Abstract
Lake Baikal’s ecosystem is among the oldest lacustrine ecosystems on the planet with a diverse and unique flora and fauna. Amphipods (Amphipoda, Crustacea) are one of the most diverse groups among the lake’s invertebrate animals. The lake contains more than 350 species and subspecies of amphipods, and each of these species play a significant role in the lake’s food webs. However, their relationships with microorganisms are poorly investigated, which leaves gaps in understanding the relations between aquatic crustaceans and bacteria. We studied the diversity of a specific group of culturable microorganisms (Actinobacteria), and their possible relations with deepwater endemic amphipods present in Lake Baikal. We found that while 73% of the isolated strains belong to the widely distributed genus of Streptomyces, 27% of the isolates belong to rare genera such as Amycolatopsis, Micromonospora, and Pseudonocardia. Fifty-three percent of the studied strains expressed antibiotic activity against Gram-positive bacteria and possess polyketide synthase (PKS) genes. Thus, Actinobacteria associated with Baikal’s endemic deepwater amphipods might both protect amphipods against pathogenic microorganisms and be an untapped source of new natural products with biosynthetic potential.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bacteria are widely distributed in nature, in both free-living forms and biofilms. The latter is a community of microbes consisting often of different species that are attached to various surfaces, and coated by secreted polymers (Nadell et al. 2009). The transition from one form to another, i.e., free living to the community, requires dramatic shifts in gene transcription, cellular processes, and physiology that are accompanied by diversification among the progeny (Kolter and Greenberg 2006). In aquatic environments, bacteria can be associated with phyto- and zoo-plankton, as well as animals of higher taxa. Attached forms are characterized by different physical appearance and metabolic activity, in comparison with free-living forms (Grossart et al. 2007). Additionally, attached forms are more prone to exhibit interference competition (direct intra- and interspecies negative interactions via predation, chemical competition, etc.) than their free-living counterparts, at least in marine environments (Long and Azam 2001).
Aquatic crustaceans, their gut and chitin surfaces represent nutrient-rich microenvironments in comparison to the surrounding water (Tang 2005). It is especially true for oligotrophic water bodies where resources are scarce. For example, bacteria associated with the copepod Thermocyclops oithonoides showed higher affinity to the copepod surfaces in oligotrophic rather than in eutrophic waters (Grossart et al. 2009). In addition to providing nutrients, crustaceans are “safe harbors”, protecting bacteria from UV and ozone (Tang et al. 2011). Also, migration of zooplankton in the water column allows bacteria to move among areas with different nutrient content, oxygen level, or temperature (Grossart et al. 2010). Chitin, the most abundant natural polymer in aquatic environments, is the main component of the crustacean exoskeleton. It provides a surface for the attachment of microorganisms and it can be used as a source of carbon and nitrogen. Another niche inhabited by bacteria is the digestive tract of crustaceans, which provides both oxygenated and anoxic conditions. For a long time, it has been known that marine crustaceans possess specific microbiota associated with both digestive tracts (Nagasawa and Nemoto 1988; Dempsey et al. 1989; Harris 1993; Ampe and Thiery 1998) and chitin surfaces (Sochard et al. 1979; Holland and Hergenrader 1981; Carman and Dobbs 1997). The role of this microbiota in the life of crustaceans is still not well understood. However, a few examples suggest nutritional (Schmidt et al. 2008) and protective hypotheses (Gil-Turnes et al. 1989; Gil-Turnes and Fenical 1992).
Amphipods (Amphipoda, Crustacea) are a large group of aquatic crustaceans characterized by direct development and the lack of a larval stage (Väinölä et al. 2008). As mesograzers, they play a significant role in aquatic food webs. Most amphipod species inhabit oceans while about 20% of all amphipod diversity is found in freshwaters. There are a few hot spots where amphipods are most diverse—Southern Europe and Southern Australia, the Ponto-Caspian basin, and Lake Baikal. The latter accounts for 4.3% of the world’s amphipod fauna including 276 species and 78 subspecies (Takhteev et al. 2015). Amphipods are one of the most evolutionary successful groups among Lake Baikal’s animals inhabiting all depths of the lake (down to 1642 m) (Sitnikova and Mekhanikova 2014). About one third of amphipods inhabiting Lake Baikal undertake daily vertical migration (Karnaukhov et al. 2016; Takhteev et al. 2019). While the ecology, taxonomy, physiology, and biochemistry of the Lake Baikal amphipods are well studied, little is known about their symbiotic relations with microorganisms. However, long evolution and geographic isolation in a stable environment might have led to the development of unusual biochemical traits such as protective secondary metabolites synthesized by bacteria. Those metabolites may be involved in host–symbiont interactions that play an important role in the crustacean’s life history.
Colonization of chitin surfaces of the crustaceans typically occurs unevenly. Urothoe posedonis amphipods living in marine sediments or in burrows of other invertebrates have epibiotic bacteria (genus Thiothrix) on their walking appendices (Gillan and Dubilier 2004; Gillan et al. 2004). The main sites of marine isopods inhabited by epibionts are the anal and oral regions (Carman and Dobbs 1997). Previously it was shown that some Lake Baikal amphipods had cuticular nonsensory microstructures and the surface of their bodies was covered by colonies of microorganisms (Mekhanikova and Takhteev 2008). Recent electron microscopy scanning of deepwater amphipods inhabiting the Frolikha underwater hydrothermal vent (the northeastern part of the lake Baikal) revealed a high number of ciliates on their exoskeleton along with unknown bacteria (Khalzov et al. 2018).
Actinobacteria are one of the most abundant taxa in freshwaters (Glockner et al. 2000). They are prolific producers of multiple natural products, many of which are widely used as therapeutics (Bérdy 2012). While symbiotic interactions between terrestrial arthropods (e.g., insects) and Actinobacteria are well studied, understanding of the relationships between aquatic crustaceans and Actinobacteria is still in the infant stage. Examples of insect–actinobacteria interactions include fungus-growing ants and Pseudonocardia sp., and solitary wasps and Streptomyces spp. These exemplify the widespread symbiotic relations between insects and Actinobacteria (Cafaro et al. 2011; Seipke et al. 2012). At the same time, examples of defensive symbiosis between crustaceans and bacteria in aquatic environments are scarce, e.g., embryos of the shrimp Palaemon macrodactylus and the lobster Homarus americanus are protected by small molecules produced on the egg surface by surface-associated Gram-negative bacteria (Gil-Turnes et al. 1989; Gil-Turnes and Fenical 1992). The protective compounds showed moderate activity against the main parasitic agent of both crustaceans, the fungus Lagenidium callinectes. The bacterial population density might be responsible for high concentrations of antifungal compounds or exclusive activity against particular fungi species. To our knowledge, the biosynthetic genes responsible for the synthesis of these protective compounds active are unknown, despite numerous studies investigating biosynthetic genes in Actinobacteria (Bilyk and Luzhetskyy 2016).
Antifungal nystatin-like compounds synthesized by some Pseudonocardia strains associated with leafcutter ants were encoded by polyketide synthase (PKS) genes (Holmes et al. 2016; Van Arnam et al. 2016). Holmes and colleagues showed that the Pseudonocardia genomes also contain non-ribosomal peptide synthetase (NRPS) clusters as well as hybrid NRPS–PKS ones. PKS and NRPS clusters are responsible for the synthesis of secondary metabolites, some of which are widely used in medicine and veterinary practice as antibiotics, anticancer drugs, antivirals, and immunosuppressive agents. However, the analysis of the secondary metabolite gene clusters is not readily available in many cases, so a PCR detection method might be applied for a first screening (Sun et al. 2015; Passari et al. 2018).
The aim of the current study was to investigate the biotechnological potential of culturable Actinobacterial strains associated with different species of endemic amphipods from a wide range of depth (126–300 m) and two different locations in Lake Baikal. Previously, we isolated Actinobacteria strains from deepwater amphipods of the genus Ommatogammarus (Protasov et al. 2017). In our current study, we focused on the secondary metabolite genes that might be responsible for antibiotic activities. Also, we collected different species of amphipods to compare the diversity of culturable Actinobacteria strains and their antibiotic activity.
Materials and methods
Animals and sampling
Several amphipod species were collected from the southern part of Lake Baikal on the opposite sides of the lake between July 2014 and March 2015 (Fig. 1, Table 1). Amphipods of the species Pallasea brandtii flaviceps (Dorogostaisky, 1922), Crypturopus tuberculatus (Dybowsky, 1874), and Acanthogammarus godlewskii (Dybowsky, 1874) were collected by bottom trawling on the southeastern shore near Mishikha village (51.4147°N, 105.40083°E). Eulimnogammarus ussolzewii (Dybowsky, 1874), E. aheneus (Dybowsky, 1874), Odontogammarus calcaratus pulcherrimus (Dorogostaisky, 1930), Ommatogammarus carneolus malanophthalmus (Dybowsky, 1874) were collected using traps baited with dead fish on the southwestern shore near Bolshie Koty village (51.9053°N, 105.0753°E).
Lake Baikal amphipod species from the Odontogammarus and Ommatogammarus genera are mostly benthic scavengers searching the bottom of the lake for dead animals (Takhteev 2000a). Different Acanthogammarus species are also bottom dwellers, but they can regularly migrate into the pelagic zone. These species have a niche similar to the marine scavenger amphipod species Hirondellea gigas and many specimens of the family Lysianassidae (Takhteev 2000b). Pallasea brandtii is an herbivorous species, usually inhabiting the littoral zone (Takhteev 2000a). Crypturopus tuberculatus is a detritivore mostly living in the 1.5–99 m depth range (Romanova et al. 2016). Specimens of the genus Eulimnogammarus inhabit both littoral/sublittoral and deepwater areas of the lake (Bedulina et al. 2014).
The sampling depth varied from 126 to 300 m. After the samples were collected, the amphipods were rinsed with 70% ethanol followed by sterile water (three times) to remove sediment particles and unattached microorganisms. The samples were then transferred into a 20% sterile glycerol solution and homogenized. In the case of small amphipods, we combined specimens of the same species in one sample for further inoculation. The glycerol stock was kept in the refrigerator at − 20 °C until inoculation.
Isolation of actinobacteria
Each sample (100 µL) was plated onto several selective media: MS (soy flour—20 g/L, d-mannitol—20 g/L, agar—20 g/L, pH 7.2), ISP 2 (yeast extract—4 g/L, malt extract—10 g/L, dextrose 4 g/L, agar—20 g/L, pH 7.2), starch–ammonia agar SAA ((NH4)2SO4)—2 g/L, K2HPO4—1 g/L, MgSO4—1 g/L, NaCl—1 g/L, CaCO3—3 g/L, starch—10 g/L, agar—20 g/L), Gauze’s synthetic agar (starch—20 g/L, KNO3—1 g/L, NaCl—0.5 g/L, MgSO4*7H2O—0.5 g/L, K2HPO4—0.5 g/L, FeSO4*7H2O—0.01 g/L, agar—15 g/L, pH 7.4). All media were supplemented with nystatin (50 mg/L) and phosphomycin (50 mg/L) to inhibit the growth of fungi and Gram-negative bacteria (Kieser et al. 2000). The inoculated plates were incubated at 28 °C for up to 4 weeks. Actinobacteria-like strains were identified based on colony morphology and then transferred onto MS medium to obtain pure cultures.
DNA extraction and 16S rRNA gene sequencing
Total DNA was isolated using the salting out procedure as described in Kieser et al. (2000). Almost-complete 16S rRNA gene sequences were obtained by PCR amplification with the following set of primers: 24F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492R (5′-GGY TAC CTT GTT AAC GAC TT-3′). The parameters of the PCR were as follows: initial denaturation at 95 °C for 3 min, followed by 30 cycles of 95 °C for 35 s, 51 °C for 40 s, and 72 °C for 110 s, and final elongation at 72 °C for 7 min. The forward and reverse sequences were assembled with BioEdit 7.2.6.1 (Hall 1999). The amplified PCR products were visualized in 1% agarose gel, purified with Cleanup Standard PCR purification kit (Kat. BC022, Evrogen, Russia), and sequenced with primers employed for amplification by Syntol company (Moscow, Russia) using the Sanger method. Almost complete sequences were aligned with the 16S rRNA gene sequence of Escherichia coli K-12 using Clustal X (Larkin et al. 2007). Kimura’s two-parameter model was used to calculate the phylogenetic tree evolutionary distance matrices (Kimura 1980). Phylogenetic analysis based on the maximum likelihood method was performed using the MEGA 7.0 software package (Felsenstein 1981; Tamura et al. 2013).
Amplification and cloning of PKS1 and PKS2 biosynthetic genes
Three sets of degenerate primers were used to target the PKS type I gene: KSMA-F (5′-TS GCS ATG GAC CCS CAG CAG-3′) and KSMB-R (5′-CC SGT SCC GTG SGC CTC SAC-3′); K1F (5′-TSAAGTCSAACATCGGBCA-3′) and M6R (5′-CGCA GGTTSCSGTACCAG-3′) (Izumikawa et al. 2003; Ayuso-Sacido and Genilloud 2005). Also, primers targeting PKS type II were used: 540F (5′-GGITGCACSTCIGGIMTSGAC-3′) and 1100R (5′-CCGATSGCICCSAGIGAGTG-3′) (Wawrik et al. 2005). PCR parameters followed those of the original studies. The amplified PCR products were monitored in 1% agarose gel via electrophoresis.
Purified PCR products encoding the ketosynthase (KS) domain of type I PKSs were cloned using vector pAL2-T (Evrogen, Russia) with E. coli XL1-Blue host following the manufacturer’s directions. DNA sequencing of cloned genes was conducted by Syntol (Russia). The resultant DNA sequences were translated into amino acid sequences, which were then analyzed against the NCBI protein database using the BLASTp algorithm with default parameters. A maximum-likelihood tree for the amino acid sequences of KS domains was constructed using the MEGA 7.0 software package applying 1000 bootstrap resampling with beta-ketoacyl–ACP synthase sequence from E. coli as an outgroup.
Extraction of secondary metabolites
For extraction of secondary metabolites, isolated strains were cultivated in 10 mL of TSB medium for 2 days at 28 °C to obtain a pre-culture for further cultivation in liquid production medium. We used 5 mL of pre-culture to inoculate 50 mL of production medium (NL-19, SG, or ISP 2). Streptomyces strains were cultivated for 5 days while Amycolatopsis, Micromonospora, and Pseudonocardia strains were cultivated for 10 days at 28 °C. After the supernatant and biomass were separated by centrifugation at 3000 rpm for 5 min, metabolites from the supernatant were extracted with an equal volume of ethyl acetate, while metabolites from the biomass were extracted with 10 mL of acetone:methanol mixture (1:1). All of the dry extracts obtained were weighed and then solubilized in appropriate amount of DMSO to achieve a 50 µg/mL concentration.
Antimicrobial bioassays
Antimicrobial assays were conducted in 96-well plates with Bacillus subtilis ATCC6633, Pseudomonas putida KT2440, Escherichia coli ATCC25922, Saccharomyces cerevisiae BY4742, and Candida albicans ATCC 90027.
Overnight cultures of bacteria in TSB medium were diluted by a factor of 3 × 10−3 in TSB, and 100 µL was seeded into each well of a 96-well plate. Chemical extracts solubilized in DMSO were added to each well at concentration of 50, 25, 12.5, and 6.25 µg/mL. Gentamycin (50 µg/mL) was used as a positive control, while wells containing the DMSO vehicle acted as a negative control. All assays were conducted in duplicate. Plates were incubated for 24 h at 37 °C. Absorbance at 630 nm was measured for each well, and the absorbance values were compared between treatments and vehicle-only controls to determine growth inhibition with response to each chemical extract. Antimicrobial assays with C. albicans and S. cerevisiae were conducted in the same way, except that YPD medium was used instead of TSB, and nystatin (4 µg/mL) was used as positive control.
Results
Isolation and diversity of culturable Actinobacteria associated with deepwater endemic amphipods
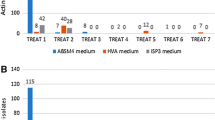
To assess the diversity of culturable Actinobacteria associated with deepwater endemic amphipods of Lake Baikal, selective isolation was carried out for seven amphipod species—Pallasea brandtii flaviceps, Crypturopus tuberculatus, Acanthogammarus godlewskii collected near the southeastern shore. Eulimnogammarus ussolzewii, E. aheneus, Odontogammarus calcaratus pulcherrimus, Ommatogammarus carneolus melanophthalmus were collected near the southwestern shore (Table 1, Fig. 1). A total of 15 morphologically distinct actinobacterial isolates were obtained from deepwater Baikal endemic amphipods. The maximum number of strains were isolated using ISP 2 medium (n = 6), followed by MS (n = 5), SAA (n = 2) and Gauze’s synthetic medium (n = 2).
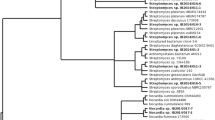
Phylogeny of the strains was evaluated based on their 16S rRNA gene sequences. Most of the strains (11 isolates) belonged to the genus Streptomyces (Table 2). Other strains were represented by Amycolatopsis (1), Micromonospora (1), and Pseudonocardia (2) genera. However, only four strains had more than 98.7% similarity to the closest known species from the NCBI database. Thus, the threshold value 98.7% was set as a cut-off value for defining different species (Stackebrandt and Ebers 2006). The constructed phylogenetic tree combined 16S rRNA sequences of the isolated strains, sequences of the closest species obtained from the NCBI database and some sequences of Actinobacteria previously isolated from Lake Baikal and its inhabitants (Fig. 2). The 16S rRNA sequences formed a tight clade with several representatives of the respective genera. We did not find any strict clades of Streptomyces sp. that could be described by the depth of sampling. However, we surprisingly found that all samples collected near the southeastern shore contained representatives of rare Actinobacteria strains.
PCR-based screening for PKSI and PKSII genes
The biosynthetic potential of the isolated strains was examined using PCR-based screening for PKS types I and II. Out of 15 isolated strains only 2 (13%), Pseudonocardia sp. IB2014P11-4 and Streptomyces sp. IB2015P140-1, did not possess any PCR signal (Table 3). Among 15 strains, 10 (66%) were positive for amplification of KS-domain using the primer pair KSMA/KSMB that was designed based on the highly conserved region of macrolide PKS. The translated deduced amino acid sequences of these PKS type I genes showed mostly low (less than 50%) similarity with the top pBLAST match. However, two sequences from Pseudonocardia sp. IB2014P10-1, and Streptomyces sp. IB2015P143-2 showed high (more than 90%) similarity with known KS domains (Tables 3 and 4, Fig. 3).
Maximum-likelihood phylogenetic tree based on the translated amino acid sequences of the KS domain of PKS type I (KSMA/KSMB primers) from the strains isolated from deepwater lake Baikal amphipods (shown in bold) with closest neighbors from the GenBank database. Percentage bootstrap values from 1000 resamplings are indicated at nodes
Ten positive PCR signals were obtained using the primer pair K1/M6 (PKSI type); however, not all strains that possessed the KSMA/KSMB PCR signal also had the K1/M6 PCR signal and vice versa (Figs. 3 and 4). Also, 8 out of 15 strains (53%) showed positive PCR signal for mentioned PKSII type gene.
Maximum-likelihood phylogenetic tree based on the translated amino acid sequences of the KS domain of PKS type II (540/1100 primers) from the strains isolated from deepwater lake Baikal amphipods (shown in bold) with closest neighbors from the GenBank database. Percentage bootstrap values from 1000 resamplings are indicated at nodes
Antimicrobial assay
Among all the isolates obtained, 53% of strains (8 out of 15 isolates) demonstrated activity against either B. subtilis or S. carnosus or both of them (Table 5). None of the strains was active against P. putida, S. cerevisiae, or C. albicans. Only one strain (Streptomyces sp. IB2015P143-3) was active against E. coli at a concentration of 50 µg/mL. Antimicrobial activity of other isolated Actinobacteria strains at 25 µg/mL concentration is presented in Table 5.
Several strains (Streptomyces sp. IB2014P11-3, Pseudonocardia sp. IB2014P11-4, Amycolatopsis sp. IB2014P14-2, Streptomyces sp. IB2015P138-2, Streptomyces sp. IB2015P139-1, Streptomyces sp. IB2015P139-2, and Streptomyces sp. IB2015P143-1) did not demonstrate any inhibitory activity against the test cultures used.
Discussion
In the current study, we isolated 15 strains of Actinobacteria from 7 species of Baikal endemic deepwater amphipods collected in the southern part of the lake (Table 1). Among isolated strains, 73% belonged to the Streptomyces genus leaving remaining 23% to the so-called “rare” genera of Amycolatopsis, Micromonospora, and Pseudonocardia (Table 2, Fig. 2). Previously, I. Terkina and her colleagues showed that lake’s sediments were slightly dominated by Micromonospora specimens (51%), while in water, their number decreased to 26% (Terkina et al. 2002). Another study revealed more striking dominance of the Micromonospora genus for Actinobacteria specimens isolated from sediments in the Selenga river mouth, in the central part of Lake Baikal (Parfenova et al. 2005). Among thirteen studied sites, only one had a fraction of Micromonospora less than 76% (one site lacked any Actinobacteria isolates) and eight had only Micromonospora isolates.
Based on the previous studies, we hypothesized that Actinobacteria isolated from amphipods were derived from either sediments or water and their distribution pattern should be somehow similar to those in the water or sediments. However, in our current research and previous studies devoted to the Actinobacteria isolated from deepwater genus Ommatogammarus and littoral genera Brandtia and Pallasea, we found a high proportion of Streptomyces strains among the isolated Actinobacteria (Axenov-Gribanov et al. 2016; Protasov et al. 2017). The discrepancy between our data and materials obtained by I. Terkina et al. (2002) might be attributed to the fact that we used a different isolation media composition, i.e., we used nutrient-rich media while previous studies relied on low-nutrient media mimicking the lake’s oligotrophic environment. Previously, it was shown that among Actinobacteria isolated from freshwater lake sediments, representatives of the genus Streptomyces prevail (Sanasam et al. 2011; Gebreyohannes et al. 2013; Zothanpuia et al. 2017). However, only nutrient-rich media were used in those studies for isolation.
When nucleotide sequences of the strains isolated from Lake Baikal deepwater amphipods were compared to the NCBI top BLAST matches, our isolates showed high similarity with terrestrial strains (Table 2). Also, the sequences from Actinobacteria previously isolated from littoral amphipod species and Baikal invertebrates (Axenov-Gribanov et al. 2016) aligned closely with the sequences from the present study and some terrestrial isolates. The lack of phylogenetic specificity among the isolated strains, amphipod species, and the depth of amphipod collection might indicate that Actinobacteria are transient microorganisms derived from the surrounding environment (Fig. 2). About 80% of isolated strains have less than 98.7% of identity with the known species, so alternatively they might be new species (Table 2) (Stackebrandt and Ebers 2006).
The Actinobacteria isolated in this study displayed the presence of PKS type I and PKS type II secondary metabolite genes (Tables 3 and 4). The sequence from Pseudonocardia sp. IB2014P10-1 is almost identical (98.97% identity) to the sequence of malonyl CoA-acyl carrier protein transacylase from Pseudonocardia sp. Ae150A_Ps1 isolated from Acromyrmex echinatior working ant (Table 4). The strain Pseudonocardia sp. Ae150A_Ps1 has the nystatin-like gene cluster and plays an important role in defense of the leaf-cutter ant Acromyrmex echinatior from fungal pathogens (Holmes et al. 2016). However, the strain Pseudonocardia sp. IB2014P10-1 showed antibiotic activity only against B. subtilis but not against fungal test-organisms (Table 5). Another amino acid sequence from Streptomyces sp. IB2014P143-2 is highly similar to the sequence of polyketide synthase from Streptomyces sp. KM273126 isolated from marine sediment. The remaining amino acid sequences have low identity to the known sequences ranging from 41 to 63%.
The sequences related to PKS type II were slightly less abundant among the studied strains and showed a high level of similarity with sequences deposited in GenBank (Table 4). The sequence from Streptomyces sp. IB2015P143-2 was similar to the sequences from the strains Streptomyces sp. IB2014 011-1 and Streptomyces sp. IB2014 011-12 previously isolated from larvae Trichoptera sp. of Lake Baikal (Axenov-Gribanov et al. 2016). They form a distinct clade on the PKS type II phylogenetic tree with sequences derived from soil Streptomyces (Fig. 4).
It is worth mentioning that two strains with the lowest identity with their top BLAST matches (16S rRNA gene), Pseudonocardia sp. IB2014P11-4 and Streptomyces sp. IB2015P140-1, have no PCR signals for all three primer pairs (Tables 2 and 3). One plausible explanation is that those strains are new species (81 and 87% of identity to their top BLAST matches respectively), so their genomes diverge greatly from those used to design the PKS primers. The lack of detectable PKS genes does not indicate the absence of biosynthetic gene clusters since primers flank only specific nucleotide sequences leaving unknown genes undetectable. It was shown that the Actinobacteria genomes contain many cryptic or silent clusters (Weber et al. 2015). Those genes and clusters require specific conditions (for example, environmental factors or chemical signals from other bacteria) to be activated and expressed. PCR screening of the PKS genes might be more effective when it is combined with antibiotic tests. The next step in discovery of new compounds might be genome mining of putatively new strains. In that sense, genome mining of non-Streptomyces strains is of particular interest because of constant re-discovery of the known compounds from representatives of the Streptomyces genus (Ward and Allenby 2018; Chevrette et al. 2019).
Antibiotic tests of crude extracts from the isolated strains showed that only eight (53%) out of 15 strains show activity against at least one test culture (Table 5). None of the strains was active against fungi or Gram-negative bacteria except the extract from Streptomyces sp. IB2015P143-3 that was active against E. coli at concentration of 50 ug/mL. Previously, we found that about 70% of the strains isolated from deepwater amphipod of the genus Ommatogammarus exhibit fungicide activity (Protasov et al. 2017). This discrepancy might be attributed to the fact that we used two different antibiotic assays, the disk-diffusion method and tests in 96-well plates. The latter used equal concentrations for each crude extract. Previously we might have used an inappropriately high concentration of the extracts. However, this does not obviate the fact that among the tested strains might be some that are producers of new compounds.
Actinobacteria isolated from Lake Baikal and its inhabitants showed high biotechnological potential. Streptomyces 156A isolated from Baikal water (100 m) synthesize a wide range of ionophore antibiotics from the polynactin family (Shishlyannikova et al. 2017). We also found nonactin in a crude extract of the stain Streptomyces sp. IB2015P113-12 (Protasov et al. 2017). Both strains showed activity against Gram-positive test cultures that were the primary target of the polynactin family of antibiotics. Another strain, Streptomyces sp. IB2014011-12, isolated from Lake Baikal Trichoptera sp. larvae showed activity against Gram-positive bacteria. The NRPS–trans-AT–PKS enzyme from this strain is involved in the synthesis of new derivatives of alpiniamide, and the genome of this strain contains 29 biosynthetic gene clusters (Paulus et al. 2018). Another strain also isolated from Trichoptera sp. larvae, Streptomyces sp. IB2014011-1, contains 30 biosynthetic gene clusters (Axenov-Gribanov et al. 2017). During our preliminary studies, both strains revealed high level of activity against Gram-positive bacteria (Axenov-Gribanov et al. 2016).
Conclusions
Lake Baikal amphipods were shown to be a valuable source of Actinobacteria strains with a high proportion of putatively new and rare species. Prevalence of the Streptomyces specimens among the isolated strains can be attributed to the use of nutrient-rich media for isolation. A relatively large number of strains exhibited antibiotic activity and the presence of secondary metabolite genes. Our study demonstrated, via the use of PCR, the presence of PKS type I and II genes and was used with antibiotic assays for the selection of the most promising strains. Thus, Actinobacteria associated with Baikal’s endemic deepwater amphipods are potentially an untapped source of natural products with biosynthetic potential, and they may protect amphipods against pathogenic microorganisms.
References
Ampe F, Thiery A (1998) Microflora associated with the digestive tract of the fairy shrimp Branchinella spinosa. FEMS Microbiol Lett 158:201–205
Axenov-Gribanov D, Rebets Y, Tokovenko B, Axenov-Gribanov D, Voytsekhovskaya I, Timofeyev M, Luzhetskyy A (2016) The isolation and characterization of actinobacteria from dominant benthic macroinvertebrates endemic to Lake Baikal. Folia Microbiol (Praha) 61:159–168. https://doi.org/10.1007/s12223-015-0421-z
Axenov-Gribanov DV, Tokovenko BT, Rebets YV, Voytsekhovskaya IV, Shatilina ZM, Protasov ES, Luzhetskyy AN, Timofeyev MA (2017) Draft genome sequence of Streptomyces sp. strain IB2014011-1, isolated from Trichoptera sp. larvae of lake Baikal. Genome Announc 5(17): e00062-17. https://doi.org/10.1128/genomeA.00062-17
Ayuso-Sacido A, Genilloud O (2005) New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb Ecol 49:10–24. https://doi.org/10.1007/s00248-004-0249-6
Bedulina DS, Takhteev VV, Pogrebnyak SG, Govorukhina EB, Madyarova EV, Lubyaga YA (2014) On Eulimnogammarus messerschmidtii, sp. n. (Amphipoda: Gammaridea) from Lake Baikal, Siberia, with redescription of E. cyanoides (Sowinsky) and remarks on taxonomy of the genus Eulimnogammarus. Zootaxa 3838:518–544
Bérdy J (2012) Thoughts and facts about antibiotics: where we are now and where we are heading. J Antibiot (Tokyo) 65:385–395. https://doi.org/10.1038/ja.2012.54
Bilyk O, Luzhetskyy A (2016) Metabolic engineering of natural product biosynthesis in actinobacteria. Curr Opin Biotechnol 42:98–107. https://doi.org/10.1016/j.copbio.2016.03.008
Cafaro MJ, Poulsen M, Little AE, Price SL, Gerado NM, Wong B, Currie CR (2011) Specificity in the symbiotic association between fungus-growing ants and protective Pseudonocardia bacteria. Proc Biol Sci 278:1814–1822. https://doi.org/10.1098/rspb.2010.2118
Carman KR, Dobbs FC (1997) Epibiotic microorganisms on copepods and other marine crustaceans. Microsc Res Tech 37:116–135
Chevrette MG, Carlson CM, Ortega HE, Thomas C, Ananiev GE, Barns KJ, Grubbs KJ (2019) The antimicrobial potential of Streptomyces from insect microbiomes. Nat Commun 10:516. https://doi.org/10.1038/s41467-019-08438-0
Dempsey AAC, Kitting CL, Rosson RA (1989) Bacterial variability among individual Penaeid shrimp digestive tracts. Crustaceana 56:267–278
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376. https://doi.org/10.1007/BF01734359
Gebreyohannes G, Moges F, Sahile S, Raja N (2013) Isolation and characterization of potential antibiotic producing actinomycetes from water and sediments of Lake Tana, Ethiopia. Asian Pac J Trop Biomed 3:426–435. https://doi.org/10.1016/S2221-1691(13)60092-1
Gillan DC, Dubilier N (2004) Novel epibiotic Thiothrix bacterium on a marine amphipod. Appl Environ Microbiol 70:3772–3775. https://doi.org/10.1128/AEM.70.6.3772-3775.2004
Gillan DC, Ribesse J, De Ridder C (2004) The iron-encrusted microbial community of Urothoe poseidonis (Crustacea, Amphipoda). J Sea Res 52:21–32. https://doi.org/10.1016/j.seares.2003.08.009
Gil-Turnes MS, Fenical W (1992) Embryos of Homarus americanus are protected by epibiotic bacteria. Biol Bull 182:105–108. https://doi.org/10.2307/1542184
Gil-Turnes MS, Hay ME, Fenical W (1989) Symbiotic marine bacteria chemically defend crustacean embryos from a pathogenic fungus. Science 80(246):116–118. https://doi.org/10.1126/science.2781297
Glockner FO, Zaichikov E, Belkova N, Glöckner FO, Zaichikov E, Belkova N, Denissova L, Pernthaler J, Pernthaler A, Amann R (2000) Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl Environ Microbiol 66:5053–5065. https://doi.org/10.1128/AEM.66.11.5053-5065.2000
Grossart HP, Tang KW, Kiørboe T, Ploug H (2007) Comparison of cell-specific activity between free-living and attached bacteria using isolates and natural assemblages. FEMS Microbiol Lett 266:194–200. https://doi.org/10.1111/j.1574-6968.2006.00520.x
Grossart H-P, Dziallas C, Tang KW (2009) Bacterial diversity associated with freshwater zooplankton. Environ Microbiol Rep 1:50–55. https://doi.org/10.1111/j.1758-2229.2008.00003.x
Grossart H-P, Dziallas C, Leunert F, Tang KW (2010) Bacteria dispersal by hitchhiking on zooplankton. Proc Natl Acad Sci USA 107:11959–11964. https://doi.org/10.1073/pnas.1000668107
Hall T (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Harris JM (1993) The presence, nature, and role of gut microflora in aquatic invertebrates: a synthesis. Microb Ecol 25:195–231. https://doi.org/10.1007/BF00171889
Holland R, Hergenrader G (1981) Bacterial epibionts of diaptomid copepods. Trans Am Microsc Soc 100:56–65
Holmes NA, Innocent TM, Heine D, Bassam MA, Worsley SF, Trottmann F, Wilkinson B (2016) Genome analysis of two Pseudonocardia phylotypes associated with Acromyrmex leafcutter ants reveals their biosynthetic potential. Front Microbiol 7:1–16. https://doi.org/10.3389/fmicb.2016.02073
Izumikawa M, Murata M, Tachibana K, Ebizuka Y, Fujii I (2003) Cloning of modular type I polyketide synthase genes from salinomycin producing strain of Streptomyces albus. Bioorg Med Chem 11:3401–3405. https://doi.org/10.1016/S0968-0896(03)00337-7
Karnaukhov DY, Bedulina DS, Kaus A, Prokosov SO, Sartoris L, Timofeyev MA, Takhteev VV (2016) Behaviour of Lake Baikal amphipods as a part of the night migratory complex in the Kluevka settlement region (south-eastern Baikal). Crustaceana 89:419–430. https://doi.org/10.1163/15685403-00003531
Khalzov IA, Mekhanikova IV, Sitnikova TY (2018) First data on ectosymbiotic consortia of infusoria and prokaryotes associated with amphipods inhabiting the Frolikha underwater hydrothermal vent, lake Baikal. Zool Zhurnal 97:1525–1530. https://doi.org/10.1134/S0044513418120073 (in Russian)
Kieser B, Buttner M, Charter K, Hopwood B (2000) Practical streptomyces genetics. John Innes Foundation, Norwich
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. https://doi.org/10.1007/BF01731581
Kolter R, Greenberg EP (2006) The superficial life of microbes. Nature 441:300–302. https://doi.org/10.1038/441300a
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Long RA, Azam F (2001) Antagonistic interactions among marine pelagic bacteria. Appl Environ Microbiol 67:4875–4983. https://doi.org/10.1128/AEM.67.11.4975-4983.2001
Mekhanikova IV, Takhteev VV (2008) Cuticular nonsensory microstructures of amphipods from Lake Baikal (Crustacea: Amphipoda), their taxonomic and adaptive significance. Invertebr Zool 5:17–37 (In Russian)
Nadell CD, Xavier JB, Foster KR (2009) The sociobiology of biofilms. FEMS Microbiol Rev 33:206–224. https://doi.org/10.1111/j.1574-6976.2008.00150.x
Nagasawa S, Nemoto T (1988) Presence of bacteria in guts of marine crustaceans and on their fecal pellets. J Plankton Res 10:559–564. https://doi.org/10.1093/plankt/10.3.559
Parfenova VV, Pavlova ON, Terkina IA, Suslanova MY, Kostornova TY, Nikulina IG, Sorokovikova LM (2005) Water quality and protection : microbial community of the oxidized layer of Lake Baikal bottom sediments in the Selenga mouth. Water Resour 32:227–231
Passari AK, Leo VV, Chandra P, Kumar B, Nayak C, Hashem A, Singh BP (2018) Bioprospection of actinobacteria derived from freshwater sediments for their potential to produce antimicrobial compounds. Microb Cell Fact 17:68. https://doi.org/10.1186/s12934-018-0912-0
Paulus C, Rebets Y, Zapp J, Rückert C, Kalinowski J, Luzhetskyy A (2018) New Alpiniamides from Streptomyces sp. IB2014/011-12 assembled by an unusual hybrid nonribosomal peptide synthetase trans-AT polyketide synthase enzyme. Front Microbiol 9:1959. https://doi.org/10.3389/fmicb.2018.01959
Protasov ES, Axenov-Gribanov DV, Voytsekhovskaya IV, Tokovenko BT, Shatilina ZM, Timofeyev MA (2017) The diversity and antibiotic properties of actinobacteria associated with endemic deepwater amphipods of lake Baikal. Ant Leeuw Int J Gen Mol Microbiol 110:1593–1611. https://doi.org/10.1007/s10482-017-0910-y
Romanova EV, Aleoshin VV, Kamaltynov RM, Mikhailov K, Logacheva MD, Sirotinina EA, Sherbakov DY (2016) Evolution of mitochondrial genomes in Baikalian amphipods. BMC Genom 17:1016. https://doi.org/10.1186/s12864-016-3357-z
Sanasam S, Nimaichand S, Ningthoujam D (2011) Novel bioactive actinomycetes from a niche biotope, Loktak Lake, in Manipur, India Isolation of Lake Actinomycetes. J Pharm Res 4:1707–1710
Schmidt C, Le Bris N, Gaill F (2008) Interactions of deep-sea vent invertebrates with their environment: the case of Rimicaris exoculata. J Shellfish Res 27:79–90. https://doi.org/10.2983/0730-8000(2008)27[79:IODVIW]2.0.CO;2
Seipke RF, Kaltenpoth M, Hutchings MI (2012) Streptomyces as symbionts: an emerging and widespread theme? FEMS Microbiol Rev 36:862–876. https://doi.org/10.1111/j.1574-6976.2011.00313.x
Shishlyannikova TA, Kuzmin AV, Fedorova GA, Shishlyannikov SM, Lipko IA, Sukhanova EV, Belkova NL (2017) Ionofore antibiotic polynactin produced by Streptomyces sp. 156A isolated from Lake Baikal. Nat prod res 31(6):639–644. https://doi.org/10.1080/14786419.2016.1217203
Sitnikova TY, Mekhanikova IV (2014) Amphipods (Amphipoda, Gammaridea) at the Gorevoy Utes oil and methane seep, Lake Baikal. Crustaceana 87:1500–1520. https://doi.org/10.1163/15685403-00003367
Sochard MR, Wilson DF, Austin B, Colwell RR (1979) Bacteria associated with the surface and gut of marine copepods. Appl Environ Microbiol 37:750–759
Stackebrandt E, Ebers J (2006) Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 33:152–155
Sun W, Zhang F, He L, Loganathan K (2015) Actinomycetes from the South China Sea sponges: Isolation, diversity, and potential for aromatic polyketides discovery. Front Microbiol 6:1–15. https://doi.org/10.3389/fmicb.2015.01048
Takhteev VV (2000a) Essays on the amphipods of Lake Baikal (systematics, comparative ecology, evolution), 1st edn. Irkutsk State University Press, Irkutsk (In Russian)
Takhteev VV (2000b) Trends in the evolution of Baikal amphipods and evolutionary parallels with some marine malacostracan faunas. Adv Ecol Res 31:197–220
Takhteev VV, Berezina NA, Sidorov DA (2015) Checklist of the Amphipoda (Crustacea) from continental waters of Russia, with data on alien species. Arthropoda Sel 24:335–370
Takhteev VV, Karnaukhov DY, Govorukhina EB, Misharin AS (2019) Diel vertical migrations of hydrobionts in the coastal area of Lake Baikal. Inl Water Biol 12:178–189
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Tang KW (2005) Copepods as microbial hotspots in the ocean: effects of host feeding activities on attached bacteria. Aquat Microb Ecol 38:31–40. https://doi.org/10.3354/ame038031
Tang KW, Dziallas C, Grossart HP (2011) Zooplankton and aggregates as refuge for aquatic bacteria: protection from UV, heat and ozone stresses used for water treatment. Environ Microbiol 13:378–390. https://doi.org/10.1111/j.1462-2920.2010.02335.x
Terkina IA, Driukker VV, Parfenova VV, Kostornova TI (2002) The biodiversity of actinomycetes in Lake Baikal. Microbiology 71:404–408
Väinölä R, Witt JDS, Grabowski M, Bradbury JH, Jazdzewski K, Sket B (2008) Global diversity of amphipods (Amphipoda; Crustacea) in freshwater. Hydrobiologia 595:241–255. https://doi.org/10.1007/s10750-007-9020-6
Van Arnam EB, Clardy J, Currie CR, Ruzzini AC, Sit CS, Horn H, Pinto-Tomás AA, Clardy J (2016) Selvamicin, an atypical antifungal polyene from two alternative genomic contexts. Proc Natl Acad Sci 113:12940–12945. https://doi.org/10.1073/pnas.1613285113
Ward A, Allenby N (2018) Genome mining for the search and discovery of bioactive compounds: the Streptomyces paradigm. FEMS Microbiol Lett. https://doi.org/10.1093/femsle/fny240
Wawrik B, Kerkhof L, Zylstra GJ, Kukor JJ (2005) Identification of unique type II polyketide synthase genes in soil. Appl Environ Microbiol 71:2232–2238. https://doi.org/10.1128/AEM.71.5.2232
Weber T, Charusanti P, Musiol-Kroll EM, Jiang X, Tong Y, Kim HU, Lee SY (2015) Metabolic engineering of antibiotic factories: new tools for antibiotic production in actinomycetes. Trends Biotechnol 33:15–26. https://doi.org/10.1016/j.tibtech.2014.10.009
Zothanpuia Chandra P, Leo VV, Mishra VK, Kumar B, Singh BP (2017) Production of potent antimicrobial compounds from Streptomyces cyaneofuscatus associated with fresh water sediment. Front Microbiol. https://doi.org/10.3389/fmicb.2017.00068
Acknowledgements
We are grateful to Prof. Vadim Takhteev (Irkutsk State University) for help with amphipod identification. Also, we thank Dr. Polina Drozdova (Irkutsk State University), reviewers and editors for their valuable comments that improved the article greatly. This study was carried out with partial financial support of the RSF project (18-74-00018 (DAG)), RFBR project (18-29-05051 (DAG), 18-34-00294 (EP, VE)), projects of the Ministry of Education and Science of the Russian Federation (6.12737.2018/12.2 (EP), 6.9654.2017/8.9 (ZS), 6.12738.2018/12.2 (DAG)).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Handling Editor: Marianne V. Moore.
Rights and permissions
About this article
Cite this article
Protasov, E.S., Axenov-Gribanov, D.V., Rzhechitsky, Y.A. et al. Diversity of culturable actinobacteria associated with deepwater endemic amphipods of Lake Baikal and study of their biosynthetic capabilities. Limnology 21, 35–47 (2020). https://doi.org/10.1007/s10201-019-00593-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10201-019-00593-z