profiles, ACPAs and presence of DNA from Prevotella in synovial fluid samples [79]. Unlike P. copri, another bacteria from this genus, P. histicola, was shown to induce anti-inflammatory responses in murine models of RA [80]. Therefore, physiologic roles for bacteria at the genus level should be cautiously interpreted and not generalized.
On the other hand, epitopes belonging to Prevotella spp. exhibit molecular mimicry with a citrullinated autoantigen found in RA joints (N-acetylglucosamine-6-sulfatase, GNS), providing a link between mucosal and joint immune response. Both self- and microbial-peptides were predicted to bind strongly to HLA-SE molecules, and were able to activate T and B cell responses in approximately 50% of RA patients [81]. Based on this, the authors speculated that T cells, especially those from patients carrying the SE alleles, are activated at the gut mucosa after recognition of Prevotella-derived epitopes and can subsequently migrate to inflamed joints—where there is a high expression of similar autoantigens—thereby mounting a cross-reaction to self-epitopes.
Another mechanism by which gut microbiota alterations can promote autoimmunity or RA progression is by increasing intestinal permeability [9, 12]. In a murine model of AIC, treatment with Collinsella aerofaciens (which is abundant in some RA patients) augmented the incidence and severity of arthritis [12]. C. aerofaciens promoted intestinal permeability in the epithelial cell line CACO-2 by reducing the expression of tight junction proteins (ZO-1 and occludin) while increasing the expression of the proinflammatory mediators IL-17A, CXCL1, CXCL5 and NF-κB1 [12]. In a similar way, C57BL/6 mice orally treated with P. copri exhibited increased numbers of T-CD4+ (IFNγ+) lymphocytes and developed severe colitis after treatment with dextran sulfate sodium [9]. Interestingly, individuals at risk of developing RA had reduced numbers of IFN-γ-producing ILC1 cells, whereas early RA patients showed increased numbers of IL-17-producing ILC3 cells, thus evidencing immunophenotypic alterations at the intestinal level in early stages of disease [82]. Therefore, intestinal bacteria may promote inflammation and permeability of the intestine by modulating the GALT. Loss of gut barrier integrity allows translocation of bacterial antigens to systemic circulation and may consequently activate inflammatory immune responses in extra-intestinal sites [83]. Several butyrate-producing bacteria, such as Eubacterium, Fusobacterium, Anaerostipes, Roseburia and Faecalibacterium, have shown to promote the maintenance of gut epithelial barrier integrity and anti-inflammatory response both in vitro and in vivo studies [84, 85]; therefore, represent potential candidates for restoring of gut integrity in RA.
Gut microbiota as diagnostic, severity or therapeutic response biomarker
Early RA diagnosis is of significant importance as it allows opportune therapeutic interventions to improve long-term outcomes such as joint damage. In this context, metagenome wide association studies (MWAS) are flourishing tools for the search of microbial biomarkers for disease diagnosis or prognosis [43]. Zhang and colleagues estimated the diagnostic utility of the RA-associated microbiome by using prediction models; they provided a model with an area under the curve of 0.940, a specificity of 0.922 and sensitivity of 0.838. The performance of this microbiome-based diagnostic model for RA was comparable to those based on serum biomarkers [13]. Meanwhile, another study by Scher et al. [9] identified several genes of the microbiome differentially expressed in early RA and pointed out their potential for identifying individuals at risk of developing RA.
The potential of some intestinal bacteria as disease activity or severity biomarkers is supported by their relationship with clinical parameters of RA. For instance, the phylum of Euryarchaeota was directly correlated with disease activity and was proposed as an independent risk factor for RA by multivariate analysis [47], whereas Prevotella-2 and Alloprevotella were positively correlated with the inflammatory biomarker C-reactive protein [86]. A recent study identified an increased abundance of Gammaproteobacteria and less abundance of Bifidobacterium in patients with elevated levels of inflammatory cytokines (TNF-α and IL-17) than those with lower levels, and Collinsella and Akkermansia abundance was higher in patients with active RA than those with inactive RA [45]. In another study, the abundance of Haemophilus spp. showed a negative correlation with serum anti-CCP and RF antibodies levels, while expansion of Lactobacillus salivarius was higher in the active disease RA group than in the other groups [13]. Since these studies reveal variations in gut microbiota composition in subsets of RA patients with distinct clinical features, such findings may have clinical implications for the development of new therapeutic strategies focused on modifying specific intestinal microbes or their metabolites.
Osteomicrobiology is an emerging research field that studies the relationship between microbiota and bone metabolism [87]. Bacterial antigens activate the immune system and can induce osteoclast differentiation and bone erosion by production of TNF-α, IL-17 and RANK [88]. In a mice model of CIA, P. gingivalis oral infection was shown to produce oral dysbiosis, higher bone loss and joint destruction [64]. However, to date there are no studies in RA patients demonstrating the impact of gut alterations on bone catabolism or radiographic progression.
Meanwhile, pharmacomicrobiomics is focused on studying the relationship between gut microbiota and the therapeutic response, absorption, distribution and metabolism of drugs [89]. Despite it has been reported that DMARD treatment partially reestablishes RA gut dysbiosis [13], and significant differences on gut microbiota composition were reported when comparing patients under different treatment schemes [47] (see Table 1), several aspects remain to be explored in relation to RA drugs and gut microbiome, for example, how gut microbial composition and function impact the bioavailability, toxicity or therapeutic response, which is highly heterogeneous among patients.
In summary, the usefulness of the microbiome as diagnostic, severity or therapeutic response biomarker remains scarcely explored. It should be considered that currently, the study of the microbiome is restricted to a few laboratories and its analysis requires highly trained personnel. Therefore, in the long-term, it will be necessary to replicate findings in other populations, as well as perform larger longitudinal studies and evaluate its usefulness to the clinical practice considering benefit–cost analysis.
Implications and potential of the gut microbiota for RA treatment
The gut microbiome is highly dynamic and responds rapidly to intrinsic and extrinsic stimuli of the host. Hence, it can be modulated by strategies such as the use of antibiotics, prebiotics and probiotics intake, fecal microbiota transplant (FMT) or simply, trough dietary intake [18]. In recent years, the pharmaceutical industry has shown substantial interest on the development of therapeutic products based on bacterial-cocktails or on microbiota-derived molecules for the treatment of chronic diseases associated with gut dysbiosis.
Use of probiotics in RA
Evidence about the effects of supplementation of RA patients with bacterial strains that provide health benefits, namely probiotics, is modest in general terms, as the majority of studies performed to date are limited in sample size and are heterogenous with respect to supplementation duration, the administered strain or the clinical and inflammatory outcomes measured. There is evidence of significant reductions on clinical, inflammatory biomarkers and metabolic profiles on RA patients after supplementation with Lactobacillus rhamnosus GR-1, Lactobacillus reuteri RC-14 or Lactobacillus casei 01 [50, 52], while other clinical trials did not show significant changes on clinical activity parameters [52, 53]. A recent study in adjuvant-induced arthritis (AIA) rats showed that treatment with a single bacterium (L. casei ATCC334) was able to suppress the induction of arthritis and protect bones from destruction in AIA rats by restoring the microbiome dysbiosis in the gut [90].
In general terms, probiotics supplementation in RA patients has proven to be secure but further research is still required to provide a solid rationale regarding which strains are more appropriate, taking into consideration the clinical features of the patients. Few studies have explored the effect of probiotic supplementation on the management and prevention of RA comorbidities such as depression, infections, body composition, metabolic alterations and oxidative stress [2, 91]; therefore, this creates an area of opportunity that should be further addressed in the future. Finally, characterization and evaluation of probiotics at strain level, not only genus and species, is a relevant aspect since it may yield completely opposite effects [54]. For example, E. coli includes strains that are commensal, pathogenic or carcinogenic, while P. histicola -in contrast to P. copri- has been identified as a potential probiotic in RA, as it induces an expansion of Treg cells and suppresses the development of arthritis in experimental models [80].
Diet, microbiota and RA
Epidemiologic associations between intake of specific nutrients or foods (e.g., intake of saturated fats, red meat, alcohol or sugars) with the development of RA remain controversial [92]. An explanation for such inconsistent findings may be that isolated food or nutrients only confer modest effects that are difficult to detect in epidemiologic studies, as in real context, individuals rather consume food groups that are better depicted by dietary pattern analysis [93]. A generally regarded healthy diet pattern is the Mediterranean diet, characterized by high consumption of fruit, vegetables, whole grains, legumes, fish and olive oil, low intake of red meat and moderate consumption of alcohol [94, 95]. However, even the relationship of this diet with reduced RA risk shows conflicting results [92, 94, 96]. A recent large population-based case–control study revealed lower RA risk in women who adhere to a healthy dietary pattern [93]. This can be explained by the immunomodulatory effects of fiber as well as the anti-inflammatory effects of omega-3 fatty acids intake [92]. Conversely, western diets and high intake of saturated fats have shown to promote intestinal permeability and inflammation, thereby allowing the translocation of bacterial proinflammatory components such as lipopolysaccharide (LPS) to systemic circulation [95]. During obesity, the adipose tissue releases numerous biologically active factors; including adipokines such as leptin, and inflammatory cytokines such as TNF-α, IL1-β e IL-6, that may contribute to RA pathogenesis by heightening the inflammatory response, accelerating metabolic alterations and the development of CV comorbidities [42, 97].
Diet is a source of multiple bioactive compounds with potent immunomodulatory and antioxidant effects; for example, capsaicin derived from chili has shown to bind TRPV1 receptors expressed on T lymphocytes and regulate its activation [98], while curcumin, derived from the Asian spice Curcuma longa, exerts potent antioxidant and anti-inflammatory effects trough the inhibition of nuclear transcription factor kB (NF-kB), cyclooxygenase-2 (COX-2) and inducible nitric oxide synthase (iNOS) [99]. 1,25-dihydroxyvitamin D, the active form of vitamin D; regulates the expression of hundreds of target genes and has shown to suppress B cell proliferation and antibody production, inhibit the release of Th1 and Th17 cytokines [100] and downregulate proinflammatory cytokine secretion such as IL-1α, IL-1β and IL-6 in monocyte-derived macrophages from RA patients [101]. Likewise, dietary fiber promotes intestinal SCFAs generation by gut microbiota, and these metabolites have demonstrated extensive anti-inflammatory and beneficial metabolic effects [95].
In this context, a recent clinical trial reported that dietary supplementation of RA patients with high-fiber bars during 28 days, reduced the number of circulating Treg cells, improved the Th1/Th17 ratio and reduced the levels of CTX-1, a biomarker of bone erosion [102]. Currently, the evidence of specific diets (e.g., vegetarian or restrictive diets) or nutritional supplementation with specific compounds is still limited, but clinical trials are on course to evaluate the effect of anti-inflammatory diets or Mediterranean diets on clinical activity parameters and quality of life in RA patients (NCT02941055; NCT04262505). Therefore, dietary interventions as a complement to RA therapy may be useful not only to help reestablish the dysbiosis associated to the disease, but also to improve the metabolic profile of the patients and to reduce the CV risk, which represents the major comorbidity of RA [1, 3].
Fecal microbiota transplant on RA
FMT is the transfer of fecal bacteria from a healthy donor into the gastrointestinal tract of a recipient in order to change the recipient’s microbial composition and confer a health benefit [103]. This procedure is highly efficient for the treatment of intestinal diseases, in specific, infections by Clostridium difficile. Although the regulatory aspects of FMT are still on progress on several countries, the FDA already classified it as a procedure in research phase. Nowadays, its therapeutic potential is being explored increasingly on a wide range of chronic- and extra-intestinal diseases, including autism, depression, epilepsy and metabolic syndrome [103]. As for RA, there are no data available regarding its effects on the clinical parameters or disease activity in the patients; however, a clinical trial is on course to evaluate the efficacy and security of FMT in Chinese patients with RA refractory to methotrexate treatment (registry: NCT03944096).
Conclusions and perspectives
The evidence about the role of gut microbiota on RA is constantly increasing and has led to a better understanding of the etiopathogenesis of the disease. Metagenomic studies performed to date in RA are heterogeneous, but all of them have demonstrated that gut microbiota alterations are present both in early and established stages of the disease. A prevailing challenge is to expand gut microbiota studies to other populations and further explore its potential as biomarker for clinical activity or therapeutic response. Emerging studies are also shedding light on the functional and molecular underlying mechanisms by which gut microbiota contributes to RA; however, several research areas remain barely explored, for instance, the interaction between the genome and the microbiome of RA patients, and the role of other microbiota constituents in RA etiopathogenesis, such as viruses. Since some phages can infect bacteria, it is predictable that virome alterations contribute to intestinal dysbiosis.
Gut microbiota modifications of the RA patient by the use of probiotics, prebiotics, diet or FMT as a complementary approach to drug therapy have the potential to restore gut dysbiosis, improve clinical activity parameters and metabolic profile, and reduce the CV risk, which is one of the major RA comorbidities. As we advance on understanding host–microbial interactions, and we further elucidate the molecular mechanisms by which gut microbiota contributes to RA development, progression or even therapeutic response, it will be possible to develop intervention strategies tailored to each patient, considering their clinical characteristics, genetics and environmental exposures, in order to restore not only gut microbiota composition, but to modulate specific functional and metabolic pathways with therapeutic goals.
Data availability
Not applicable.
Code availability
Not applicable.
References
McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med. 2011;365:2205–19.
Dougados M, Soubrier M, Antunez A, Balint P, Balsa A, Buch MH, et al. Prevalence of comorbidities in rheumatoid arthritis and evaluation of their monitoring: results of an international, cross-sectional study (COMORA). Ann Rheum Dis. 2014;73:62–8.
Scherer HU, Häupl T, Burmester GR. The etiology of rheumatoid arthritis. J Autoimmun. 2020;110:102400.
Okada Y, Eyre S, Suzuki A, Kochi Y, Yamamoto K. Genetics of rheumatoid arthritis: 2018 status. Ann Rheum Dis. 2019;78:446–53.
Holers VM, Demoruelle MK, Kuhn KA, et al. Rheumatoid arthritis and the mucosal origins hypothesis: protection turns to destruction. Nat Rev Rheumatol. 2018;14:542–57.
Benedek TG. The history of bacteriologic concepts of rheumatic fever and rheumatoid arthritis. Semin Arthritis Rheum. 2006;36:109–23.
Sakkas LI, Daoussis D, Liossis S-N, Bogdanos DP. The infectious basis of ACPA-positive rheumatoid arthritis. Front Microbiol. 2017. https://doi.org/10.3389/fmicb.2017.01853/full.
Balandraud N, Roudier J. Epstein-Barr virus and rheumatoid arthritis. Joint Bone Spine. 2018;85:165–70.
Scher JU, Sczesnak A, Longman RS, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife. 2013. https://doi.org/10.7554/eLife.01202.
Maeda Y, Kurakawa T, Umemoto E, Motooka D, Ito Y, Gotoh K, et al. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol. 2016;68:2646–61.
Alpizar-Rodriguez D, Lesker TR, Gronow A, Gilbert B, Raemy E, Lamacchia C, et al. Prevotella copri in individuals at risk for rheumatoid arthritis. Ann Rheum Dis. 2019;78:590–3.
Chen J, Wright K, Davis JM, et al. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016. https://doi.org/10.1186/s13073-016-0299-7.
Zhang X, Zhang D, Jia H, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21:895–905.
Ahluwalia B, Magnusson MK, Öhman L. Mucosal immune system of the gastrointestinal tract: maintaining balance between the good and the bad. Scand J Gastroenterol. 2017;52:1185–93.
Geuking MB, Köller Y, Rupp S, McCoy KD. The interplay between the gut microbiota and the immune system. Gut Microbes. 2014;5:411–8.
Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. 2016;535:65–74.
Jiao Y, Wu L, Huntington ND, Zhang X. Crosstalk between gut microbiota and innate immunity and its implication in autoimmune diseases. Front Immunol. 2020. https://doi.org/10.3389/fimmu.2020.00282/full.
Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome Med. 2016. https://doi.org/10.1186/s13073-016-0307-y.
Bokulich NA, Chung J, Battaglia T, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8:343ra82.
Li J, Jia H, MetaHIT Consortium, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32:834–41.
Tomkovich S, Jobin C. Microbiota and host immune responses: a love-hate relationship. Immunology. 2016;147:1–10.
Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–23.
Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol. 2007;19:59–69.
Östman S, Rask C, Wold AE, Hultkrantz S, Telemo E. Impaired regulatory T cell function in germ-free mice. Eur J Immunol. 2006;36:2336–46.
Ishikawa H, Tanaka K, Maeda Y, et al. Effect of intestinal microbiota on the induction of regulatory CD25 CD4 T cells. Clin Exp Immunol. 2008;153:127–35.
Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–5.
Corrêa-Oliveira R, Fachi JL, Vieira A, Sato FT, Vinolo MA. Regulation of immune cell function by short-chain fatty acids. Clin Transl Immunol. 2016;5:e73.
Singh N, Gurav A, Sivaprakasam S, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40:128–39.
Shimada Y, Kinoshita M, Harada K, et al. Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PLoS ONE. 2013;8:e80604.
Berndt BE, Zhang M, Owyang SY, et al. Butyrate increases IL-23 production by stimulated dendritic cells. Am J Physiol-Gastrointest Liver Physiol. 2012;303:G1384–92.
Wen L, Ley RE, Volchkov PY, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–13.
Picchianti-Diamanti A, Rosado MM, D’Amelio R. Infectious agents and inflammation: the role of microbiota in autoimmune arthritis. Front Microbiol. 2018. https://doi.org/10.3389/fmicb.2017.02696.
Kalinkovich A, Gabdulina G, Livshits G. Autoimmunity, inflammation, and dysbiosis mutually govern the transition from the preclinical to the clinical stage of rheumatoid arthritis. Immunol Res. 2018;66:696–709.
Ege MJ. The hygiene hypothesis in the age of the microbiome. Ann Am Thorac Soc. 2017;14:S348–53.
Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299:1259–60.
Bach JF. The hygiene hypothesis in autoimmunity: the role of pathogens and commensals. Nat Rev Immunol. 2018;18:105–20.
Savin Z, Kivity S, Yonath H, Yehuda S. Smoking and the intestinal microbiome. Arch Microbiol. 2018;200:677–84.
OToole PW, Jeffery IB. Gut microbiota and aging. Science. 2015;350:1214–5.
Kolde R, Franzosa EA, Rahnavard G, et al. Host genetic variation and its microbiome interactions within the Human Microbiome Project. Genome Med. 2018. https://doi.org/10.1186/s13073-018-0515-8.
Molina-Torres G, Rodriguez-Arrastia M, Roman P, Sanchez-Labraca N, Cardona D. Stress and the gut microbiota-brain axis. Behav Pharmacol. 2019;30:187–200.
Ogrendik M. Antibiotics for the treatment of rheumatoid arthritis. Int J Gen Med. 2013;7:43–7.
Daïen CI, Sellam J. Obesity and inflammatory arthritis: impact on occurrence, disease characteristics and therapeutic response. RMD Open. 2015;1:e000012.
Kishikawa T, Maeda Y, Nii T, et al. Metagenome-wide association study of gut microbiome revealed novel aetiology of rheumatoid arthritis in the Japanese population. Ann Rheum Dis. 2020;79:103–11.
Breban M, Tap J, Leboime A, et al. Faecal microbiota study reveals specific dysbiosis in spondyloarthritis. Ann Rheum Dis. 2017;76:1614–22.
Chiang HI, Li JR, Liu CC, et al. An association of gut microbiota with different phenotypes in chinese patients with rheumatoid arthritis. J Clin Med. 2019;8:1770.
Liu X, Zou Q, Zeng B, Fang Y, Wei H. Analysis of fecal Lactobacillus community structure in patients with early rheumatoid arthritis. Curr Microbiol. 2013;67:170–6.
Picchianti-Diamanti A, Panebianco C, Salemi S, et al. Analysis of gut microbiota in rheumatoid arthritis patients: disease-related dysbiosis and modifications induced by etanercept. Int J Mol Sci. 2018;19:2938.
Liu X, Zhang J, Zou Q, et al. Lactobacillus salivarius isolated from patients with rheumatoid arthritis suppresses collagen-induced arthritis and increases Treg frequency in mice. J Interferon Cytokine Res. 2016;36:706–12.
Liu X, Zeng B, Zhang J, et al. Role of the gut microbiome in modulating arthritis progression in mice. Sci Rep. 2016. https://doi.org/10.1038/srep30594.
Vaghef-Mehrabany E, Alipour B, Homayouni-Rad A, Sharif S-K, Asghari-Jafarabadi M, Zavvari S. Probiotic supplementation improves inflammatory status in patients with rheumatoid arthritis. Nutrition. 2014;30:430–5.
Zamani B, Golkar HR, Farshbaf S, et al. Clinical and metabolic response to probiotic supplementation in patients with rheumatoid arthritis: a randomized, double-blind, placebo-controlled trial. Int J Rheum Dis. 2016;19:869–79.
de los Angeles Pineda M, Thompson SF, Summers K, de Leon F, Pope J, Reid G. A randomized, double-blinded, placebo-controlled pilot study of probiotics in active rheumatoid arthritis. Med Sci Monit. 2011;17:CR347–54.
Hatakka K, Martio J, Korpela M, et al. Effects of probiotic therapy on the activity and activation of mild rheumatoid arthritis—a pilot study. Scand J Rheumatol. 2003;32:211–5.
Sánchez B, Delgado S, Blanco-Míguez A, Lourenço A, Gueimonde M, Margolles A. Probiotics, gut microbiota, and their influence on host health and disease. Mol Nutr Food Res. 2017;61:1600240.
Kalinkovich A, Livshits G. A cross talk between dysbiosis and gut-associated immune system governs the development of inflammatory arthropathies. Semin Arthritis Rheum. 2019;49:474–84.
Darrah E, Andrade F. Rheumatoid arthritis and citrullination. Curr Opin Rheumatol. 2018;30:72–8.
Volkov M, Schie KA, Woude D. Autoantibodies and B cells: The ABC of rheumatoid arthritis pathophysiology. Immunol Rev. 2020;294:148–63.
Farquharson D, Butcher JP, Culshaw S. Periodontitis, Porphyromonas, and the pathogenesis of rheumatoid arthritis. Mucosal Immunol. 2012;5:112–20.
Wegner N, Wait R, Sroka A, et al. Peptidylarginine deiminase from Porphyromonas gingivalis citrullinates human fibrinogen and α-enolase: Implications for autoimmunity in rheumatoid arthritis. Arthritis Rheum. 2010;62:2662–72.
Goulas T, Mizgalska D, Garcia-Ferrer I, et al. Structure and mechanism of a bacterial host-protein citrullinating virulence factor, Porphyromonas gingivalis peptidylarginine deiminase. Sci Rep. 2015. https://doi.org/10.1038/srep11969.
Konig MF, Abusleme L, Reinholdt J, et al. Aggregatibacter actinomycetemcomitans-induced hypercitrullination links periodontal infection to autoimmunity in rheumatoid arthritis. Sci Transl Med. 2016;8:369ra176.
Rosenstein ED, Greenwald RA, Kushner LJ, Weissmann G. Hypothesis: the humoral immune response to oral bacteria provides a stimulus for the development of rheumatoid arthritis. Inflammation. 2004;28:311–8.
du Teil Espina M, Gabarrini G, Harmsen HJM, Westra J, van Winkelhoff AJ, van Dijl JM. Talk to your gut: the oral-gut microbiome axis and its immunomodulatory role in the etiology of rheumatoid arthritis. FEMS Microbiol Rev. 2019;43:1–18.
Jeong SH, Nam Y, Jung H, et al. Interrupting oral infection of Porphyromonas gingivalis with anti-FimA antibody attenuates bacterial dissemination to the arthritic joint and improves experimental arthritis. Exp Mol Med. 2018;50:e460.
Carrion J, Scisci E, Miles B, et al. Microbial carriage state of peripheral blood dendritic cells (DCs) in chronic periodontitis influences DC differentiation, atherogenic potential. J Immunol. 2012;189:3178–87.
El-Awady AR, Miles B, Scisci E, et al. Porphyromonas gingivalis evasion of autophagy and intracellular killing by human myeloid dendritic cells involves DC-SIGN-TLR2 crosstalk. PLoS Pathog. 2015;11:e1004647.
Totaro M, Cattani P, Ria F, et al. Porphyromonas gingivalis and the pathogenesis of rheumatoid arthritis: analysis of various compartments including the synovial tissue. Arthritis Res Ther. 2013;15:R66.
Reichert S, Haffner M, Keyßer G, et al. Detection of oral bacterial DNA in synovial fluid. J Clin Periodontol. 2013;40:591–8.
Nakajima M, Arimatsu K, Kato T, et al. Oral administration of P. gingivalis induces dysbiosis of gut microbiota and impaired barrier function leading to dissemination of enterobacteria to the liver. PLoS ONE. 2015;10:e0134234.
Flak MB, Colas RA, Muñoz-Atienza E, Curtis MA, Dalli J, Pitzalis C. Inflammatory arthritis disrupts gut resolution mechanisms, promoting barrier breakdown by Porphyromonas gingivalis. JCI Insight. 2019;4:e125191.
Bennike TB, Ellingsen T, Glerup H, et al. Proteome analysis of rheumatoid arthritis gut mucosa. J Proteome Res. 2017;16:346–54.
Kinslow JD, Blum LK, Deane KD, et al. Elevated IgA plasmablast levels in subjects at risk of developing rheumatoid arthritis. Arthritis Rheumatol. 2016;68:2372–83.
Clavel C, Nogueira L, Laurent L, et al. Induction of macrophage secretion of tumor necrosis factor α through Fcγ receptor IIa engagement by rheumatoid arthritis–specific autoantibodies to citrullinated proteins complexed with fibrinogen. Arthritis Rheum. 2008;58:678–88.
Harre U, Georgess D, Bang H, et al. Induction of osteoclastogenesis and bone loss by human autoantibodies against citrullinated vimentin. J Clin Invest. 2012;122:1791–802.
Juarez M, Bang H, Hammar F, et al. Identification of novel antiacetylated vimentin antibodies in patients with early inflammatory arthritis. Ann Rheum Dis. 2016;75:1099–107.
Christensen DG, Xie X, Basisty N, et al. Post-translational protein acetylation: an elegant mechanism for bacteria to dynamically regulate metabolic functions. Front Microbiol. 2019. https://doi.org/10.3389/fmicb.2019.01604.
Simon GM, Cheng J, Gordon JI. Quantitative assessment of the impact of the gut microbiota on lysine -acetylation of host proteins using gnotobiotic mice. Proc Natl Acad Sci. 2012;109:11133–8.
Kim GW, Gocevski G, Wu CJ, Yang XJ. Dietary, metabolic, and potentially environmental modulation of the lysine acetylation machinery. Int J Cell Biol. 2010;2010:1–14.
Pianta A, Arvikar S, Strle K, et al. Evidence of the immune relevance of Prevotella copri, a gut microbe, in patients with rheumatoid arthritis. Arthritis Rheumatol. 2017;69:964–75.
Marietta EV, Murray JA, Luckey DH, et al. Suppression of inflammatory arthritis by human gut-derived Prevotella histicola in humanized mice. Arthritis Rheumatol. 2016;68:2878–88.
Pianta A, Arvikar SL, Strle K, et al. Two rheumatoid arthritis–specific autoantigens correlate microbial immunity with autoimmune responses in joints. J Clin Invest. 2017;127:2946–56.
Rodríguez-Carrio J, Hähnlein JS, Ramwadhdoebe TH, et al. Brief report: altered innate lymphoid cell subsets in human lymph node biopsy specimens obtained during the at-risk and earliest phases of rheumatoid arthritis. Arthritis Rheumatol. 2017;69:70–6.
Fine RL, Manfredo Vieira S, Gilmore MS, Kriegel MA. Mechanisms and consequences of gut commensal translocation in chronic diseases. Gut Microbes. 2020;11:217–30.
Zhou L, Zhang M, Wang Y, et al. Faecalibacterium prausnitzii produces butyrate to maintain Th17/Treg balance and to ameliorate colorectal colitis by inhibiting histone deacetylase 1. Inflamm Bowel Dis. 2018;24:1926–40.
Guilloteau P, Martin L, Eeckhaut V, Ducatelle R, Zabielski R, Van Immerseel F. From the gut to the peripheral tissues: the multiple effects of butyrate. Nutr Res Rev. 2010;23:366–84.
Sun Y, Chen Q, Lin P, et al. Characteristics of gut microbiota in patients with rheumatoid arthritis in Shanghai, China. Front Cell Infect Microbiol. 2019. https://doi.org/10.3389/fcimb.2019.00369.
Jones RM, Mulle JG, Pacifici R. Osteomicrobiology: the influence of gut microbiota on bone in health and disease. Bone. 2018;115:59–67.
Novince CM, Whittow CR, Aartun JD, et al. Commensal gut microbiota immunomodulatory actions in bone marrow and liver have catabolic effects on skeletal homeostasis in health. Sci Rep. 2017. https://doi.org/10.1038/s41598-017-06126-x.
Saad R, Rizkallah MR, Aziz RK. Gut pharmacomicrobiomics: the tip of an iceberg of complex interactions between drugs and gut-associated microbes. Gut Pathog. 2012;4:16.
Pan H, Guo R, Ju Y, et al. A single bacterium restores the microbiome dysbiosis to protect bones from destruction in a rat model of rheumatoid arthritis. Microbiome. 2019. https://doi.org/10.1186/s40168-019-0719-1.
Costa NT, Scavuzzi BM, Iriyoda TMV, et al. Metabolic syndrome and the decreased levels of uric acid by leflunomide favor redox imbalance in patients with rheumatoid arthritis. Clin Exp Med. 2018;18:363–72.
Philippou E, Nikiphorou E. Are we really what we eat? Nutrition and its role in the onset of rheumatoid arthritis. Autoimmun Rev. 2018;17:1074–7.
Hu Y, Sparks JA, Malspeis S, et al. Long-term dietary quality and risk of developing rheumatoid arthritis in women. Ann Rheum Dis. 2017;76:1357–64.
Hu Y, Costenbader KH, Gao X, Hu FB, Karlson EW, Lu B. Mediterranean diet and incidence of rheumatoid arthritis in women: association between a mediterranean diet and risk of RA. Arthritis Care Res. 2015;67:597–606.
Tedeschi SK, Costenbader KH. Is there a role for diet in the therapy of rheumatoid arthritis? Curr Rheumatol Rep. 2016. https://doi.org/10.1007/s11926-016-0575-y.
Johansson K, Askling J, Alfredsson L, Di Giuseppe D, EIRA Study Group. Mediterranean diet and risk of rheumatoid arthritis: a population-based case-control study. Arthritis Res Ther. 2018. https://doi.org/10.1186/s13075-018-1680-2.
Maruotti N, d’Onofrio F, Cantatore FP. Metabolic syndrome and chronic arthritis: effects of anti-TNF-α therapy. Clin Exp Med. 2015;15:433–8.
Bertin S, Aoki-Nonaka Y, de Jong PR, et al. The ion channel TRPV1 regulates the activation and proinflammatory properties of CD4 + T cells. Nat Immunol. 2014;15:1055–63.
Kunnumakkara AB, Bordoloi D, Padmavathi G, et al. Curcumin, the golden nutraceutical: multitargeting for multiple chronic diseases. Br J Pharmacol. 2017;174:1325–48.
Illescas-Montes R, Melguizo-Rodríguez L, Ruiz C, Costela-Ruiz VJ. Vitamin D and autoimmune diseases. Life Sci. 2019;233:116744.
Neve A, Corrado A, Cantatore FP. Immunomodulatory effects of vitamin D in peripheral blood monocyte-derived macrophages from patients with rheumatoid arthritis. Clin Exp Med. 2014;14:275–83.
Häger J, Bang H, Hagen M, et al. The role of dietary fiber in rheumatoid arthritis patients: a feasibility study. Nutrients. 2019;11:2392.
Antushevich H. Fecal microbiota transplantation in disease therapy. Clin Chim Acta. 2020;503:90–8.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and revision of the manuscript. ZRC performed data collection, elaborated figures and tables, and write the first draft of the manuscript. EVM, MLC and JFMV were involved on the interpretation of data and commented on all versions of the manuscript for producing the final draft. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
The authors declare that the content of the manuscript, tables and figures is original and has not been published or submitted for publication elsewhere. All authors accept the responsibility for releasing this material and agree in transferring to the Editorial Springer the respective publication rights.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Reyes-Castillo, Z., Valdés-Miramontes, E., Llamas-Covarrubias, M. et al. Troublesome friends within us: the role of gut microbiota on rheumatoid arthritis etiopathogenesis and its clinical and therapeutic relevance. Clin Exp Med 21, 1–13 (2021). https://doi.org/10.1007/s10238-020-00647-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10238-020-00647-y