Abstract
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease characterized by multiple genetic mutations. Complement 4 (C4) copy number variation (CNV) is a target-of-interest located on chromosome 6. C4 encodes for either of the two C4 paralogs, C4A or C4B, and low C4 levels have been associated with SLE activity. In this study, we conducted a meta-analysis to comprehensively understand the role of C4 CNV in SLE. Three databases (PubMed, Embase, and Web of Science) were searched for relevant studies. Two investigators independently extracted and evaluated data from eligible studies. Associations between C4 CNV and SLE were estimated by odds ratios (OR) and 95% confidence intervals (95% CI). Further analysis was conducted using the STATA 12.0 software. A total of eight case-control studies were included in the analysis with 4107 SLE patients and 5889 healthy controls. Six studies used TaqMan real-time PCR to genotype C4 CNV, with 1 study used paralog ratio test and other one used multiplex ligation-dependent probe amplification (MLPA). Lower total C4 CNV and C4A CNV were associated with SLE in the overall analysis (pooled OR: 1.55, 95% CI: 1.23–1.95; pooled OR: 1.86, 95% CI: 1.51–2.29). The subgroup analysis found that total C4 CNV and lower C4A CNV were significantly associated with SLE in Caucasians (pooled OR: 1.84, 95% CI: 1.60–2.12; pooled OR: 2.23, 95% CI:1.92–2.59). However, the association was not detected in East Asians. Lastly, SLE was not associated with C4B CNV, long C4 CNV, or short C4 CNV. The meta-analysis confirmed that lower total C4 CNV and lower C4A CNV are associated with SLE in certain populations. Future studies should consider other ethnic groups to further investigate the relationship between the C4 gene and SLE.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease characterized by the excessive production of autoantibodies directed against cell nuclear antigens. The clinical symptoms of SLE vary substantially among patients [1]. SLE is a complex disease as it affects several central components of the immune system [2]. For this reason, its exact pathogenic mechanism remains to be elucidated. However, it is believed that multiple genes likely play roles in the etiology of SLE.
In previous reports, genome-wide association studies (GWAS) had identified several genes that predispose patients to SLE when activated, such as STAT4, IRF5, TNFAIP3, PTPN22 and IL-10 [3,4,5,6]. As the principle of GWAS is based on the hypothesis of “common disease, common variations,” it is essential to remember that GWAS only detect common variations with minor variation frequencies (MAF) greater than 0.05, while rare variations and copy number variations (CNV) were ignored, which provides insight into the “missing hereditary” phenomenon [7]. The complement pathway is consisted of three pathways, for instance: the classical pathway, lectin pathway, and alternative pathway. All three pathways are involved in the SLE's pathogenesis [8, 9]. C1q is an initial active molecule of the classical pathway of complement activation. Genetic deficiencies of the C1q genes are associated with development of SLE [10]. Both complete and partial deficiencies of complement 4 (C4), C3 are risk factors for SLE. Serum C4, C3 levels have been associated with the activity of SLE [8, 11]. C4 is essential for the activation of the classical and mannose-binding lectin complement pathways. As such, C4 exerts a vital role in the integrity of innate and adaptive immune responses, including the protection against bacterial pathogens through the opsonization of target antigens and cell lysis. This is accomplished through the formation of the membrane attack complex, clearance of immune complexes and apoptotic cell debris, and the negative selection of autoreactive B cells [12]. C4 CNV is located on chromosome 6 and encodes for either of the two C4 paralogs, C4A or C4B has been the central target-of-interest in the study of SLE in recent years [13,14,15,16,17,18,19,20].
Although several research groups have tried to unveil the connection between C4 CNV and SLE, the association remains to be elucidated [13,14,15,16,17,18,19,20]. One study comes to the conclusion that lower total C4 CNV and C4A CNV were the protective factor for SLE [19], while other 7 studies have the opposite conclusion. Four studies do not find the association between C4B CNV and SLE [14, 15, 17, 18], other 2 studies think lower C4B CNV was the risk factor for SLE [16, 17], but the others have the opposite views. Hence, in this study, we have performed a meta-analysis to comprehensively understand the role of C4 CNV, including total C4 CNV, C4A CNV, C4B CNV, long C4 CNV and short C4 CNV, in SLE.
Methods
Data source
A comprehensive search of studies, using the search terms C4 CNV and SLE, was conducted in three databases, including Pubmed, Embase, and Web of Science databases, through February 1, 2019. No limitations were placed on the type of study.
Inclusion and exclusion criteria
To be included in the meta-analysis, the studies had to meet the following inclusion criteria: published prior to February 1, 2019; case-control study evaluating the relation between C4 CNV and SLE; sufficient data to estimate the odds ratio (OR) with 95% confidence intervals (95% CI); and written in English. Studies were excluded from the meta-analysis for the following reasons: case report or family-based study; lack of usable data; and studies with duplicate data.
Data extraction
In accordance with the inclusion criteria, the following items were extracted independently by two investigators. First, the study characteristics, including the first author, publication study, title, sample size, study population, and genotyping methodology, were recorded. Secondly, the number of C4 CNV in SLE patients and healthy controls was noted. Any disagreement between the investigators was resolved by discussion until a consensus was reached. If they failed to reach an agreement, a third investigator was consulted to resolve the discrepancy.
Study quality assessment
Two investigators assessed the studies that met the inclusion criteria independently using the Newcastle Ottawa Scale (NOS) [21]. Three factors were assessed in the NOS, including selection (0–4 points), comparability (0–2 points), and outcome (0–3 points). NOS scores > 6 was considered high quality.
Statistical analysis
The meta-analysis was conducted using the STATA 12.0 software (Stata Corp., College Station, TX, USA). Associations between C4 CNV and SLE were estimated by pooled OR and 95% CI. Between-study heterogeneity was assessed by the Q-test, with p value [phet] < 0.10 regarded as statistically significant heterogeneity, and I2 statistics (I2 > 50%). In the absence of significant between-study heterogeneity, the analysis was conducted using a fixed-effects model. For statistically significant heterogeneity, the random-effects model was applied. Potential publication bias was estimated by the Begg’s funnel plot and Egger’s test with the threshold p value set to 0.10. A sensitivity analysis was performed to assess the influence of each study on the pooled OR. All p values less than 0.05 were considered statistically significant.
Results
Study selection and characteristics
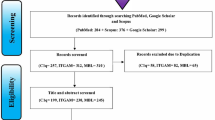
By searching the three databases, we identified 115 potential studies for the meta-analysis, including 19 from PubMed, 42 from Embase, and 54 from Web of Science. After removing duplicates and articles not relevant to our meta-analysis, 19 full-text articles were retrieved for further analysis. From the 19 articles, nine were excluded for failing to satisfy the inclusion criteria, and two were excluded for lacking usable data. Hence, a total of eight case-control studies, containing 4107 patients with SLE and 5889 healthy controls, were finally included in the meta-analysis (Fig. 1).
All the included articles were published in English. The baseline study characteristics are shown in Table 1. Six studies used TaqMan real-time PCR to genotype C4 CNV, with 1 study used paralog ratio test and other one used multiplex ligation-dependent probe amplification (MLPA).The NOS was to evaluate the quality of the enrolled studies. Among the eight studies included in the meta-analysis, two scored 8, four scored 7, one scored 6, and one scored 5 (Fig. 2).
Total C4 CNV and SLE
A total of eight studies, which contained 4107 patients with SLE and 5889 healthy controls, were included in this meta-analysis. In summary, 1152 participants had lower total C4 CNV (mutation rate: 33.96%), while that of the control group was 1152 (mutation rate: 21.83%). As the between-study heterogeneity was significant (I2 = 79.0%, phet = 0.0001), the random-effects model was used for the analysis. Next, lower total C4 CNV was found to be associated with SLE in the overall analysis (pooled OR: 1.55, 95% CI: 1.23–1.95). Four studies were conducted in Caucasians, while the others were conducted in East Asians. The subgroup analysis showed a significant association between lower total C4 CNV and SLE in Caucasians (pooled OR: 1.84, 95% CI: 1.60–2.12), yet not East Asians.
C4A CNV and SLE
Eight studies with 3393 cases and 5282 healthy controls were involved in this meta-analysis. In total, 886 participants had lower total C4 CNV (mutation rate: 26.11%), while that of the control group was 744 (mutation rate: 14.09%). As the between-study heterogeneity was significant (I2 = 67.5%, phet = 0.002), a random-effects model was used for the analysis. Next, lower C4A CNV was not found to be associated with SLE in the overall analysis (pooled OR: 1.86, 95% CI: 1.51–2.29). Four studies were conducted in Caucasians, while the others were conducted in East Asians. The subgroup analysis found the association in Caucasians (pooled OR: 2.23, 95% CI: 1.92–2.59), but not East Asians (Fig. 3).
C4B CNV and SLE
Eight studies with 3393 cases and 5276 healthy controls were involved in this meta-analysis. In total, 1003 participants had lower total C4 CNV (mutation rate: 29.56%), while that of the control group was 1350 (mutation rate: 25.59%). As the between-study heterogeneity was significant (I2 = 87.8%, phet = 0.0001), a random-effects model was used for the analysis. Next, lower C4B CNV was not found to be associated with SLE in the overall analysis (pooled OR: 1.06, 95% CI: 0.78–1.42). Four studies were conducted in Caucasians, while the others were conducted in East Asians. The subgroup analysis was unable to detect any associations in the Caucasians or East Asians.
Long C4 CNV and SLE
Four studies with 2381 cases and 2983 healthy controls were involved in this meta-analysis. In total, 60 participants had lower total C4 CNV (mutation rate: 2.52%), while that of the control group was 73 (mutation rate: 2.45%). As the between-study heterogeneity was significant (I2 = 85.4%, phet = 0.0001), a random-effects model was used for the analysis. Less of the long C4 CNV was not found to be associated with SLE in the overall analysis (pooled OR: 0.85, 95% CI: 0.25–2.91).
Short C4 CNV and SLE
Four studies with 2382 cases and 2983 healthy controls were involved in this meta-analysis. In total, 1557 participants had lower total C4 CNV (mutation rate: 65.37%), while that of the control group was 1731 (mutation rate: 58.03%). As the between-study heterogeneity was significant (I2 = 68.3%, phet = 0.024), a random-effects model was used for the analysis. Short C4 CNV was not found to be associated with SLE in the overall analysis (pooled OR: 1.13, 95% CI: 0.88–1.46).
Publication bias and sensitivity analyses
The funnel plot did not show any significant signs of symmetry for total C4 CNV, C4A CNV, C4B CNV, long C4 CNV, or short C4 CNV (Fig. 4). Similarly, the Egger’s test was also conducted and implied the absence of publication bias (p = 0.12, 0.47, 0.75, 0.73, 0.73, respectively). In addition, sensitivity analyses were also conducted. There was no single study that dominated the overall pooled results (Fig. 5), which further indicated the stability and reliability the results of the analyses were stable and reliable.
Discussion
There have been conflicting results about the relationship between C4 CNV and SLE in previous studies [13,14,15,16,17,18,19,20]. One study comes to the conclusion that lower total C4 CNV and C4A CNV were the protective factor for SLE [19], while other 7 studies have the opposite conclusion. Four studies do not find the association between C4B CNV and SLE [14, 15, 17, 18], other 2 studies think lower C4B CNV was the risk factor for SLE [16, 17], but the others have the opposite views. However, in the current study, our findings demonstrate that lower C4A and total C4 CNV are associated with SLE, whereas there SLE was not found to be associated with low C4B CNV, short C4 CNV, or long C4 CNV. The disparity could be attributed to several reasons, including the complexity of the C4 gene itself, differences in SLE patients, and differences in genotyping methods for C4 CNV.
The C4 gene, which is located on chromosome 6, is part of the segmental duplication of the RCCX module. The C4 genes showed significant CNV. It encodes for either of the two C4 paralogs, C4A or C4B, which has 99% sequence similarity in 41 exons and is differentiated by five conserved nucleotide changes in exon 26, causing four isotype-specific amino acid substitutions at positions 1101 to 1106: PCPVLD for C4A and LSPVIH for C4B. Also, the amino acid substitutions result in different biochemical activities for C4A and C4B [22]. C4A has a longer half-life and higher affinity for amino groups, suggesting a role in the clearance of immune complexes. However, C4B binds more effectively to hydroxyl groups, resulting in a reduced half-life, as compared to C4A, and implies a possible role in the membrane attack complex formation and defense against bacterial pathogens [23]. The current study detected an association between low C4A CNV and SLE, but not C4B. However, the exact molecular mechanism still requires further investigation.
It is important to mention the several caveats of this meta-analysis. First, publication bias due to the tendency of researchers to publish positive results and disregard negative findings is a well-established and inherent problem when interpreting the results of any meta-analysis [24]. Although no significant publication bias for individual mutations was apparent in this study, a selective reporting bias could affect the results, despite the evaluation at multiple genetic markers if only selected subgroups with the most significant results were reported. Secondly, the quality of the studies included in this meta-analysis could influence the reliability of the study. The NOS scores, which were used to estimate the quality of each study, ranged from five to eight, indicating that the studies were of sufficient quality. Thirdly, the study was performed by combining the results of retrospective case-control studies. Therefore, although the results of the study are highly provocative, they must be interpreted carefully.
In summary, our study demonstrates that low total C4 CNV and C4A CNV are associated with SLE in Caucasians, but not in East Asians. In addition, C4B CNV, short C4 CNV, and long C4 CNV were not associated with SLE. However, the exact mechanism by which SLE is associated with C4 CNV and low serum C4 requires further investigation.
References
Javinani A, Ashraf-Ganjouei A, Aslani S, et al. Exploring the etiopathogenesis of systemic lupus erythematosus: a genetic perspective. Immunogenetics. 2019;71:283–97.
Dammacco R. Systemic lupus erythematosus and ocular involvement: an overview. Clin Exp Med. 2018;18(2):135–49.
Chen L, Morris DL, Vyse TJ. Genetic advances in systemic lupus erythematosus: an update. Curr Opin Rheumatol. 2017;29(5):423–33.
Teruel M, Alarcon-Riquelme ME. The genetic basis of systemic lupus erythematosus: what are the risk factors and what have we learned. J Autoimmun. 2016;74:161–75.
Machado-Contreras JR, Muñoz-Valle JF, Cruz A, et al. Distribution of PTPN22 polymorphisms in SLE from western Mexico: correlation with mRNA expression and disease activity. Clin Exp Med. 2016;16(3):399–406.
Palafox-Sánchez CA, Oregon-Romero E, Salazar-Camarena DC, et al. Association of interleukin-10 promoter haplotypes with disease susceptibility and IL-10 levels in Mexican patients with systemic lupus erythematosus. Clin Exp Med. 2015;15(4):439–46.
Bomba L, Walter K, Soranzo N. The impact of rare and low-frequency genetic variants in common disease. Genome Biol. 2017;18(1):77.
Macedo AC, Isaac L. Systemic lupus erythematosus and deficiencies of early components of the complement classical pathway. Front Immunol. 2016;7:55.
Chen M, Daha MR, Kallenberg CG. The complement system in systemic autoimmune disease. J Autoimmun. 2010;34(3):J276–286.
Son M, Diamond B, Santiago-Schwarz F. Fundamental role of C1q in autoimmunity and inflammation. Immunol Res. 2015;63(1–3):101–6.
Sandhu V, Quan M. SLE and serum complement: causative, concomitant or coincidental? The Open Rheumatol J. 2017;11:113–22.
Mortensen S, Kidmose RT, Petersen SV, et al. Structural basis for the function of complement component C4 within the classical and lectin pathways of complement. J Immunol. 2015;194(11):5488–96.
Juptner M, Flachsbart F, Caliebe A, et al. Low copy numbers of complement C4 and homozygous deficiency of C4A may predispose to severe disease and earlier disease onset in patients with systemic lupus erythematosus. Lupus. 2018;27(4):600–9.
Tsang A, Sjoe MWP, Bultink IEM, Korswagen LA, et al. Comprehensive approach to study complement C4 in systemic lupus erythematosus: Gene polymorphisms, protein levels and functional activity. Mol Immunol. 2017;92:125–31.
Chen JY, Wu YL, Mok MY, et al. Effects of complement C4 gene copy number variations, size dichotomy, and C4A deficiency on genetic risk and clinical presentation of systemic lupus erythematosus in East Asian populations. Arthritis Rheumatol (Hoboken, NJ). 2016;68(6):1442–533.
Kim JH, Jung SH, Bae JS, et al. Deletion variants of RABGAP1L, 10q21.3, and C4 are associated with the risk of systemic lupus erythematosus in Korean women. Arthritis Rheum. 2013;65(4):1055–63.
Boteva L, Morris DL, Cortes-Hernandez J, et al. Genetically determined partial complement C4 deficiency states are not independent risk factors for SLE in UK and Spanish populations. Am J Hum Genet. 2012;90(3):445–56.
Lv Y, He S, Zhang Z, et al. Confirmation of C4 gene copy number variation and the association with systemic lupus erythematosus in Chinese Han population. Rheumatol Int. 2012;32(10):3047–53.
Kamatani Y, Matsuda K, Ohishi T, et al. Identification of a significant association of a single nucleotide polymorphism in TNXB with systemic lupus erythematosus in a Japanese population. J Hum Genet. 2008;53(1):64–73.
Yang Y, Chung EK, Wu YL, et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am J Hum Genet. 2007;80(6):1037–54.
Lo CK, Mertz D, Loeb M. Newcastle-Ottawa Scale: comparing reviewers' to authors' assessments. BMC Med Res Methodol. 2014;1(14):45.
Yu CY, Belt KT, Giles CM, et al. Structural basis of the polymorphism of human complement components C4A and C4B: gene size, reactivity and antigenicity. The EMBO J. 1986;5(11):2873–81.
Dodds AW, Ren XD, Willis AC, et al. The reaction mechanism of the internal thioester in the human complement component C4. Nature. 1996;379(6561):177–9.
Carroll HA, Toumpakari Z, Johnson L, et al. The perceived feasibility of methods to reduce publication bias. PLoS ONE. 2017;12(10):e0186472.
Acknowledgements
This research was supported by grants from the National Natural Science Foundation of China Grants (81671618, 81871302, 81801631, and 81771661), the Capital Health Research and Development of Special Grants (2014-1-4011), and the CAMS Initiative for Innovative Medicine (2017-I2M-3-001 and 2017-I2M-B&R-01).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wu, Z., Zhang, S., Li, P. et al. Association between complement 4 copy number variation and systemic lupus erythematosus: a meta-analysis. Clin Exp Med 20, 627–634 (2020). https://doi.org/10.1007/s10238-020-00640-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10238-020-00640-5