Abstract
Background
Multiple studies have revealed that long non-coding RNA (lncRNA) NR2F2-AS1 plays a role in affecting cancer cell proliferation and metastasis. Here, both in vitro and in vivo experiments were performed for investigating the function and mechanism of NR2F2-AS1 in human osteosarcoma (OS).
Methods
The NR2F2-AS1 level in human OS tissues and adjacent non-tumor tissues was examined by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). The NR2F2-AS1 overexpression model was constructed in OS cells, then cell proliferation, invasion, and apoptosis were monitored. The OS xenograft model was established in nude mice using NR2F2-AS1-overexpressed OS cells. The downstream target genes of NR2F2-AS1 were predicted. qRT-PCR and Western blot were implemented to validate the profiles of miR-425-5p and HMGB2. The targeting link between NR2F2-AS1 and miR-425-5p, miR-425-5p and HMGB2 was further probed by dual-luciferase reporter experiment.
Results
In comparison to adjacent non-tumor tissues, OS tissues showed upregulated NR2F2-AS1 expression. Higher NR2F2-AS1 level was predominantly correlated with worse clinical stages. In vivo and in vitro tests corroborated that NR2F2-AS1 overexpression spurred OS cell proliferation, growth, invasion, and choked apoptosis. Mechanistically, NR2F2-AS1 hampered miR-425-5p expression as its competitive endogenous RNA (ceRNA). Thus, NR2F2-AS1 facilitated the HMGB2 expression. However, miR-425-5p inhibited HMGB2 expression by targeting the latter.
Conclusion
NR2F2-AS1 expedited the evolution of OS by elevating HMGB2 levels through sponging miR-425-5p. The NR2F2-AS1/miR-425-5p/HMGB2 regulatory axis is a promising target in treating human OS.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteosarcoma (OS) is a primary malignant bone tumor that occurs most often in children and adolescents. Early lung metastasis is the primary risk factor for OS patients’ death [1]. Although surgical resection and adjuvant chemotherapy against OS have made much progress, OS patients’ prognosis remains poor owing to metastases or recurrence [2]. Hence, studying the molecular mechanism of OS is expected to bring hope for the OS’s diagnosis and therapy at the early stage.
Long non-coding RNAs (lncRNAs) are over 200 nucleotides (nt) in transcript length and do not have protein-coding capacities. A growing number of investigations have demonstrated that lncRNAs are key regulators of OS development and progression [3, 4]. For example, lncRNA BE503655 is overexpressed in OS tissues and OS cell lines. Knocking down lncRNA BE503655 restrains OS cells’ proliferation, invasion and migration [5]. The above studies suggest that lncRNAs’ expression is dysregulated in OS, and that the dysregulated lncRNAs affect the progression of OS. NR2F2 antisense RNA 1 (NR2F2-AS1) is a new-found lncRNA aberrantly expressed in diversified cancers. For instance, NR2F2-AS1 expression is elevated in prostate cancer (PC) cell lines, and high levels of NR2F2-AS1 reinforce PC cell proliferation and metastasis by positively regulating CDK4 [6]. Additionally, high expression of NR2F2-AS1 accelerates cervical cancer evolvement by enhancing cell proliferation, migration, invasion and epithelial–mesenchymal transition (EMT) [7]. Nevertheless, the NR2F2-AS1 profile in OS and its mechanism of action remain largely unknown.
MicroRNAs (miRNAs) are endogenous single-stranded non-coding RNAs that are 20–24 nucleotides long. They not only modify cell evolvement and differentiation but are also underlying molecular markers for tumors, with promising applications in early diagnosis, treatment, prognosis and chemoresistance, including OS [8,9,10]. As an essential microRNA, miR-425-5p has abnormal expression in various cancers. The miR-425-5p expression is higher in pancreatic cancer tissues [11] and breast cancer (BC) tissues [12] than that in adjacent tissues and is positively linked to tumor patients’ lymph node metastasis, clinical stage and tumor differentiation. More importantly, diminished expression of miR-425-5p is observed in OS tissues and cells. miR-425-5p overexpression abates OS cell proliferation, invasion and migration in vitro [13]. Also, Li et al. [14] concluded that miR-425-5p is a potential biomarker for the diagnosis of OS. Therefore, it is essential to probe the expression and role of miR-425-5p in OS.
High-mobility group box 2 (HMGB2) belongs to the non-histone chromosomal high mobility group protein family. It is chromatin-associated and ubiquitously distributed in the nucleus of higher eukaryotic cells. Recently, HMGB2 has been found heightened in several cancers, such as breast cancer [15], gastric cancer (GC) [16, 17], melanoma [18], prostate cancer [19] and cervical cancer [20]. In vitro experiments have revealed that HMGB2 overexpression facilitates the proliferation, migration and invasive ability of OS cells. The survival time of OS patients with enhanced HMGB2 expression is markedly reduced [21]. Yang et al. [22] have suggested that HMGB2 is differentially expressed in OS cells. Interestingly, classical theories hold that miRNAs cause target molecule silencing by specifically binding to the 3' untranslated region of mRNAs. As an example, up-regulating miR-130a reduces glioma cell growth and invasion ex vivo and in vivo through HMGB2 attenuation [23].
Based on the above findings, we probed the expression of NR2F2-AS1, miR-425-5p and HMGB2 in OS tissues and OS cells. As a result, NR2F2-AS1 and HMGB2 were highly expressed, and miR-425-5p was lowly expressed in OS tissues and cells. Meanwhile, online websites and a dual-luciferase reporter assay uncover a targeting relationship between the three. Therefore, we hypothesize that NR2F2-AS1 modulates OS evolution through the miR-425-5p/HMGB2 axis. This study provides a fresh theoretical basis for the treatment of OS by revealing a new molecular mechanism in the evolution of OS.
Materials and methods
Clinical specimen collection and processing
Cancerous and adjacent non-tumor tissues of 32 OS patients who were subjected to OS resection in Renmin Hospital of Wuhan University from May 2015 to May 2019 were gathered. Prior to operation, patients were not treated with chemotherapy, radiotherapy, or other adjuvant treatment. Control specimens were obtained from the same patients (3 cm from the resection margin at minimum), and no tumor cells were observed in the pathological examination following the operation (Table 1). All tissues were deposited in liquid nitrogen at −196 °C after removal until they could be utilized for RNA isolation. This research was authorized by the research ethics committee of Renmin Hospital of Wuhan University, with the informed consent signed by the patients involved.
Cell culture
The human normal bone cell line hFOB1.19 and the OS cell lines (Saos-2, MNNG/HOS, MG-63, and U2OS) were purchased from ATCC (Rockville, USA). They were treated with RPMI-1640 medium (Thermo Fisher Scientific, USA) comprising 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, USA) and 1% penicillin–streptomycin (Beyotime, Shanghai, China) at 37 °C in a 5% CO2 incubator with saturated humidity. The medium was substituted every 2–3 days, the cells were digested using 0.25% trypsin (Beyotime, Shanghai, China).
Cell transfection
GenePharma (Shanghai, China) provided NR2F2-AS1 overexpression plasmids, control vectors (NC-vector), NR2F2-AS1 siRNA and corresponding negative counterpart si-NC, miR-425-5p mimics, miRNA controls (miR-NC), miR-425-5p inhibitors and corresponding negative counterpart miR-in. By observing the supplier’s instructions, we employed Lipofectamine® 3000 (Invitrogen; ThermoFisher Scientific, Inc.) to transfect NR2F2-AS1 overexpression plasmids, NR2F2-AS1 siRNA, miR-425-5p mimics and miR-425-5p-in into Saos-2, MG-63 and U2OS cells. 24 h after transfection, the medium was exchanged by new fresh medium. qRT-PCR was performed for confirming the transfection efficiency.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Expression of NR2F2-AS1, miR-425-5p, and HMGB2 in hFOB1.19, Saos-2, MNNG/HOS, MG-63, and U2OS was tested by qRT-PCR. Extraction of all RNA from cultured cells was performed by utilizing the TRIzol reagent (Invitgen, Carlsbad, CA). With the use of the reverse transcription kit (Thermo Fisher Scientific), the first-strand complementary DNA (cDNA) was synthesized out of lncRNAs and mRNAs. For miRNA analysis, the microRNA first-strand cDNA synthesis kit (Sangon Biotech, Shanghai, China) was utilized for reverse transcription. We adopted the SYBR-Green PCR Master Mix Kit (Applied Biosystems, Foster City, CA) to amplify NR2F2-AS1, while the TaqMan miRNA Analysis Kit (Applied Biosystems) was employed for miR-425-5p amplification. All reactions were made on a 7900HT fast real-time system (applied biological system). GADPH served as the endogenous control of NR2F2-AS1 and HMGB2, while U6 was used as internal control of miR-425-5p. The levels of NR2F2-AS1, HMGB2 and miR-425-5p were enumerated by the 2 −ΔΔCT method. Table 2 is the list of PCR primer sequences.
Cell counting kit-8 (CCK-8) experiment
Single-cell suspensions of OS cells (Saos-2, MG-63 and U2OS) in the logarithmic growth phase were digested by 0.25% trypsin. After cell number counting, the cells were inoculated in 96-well plates with 2000 cells per well and maintained at 37 °C with 5% CO2 for 24, 48, and 72 h, respectively before removal. Next, 10 μL of the CCK-8 reagent (Beyotime Biotechnology, Shanghai, China) was added to each well for 2 h of incubation. The absorbance at 450 nm was measured on a microplate reader.
Transwell assay
The above OS cell lines were treated with 0.25% trypsin before being subjected to centrifugation, resuspension, and seeding in 24-well plates. The invasion experiment was performed using 8 µM pore size Matrigel Chambers (Corning, Beijing, China). 5 × 104 transfected cells were placed in the upper chamber, and Matrigel was added. In parallel, the lower chamber was filled with 10% FBS, with the supplementation of 400 μL RPMI-1640. After 24 h of incubation at 37 °C, the cells that failed to migrate were wiped off. The membranes were subjected to 4% paraformaldehyde immobilization for 10 min and 0.5% crystal violet dyeing. After the membranes were cleaned, the migrated cells were calculated with an inverted microscope. We did all tests in triplicate and repeated them three times.
Flow cytometry
Twenty-four hours following the transfection, OS cell lines were trypsinized, collected, and dispersed in 6-well plates (2 × 106 cells/well) for another 24 h of incubation. After supernatant removal, the cells were cleaned twice in pre-chilled PBS and resuspended in 1× binding buffer. 5 μL AnnexinV-FITC and 5 μL PI were put in the cell suspension, mixed well and maintained for 15 min at room temperature (RT). The apoptotic rate was gauged by flow cytometry within 1 h, in line with the Annexin V-FITC/PI Apoptosis Detection Kit instructions.
BrdU assay
Cell proliferation was testified by employing the BrdU cell proliferation Detection Kit (AmyJet Scientific Inc. Wuhan, China). After transfection, Saos-2, MG-63 and U2OS cells were supplemented with the BrdU labeling reagent and incubated with 5% CO2 at 37 °C. Forty-eight hours later, the cells were incubated with anti-BrdU antibody. Under the microscope, the positive cell number and the total DAPI-positive cell number in three randomly chosen views were counted. Cell proliferation rate = the BrdU-positive cell number/DAPI-positive cell number, and the mean of the cell proliferation rate of three fields was taken.
Western blot (WB)
The culture medium was removed from cells and the cells were collected. Five paired tumor tissues and adjacent normal tissues were randomly selected from OS patients according to the random number table method. The tumor tissues from nude mice were also collected. Total protein was cells and treated, and RIPA protein lysate (Roche) was adopted for total protein separation. 5 μg total protein was loaded on 12% polyacrylamide gel and subjected to 2 h of electrophoresis at 100 V. It was then transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Bedford, MA, USA). After being blocked with 5% defatted milk powder at RT for 1 h, the membranes were rinsed three times with TBST (10 min each time) and maintained at 4 °C overnight with the primary antibodies (1:1000) of HMGB2 (Abcam, ab124670, MA, USA), cyclin D1 (Abcam, ab16663), cyclin E2 (Thermo Fisher, PA5-109,492), Bax (Abcam, ab32503), Bad (Abcam, ab32445), cleaved caspase3 (Abcam, ab32042), E-cadherin (Abcam, ab40772), Vimentin (Abcam, ab8978), Snail (Abcam, ab216347), and β-actin (Abcam, ab8227). After membrane washing with TBST, the horseradish peroxidase (HRP)-tagged anti-rabbit secondary antibody (1:300) was added for incubation for 1 h, followed by three times of washing (10 min each time). For color imaging, the WB reagent (Invitrogen) was applied, and each band’s gray intensity was assessed by Image J.
Dual-luciferase reporter assay
All luciferase reporter vectors (NR2F2-AS1-WT, NR2F2-AS1-MUT, HMGB2-WT, and HMGB2-MUT) were composited by Promega Corporation (Promega, Madison, WI, USA). Saos-2 and U2OS cells (4.5 × 104) were seeded in 48-well plates and maintained until they reached 70% confluence. The above-mentioned reporter vectors were co-transfected with miR-425-5p mimics or negative controls into Saos-2 and U2OS cells by utilizing Lipofectamine 2000. The luciferase activity was gauged 48 h after transfection. We conducted all tests in triplicate and repeated them three times.
Xenograft experiment in nude mice
Forty female BALB/c nude mice (ordered from the Animal Experiment Center of Wuhan University, 4–6 weeks old) were housed under sterile conditions. 0.2 mL of Saos-2 cell suspensions (containing approximately 1.0 × 106 cells) stably transfected with vectors or NR2F2-AS1 were injected subcutaneously into the proximal axilla of each mouse’s left forelimb. After that, the mice were further fed, and their general state was monitored. The long and short diameters of the nodules were gauged every week to enumerate tumor volume. Six weeks later, the mice were executed via cervical dislocation, and the subcutaneous nodules were removed, with their size and weight calculated. For the lung metastasis group, vector- or NR2F2-AS1-stably transfected Saos-2 cells (1.0 × 106) were administered to the tail vein of mice every 2 days for 18 days. Forty-two days later, the mice were executed, and their lungs were taken and photographed. The tumors visible on the lung were monitored. All animal experiments were approved by the ethics committee of Renmin Hospital of Wuhan University.
Statistical processing
For data processing, the SPSS17.0 statistical software (SPSS Inc., Chicago, IL, USA) was applied. The measurement data were presented as mean ± standard deviation (x ± s). Univariate analysis of variance was employed for the multi-factor comparison, and the pair-wise comparison between two groups was made using t test. P < 0.05 was taken as statistically meaningful.
Results
Expression and clinical value of NR2F2-AS1 in OS tissues and cells
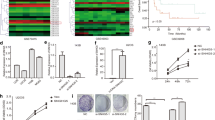
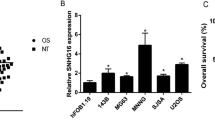
Here, we tested the NR2F2-AS1 expression in OS cells and tissues by qRT-PCR. As a result, NR2F2-AS1 level in OS tissues was upregulated in comparison to that in adjacent non-tumor tissues (P < 0.05, Fig. 1A). We also examined the NR2F2-AS1 profile in diversified OS cell lines and found that NR2F2-AS1 was substantially upregulated in OS cell lines (Saos-2, MNNG/HOS, MG-63, and U2OS) versus the human normal osteoblast lines hFOB1.19 (P < 0.05, Fig. 1B). As evidenced by clinical correlation analysis, OS patients with higher NR2F2-AS1 expression had more advanced Enneking stages (Table 1). Thus, NR2F2-AS1 played an oncogenic role in OS.
Expression and clinical significance of NR2F2-AS1 in OS tissues and cell lines. A The NR2F2-AS1 profile in OS tissues and adjacent non-tumor tissues was verified by qRT-PCR. ***P < 0.001 (vs the Normal group). B qRT-PCR was employed to monitor the NR2F2-AS1 expression in hFOB1.19, Saos-2, MNNG/HOS, MG-63, and U2OS cells. Data were expressed as mean ± SD (n = 3). ***P < 0.001 (vs group)
R2F2-AS1 overexpression heightened OS cell proliferation, invasion and repressed apoptosis
For ascertaining the influence of NR2F2-AS1 on OS, NR2F2-AS1 overexpression model was constructed in Saos-2, MG-63 and U2OS cells. As evidenced by qRT-PCR outcomes, NR2F2-AS1 expression was elevated in OS cells transfected with the NR2F2-AS1 overexpression plasmid versus the vector group (Fig. 2A). Cell viability and proliferation were checked with CCK-8 and BrdU staining. The results displayed that NR2F2-AS1 reinforced OS cell proliferation, elevated the BrdU-positive cell number (Fig. 2B–D). Flow cytometry data showed that NR2F2-AS1 upregulation dampened cell apoptosis (Fig. 2E, F), and facilitated cell invasion (Fig. 2G) versus the vector group. WB data uncovered that up-regulating NR2F2-AS1 hampered the profile of apoptosis-related proteins (including Bad, Bax and cleaved-caspase3) in OS cells (Supplementary Fig. 1A), heightened the expression of cyclin D1 and cyclin E2 (Supplementary Fig. 1B) and EMT-related genes (Vimentin and Snail) and hindered the E-cadherin level (vs. the vector group) (Supplementary Fig. 1C). These results hinted that NR2F2-AS1 exerted a carcinogenic function in OS.
NR2F2-AS1 overexpression heightened OS cell proliferation and metastasis. The NR2F2-AS1 overexpression model was constructed by transfection of the NR2F2-AS1 overexpression plasmid and its control vector into OS cell lines. A The NR2F2-AS1 profile in OS cell lines was gauged by qRT-PCR. B Cell proliferation was verified by the CCK-8 assay. C, D BrdU staining was performed with AlexaFluor594 conjugated antibody (red). The nuclei were observed by DAPI staining. The BrdU staining results were quantified. E, F: Apoptosis was testified by flow cytometry. G Transwell assay detected cell invasion. Data were expressed as mean ± SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001 (vs.vector group)
NR2F2-AS1 targeted miR-425-5p
For further probing the downstream mechanisms of NR2F2-AS1, an online database was used for predicting the targets of NR2F2-AS in ENCORI (http://starbase.sysu.edu.cn/index.php). It was discovered miR-425-5p was a potential target of NR2F2-AS1 (Fig. 3A). qRT-PCR results showed that miR-425-5p level was reduced in OS tissues compared with that in adjacent non-tumor tissues (Fig. 3B). The miR-425-5p level in Saos-2, MNNG/HOS, MG-63 and U2OS cells was lower than that in human normal osteoblast lines hFOB1.19 (Fig. 3C). Besides, the dual-luciferase reporter gene assay testified that miR-425-5p weakened the luciferase activity of NR2F2-AS1-WT, albeit with having no substantial influence on NR2F2-AS1-MUT (Fig. 3D). Furthermore, the impact of regulating the expression of NR2F2-AS1 on miR-425-5p was probed. It turned out that upregulation of NR2F2-AS1 decreased miR-425-5p expression, whereas NR2F2-AS1 knockdown increased the miR-425-5p level (Fig. 3E). These data demonstrated that miR-425-5p is a NR2F2-AS1 downstream target and is repressed by NR2F2-AS1.
NR2F2-AS1 targeted miR-425-5p. A The Starbase database was adopted to search the downstream molecules of NR2F2-AS1, and miR-425-5p was testified the one. B, C qRT-PCR was implemented to determine the miR-425-5p profile in OS tissues and cells. **P < 0.01, ***P < 0.001 (vs. Normal, vs. hFOB1.19). D The binding association between NR2F2-AS1 and miR-425-5p was ascertained by the dual-luciferase reporter gene assay. **P < 0.01, ***P < 0.001, NSP > 0.05 (vs. miR-NC). E. The miR-425-5p profile in OS cells was checked by qRT-PCR after transfecting NR2F2-AS1 overexpression plasmids and si-NR2F2-AS1, respectively. **P < 0.01, ***P < 0.001 (VS.Vector), ***P < 0.001 (vs. si-NC)
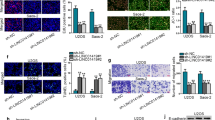
Up-regulating miR-425-5p suppressed OS cell proliferation and invasion
To validate the function of miR-425-5p in OS, we transfected miR-425-5p mimics and miR-NC in Saos-2, MG-63 and U2OS cells, respectively. qRT-PCR outcomes disclosed that the miR-425-5p expression was significantly enhanced in the miR-425-5p group (vs. the miR-NC group) (Fig. 4A). Functional assays revealed that up-regulating miR-425-5p brought about declined cell proliferation (Fig. 4B–D), intensified apoptosis (Fig. 4E, F), and reduced invasion (Fig. 4G). WB results showed that up-regulating miR-425-5p enhanced the profiles of apoptosis-related proteins (including Bad, Bax and C-caspase3) in OS cells (Supplementary Fig. 2A) and hindered cyclin D1 and cyclin E2 expression and EMT processes (Supplementary Fig. 2B, C). These results further clarified that miR-425-5p exerts an anti-tumor function in OS.
Up-regulating miR-425-5p curbed OS cell proliferation, invasion and induced apoptosis. miR-425-5p overexpression model was constructed by transfection of miR-425-5p mimics and miR-NC in OS cell lines. A The miR-425-5p profile in OS cells was tested by qRT-PCR. B CCK-8 was conducted to examine cell proliferation. C, D BrdU staining was implemented with AlexaFluor594 conjugated antibody (red). The nuclei were observed by DAPI staining. The BrdU staining results were quantified. E, F Flow cytometry was carried out for apoptosis detection. G Transwell assay examined cell invasion. Data were presented as mean ± SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001 (vs.miR-NC)
HMGB2 was the functional target of miR-425-5p
The expression and role of miR-425-5p in OS cells are understood, but the exact mechanism of miR-425-5p action remains unknown. Our search of the ENCORI website revealed that miR-425-5p binds to the 3' UTR region of HMGB2 mRNA, substantiating that HMGB2 is a likely target gene for miR-425-5p (Fig. 5A). For further verification, we implemented qRT-PCR and discovered that the HMGB2 mRNA level in OS tissues was notably facilitated (compared with adjacent normal tissues, Fig. 5B). Five paired tumor tissues and adjacent normal tissues were randomly selected from OS patients according to the random number table method. As indicated by the result of Western blot, the tumor tissues from OS patients had enhanced expression of HMGB2 compared with the paired adjacent normal tissues (Fig. 5C). Moreover, those OS cell lines (Saos-2, MNNG/HOS, MG-63, and U2OS) also had higher protein level of HMGB2 compared with the human normal osteoblast lines hFOB1.19 (Fig. 5D). As indicated by the dual-luciferase reporter results, miR-425-5p distinctly impeded the luciferase activity of HMGB2-WT while had no impact on HMGB2-MUT (Fig. 5E). WB was carried out to measure the HMGB2 profile after modulation of miR-425-5p. As a result, transfection of miR-425-5p mimics in OS cells promoted HMGB2 expression, whereas miR-425-5p inhibitors transfection led to reverse outcome (Fig. 5F). These outcomes uncovered that miR-425-5p targets HMGB2.
HMGB2 was the functional target of miR-425-5p. A By adopting Starbase database, we discovered that miR-425-5p harbored a potential binding site for HMGB2. B qRT-PCR was performed for detecting HMGB2 mRNA level in OS tissues and adjacent tissues. C, D Five paired tumor tissues and adjacent normal tissues were randomly selected from OS patients. WB was utilized for determining the HMGB2 expression in OS tissues B and cells C. ***P < 0.001 (vs.Normal, vs.hFOB1.19). E The dual-luciferase reporter assay was utilized to probe the link between miR-425-5p and HMGB2. F: The HMGB2 expression after the regulation of miR-425-5p was checked by WB and cell immunofluorescence. Data were expressed as mean ± SD (n = 3). NSP > 0.05, **P < 0.01, ***P < 0.001 (VS.miR-NC), **P < 0.01, ***P < 0.01 (vs.miR-in)
NR2F2-AS1 influenced OS cell proliferation and metastasis via the miR-425-5p/HMGB2 axis
For substantiating the role of the NR2F2-AS1/miR-425-5p/HMGB2 axis in regulating OS evolvement, we supplemented the NR2F2-AS1 overexpression plasmid, and/or si-HMGB2 after miR-425-5p overexpression and examined the altered proliferation and metastasis of MG-63 cells. As the data showed, miR-425-5p mimic transfection resulted in reduced OS cell proliferation, BrdU-positive cell rate and cell invasion. The supplementation of NR2F2-AS1 enhanced OS cell proliferation, BrdU-positive cell rate, and invasion (vs.miR-425-5p group). However, si-HMGB2 addition attenuated NR2F2-AS1-mediated promotive effects (vs.miR-425-5p + NR2F2-AS1 + si-NC group) (Fig. 6A–C). The cellular apoptosis and EMT were evaluated by Flow cytometry and WB. It was found that miR-425-5p enhanced cell apoptosis, promoted Bax, Bad, C-Caspase3 level, repressed the expression of cyclin D1 and cyclin E2 and EMT processes. NR2F2-AS1 overexpression reversed miR-425-5p-mediated effects were heightened in OS cells. Interestingly, the si-HMGB2 addition promoted cell apoptosis and reduced EMT (vs.miR-425-5p + NR2F2-AS1 + si-NC group) (Fig. 6D–G). The qRT-PCR and WB outcomes manifested that the miR-425-5p had no significant effect in NR2F2-AS1 alteration, but repressed HMGB2 expression (vs. the miR-NC group), NR2F2-AS1 overexpression inhibited miR-425-5p level and promoted HMGB2 level. si-HMGB2 had no significant effects in miR-425-5p and NR2F2-AS1 expression (Fig. 6H, I). These data illustrated that NR2F2-AS1 influences the malignant OS progression through the miR-425-5p/HMGB2 pathway.
NR2F2-AS1 influenced OS cell proliferation and metastasis via the miR-425-5p/HMGB2 pathway. miR-NC, miR-425-5p mimics, NR2F2-AS1 overexpression, and, or si-HMGB2 were transfected into MG-63 cells. A CCK-8 was adopted to test cell proliferation. B BrdU staining was made with AlexaFluor594 conjugated antibody (red). The nuclei were reviewed by DAPI staining. The BrdU staining results were quantified. C Cell invasion was evaluated by Transwell assay. D Apoptosis was assessed by flow cytometry. E–G The profiles of Bad, Bax, C-caspase3 E, cyclin D1, cyclin E2 F, and Vimentin, Snail, and E-cadherin G were testified by WB. H The cellular NR2F2-AS1 and miR-425-5p profile was gauged by qRT-PCR. I The cellular HMGB2 level was monitored by WB. Data were expressed as mean ± SD (n = 3). *P < 0.05, ***P < 0.001 (vs. miR-NC), *P < 0.05, **P < 0.01, ***P < 0.01 (vs. miR-425-5p)
Overexpressing NR2F2-AS1 amplified tumor cell growth in vivo
To ascertain the impact of NR2F2-AS1 on cancer proliferation in vivo, we set up a mouse xenograft model by overexpressing NR2F2-AS1 cells. Strikingly, NR2F2-AS1 overexpression expedited tumor volume growth in nude mice (compared with vector group, Fig. 7A–C). In addition, H&E staining of lung tissue from a nude mouse model confirmed NR2F2-AS1 overexpression enhanced the metastasis of OS cells in lung tissues (Fig. 7D). Moreover, WB results showed that overexpressing NR2F2-AS1 attenuated Bax, Bad, C-Caspase3, enhanced cyclin D1 and cyclin E2, promoted Vimentin and Snail, whereas repressed E-cadherin level (Fig. 7 E–G). We further examined the altered expression of NR2F2-AS1, miR-425-5p in tumor tissues using qRT-PCR and WB. It turned out that the NR2F2-AS1 overexpression group had elevated NR2F2-AS1 levels, impeded miR-425-5p levels, and enhanced HMGB2 expression (vs. the NC group) (P < 0.05, Fig. 7H, I). Collectively, NR2F2-AS1 promoted OS cell growth and metastasis in vivo.
NR2F2-AS1 overexpression boosted tumor cell growth in vivo. A Subcutaneous tumors were removed from nude mice at 6 weeks post tumor cell transplantation. B Tumor size was enumerated weekly. C The weight of subcutaneous tumors in nude mice was evaluated. D Number of tumor metastasis nodules in lung was calculated. E–G The profiles of Bad, Bax, C-caspase3 E, cyclin D1, cyclin E2 F, and Vimentin, Snail, and E-cadherin G were testified by WB. H and I The expression of NR2F2-AS1 and miR-425-5p in tumor tissues was assessed by qRT-PCR. J WB was adopted for testing the HMGB2 expression in tumor tissues. Data were expressed as mean ± SD (n = 5). NSP > 0.05 (vs. Blank group), **P < 0.01, ***P < 0.001 (vs. Vector group)
Discussion
With the continuous exploration of OS, emerging lncRNAs have been revealed to be implicated in OS. Here, we discovered that NR2F2-AS1 expression was heightened in OS tissues and cells. Further experiments confirmed that NR2F2-AS1 enhanced OS cell proliferation, migration and invasion.
LncRNAs have been testified to contribute to various tumors by numerous studies [24,25,26]. In addition, LncRNA SNHG3 [27], LncRNA BE503655 [5], LncRNA AFAP1-AS1 [28], lncRNA SNHG4 [29] and LncRNA TUG1 [30] expression is boosted in OS tissues, and their overexpression strengthens OS cell proliferation, migration and invasion. NR2F2-AS1, a newly identified lncRNA, have been identified as a carcinogene mediating small cell lung cancer [31], nasopharyngeal cancer [32] and colorectal cancer [33]. Presently, we discovered that NR2F2-AS1 expression was facilitated in both OS tissues and cells, reinforcing OS cell proliferation and invasion and tumor growth in vivo. NR2F2-AS1 may contribute to OS as an oncogenic gene.
The significance of miRNAs in tumor progression has been extensively studied. As reported, miRNAs regulate diversified tumor tissues. As an example, miR-491-5p overexpression hampers estrogen signaling transduction and estrogen-spurred breast cancer cell proliferation [34]. Down-regulation of miR-519a heralds unsatisfactory GC prognosis and contributes to tumor progression [35]. Also, the abnormal expression of miRNAs affects OS development. As an example, both miR-99b and miR-1296-5p are down-regulated in OS tissues, predicting poorer clinical outcomes of OS patients. In vitro, miR-99b and miR-1296-5p abates OS cell proliferation, migration and invasion [36, 37]. Moreover, miR-596 dampens the Survivin profile and facilitates OS cells’ sensitivity to anlotinib [38]. These studies suggest that in OS, aberrantly expressed miRNAs can be used not only as molecular markers of prognosis, but also as therapeutic targets. Previous studies disclosed that miR-425-5p is down-regulated in OS patients’ serum [13, 14] and that overexpression of miR-425-5p impedes OS cell proliferation, invasion and migration. This article elaborated that miR-425-5p expression was hindered in OS cells and tissues, and elevating miR-425-5p profiles hindered OS cell proliferation, invasion, and migration, suggesting that miR-425-5p functions as an antitumor miRNA. Moreover, emerging reports have stated that lncRNAs contribute to modulating miRNAs as ceRNAs. LncRNA RBMS3-AS3 sponges miR-4534 and abates prostate cancer development by up-regulating VASH1 [39]. LncRNA H19 hinder multiple myeloma (MM) tumorigenesis by targeting miR-152-3p to suppress BRD4-mediated cell proliferation [40]. As indicated by our data, overexpressing NR2F2-AS1 choked miR-425-5p's expression in OS cells and attenuated its anti-tumor effects, illustrating that NR2F2-AS1 targets miR-425-5p to affect OS cell proliferation and invasion.
The HMGB family consists primarily of HMGB1, HMGB2, and HMGB3 proteins, which are normally chromatin-associated and ubiquitously distributed in the nucleus of higher eukaryotic cells and efficiently bend DNA and form DNA circle [16]. Thus, this little group of proteins potentially enhance the activities on various gene promoters by enhancing transcription factor binding and/or bringing distant regulatory sequences into close proximity [41, 42]. Increasing studies have found that HMGB1 mediates enhanced osteosarcoma cell proliferation and invasion, but reduced apoptosis [43]. HMGB1 upregulation is induced by doxorubicin, cisplatin, and methotrexate in human osteosarcoma cells, and HMGB1 knockdown enhanced the chemosensitivity of osteosarcoma cells [44]. A recent study found that HMGB2 overexpression is closely associated with the Enneking stage, tumor volume, metastasis, and survival of OS, and it strengthens OS cell proliferation, migration and invasive capacity [21], suggesting that HMGB2 is a potential oncogene in OS. Notably, miRNAs can target and regulate HMGB2, thus affecting cellular development. As an example, down-regulating miR-130a facilitates glioma cell growth and invasion by activating HMGB2 expression [23]. Here, we discovered that HMGB2 expression was elevated in OS cells and that up- or down-regulating miR-425-5p increased or decreased HMGB2 expression in OS cells, respectively. Meanwhile, we ascertained that NR2F2-AS1 was positively linked to HMGB2 expression in OS cells, suggesting that NR2F2-AS1 and miR-425-5p can target and modulate HMGB2 in OS and thus affect cell proliferation and invasion.
Conclusion
Overall, our research confirms that NR2F2-AS1 expression is heightened in OS tissues and cells, amplifying OS cell proliferation and invasion and regulating OS evolvement by modulating the miR-425-5p-HMGB2 axis (Fig. 8). Taken together, this study provides a better insight into the molecular mechanisms modulating OS evolvement, which will help in the early detection and treatment planning of OS.
Data availability statement
The data sets used and analyzed during the current study are available from the corresponding author on reasonable request.
References
Zhao W, Qin P, Zhang D et al (2019) Long non-coding RNA PVT1 encapsulated in bone marrow mesenchymal stem cell-derived exosomes promotes osteosarcoma growth and metastasis by stabilizing ERG and sponging miR-183-5p. Aging (Albany NY) 11(21):9581–9596
Ren Z, He M, Shen T et al (2020) MiR-421 promotes the development of osteosarcoma by regulating MCPIP1 expression. Cancer Biol Ther 21(3):231–240. https://doi.org/10.1080/15384047.2019.1683331 (Epub 2019 Nov 12)
Zhao A, Liu W, Cui X et al (2021) lncRNA TUSC7 inhibits osteosarcoma progression through the miR-181a/RASSF6 axis. Int J Mol Med 47(2):583–594. https://doi.org/10.3892/ijmm.2020.4825 (Epub 2020 Dec 18)
Fu D, Lu C, Qu X et al (2019) LncRNA TTN-AS1 regulates osteosarcoma cell apoptosis and drug resistance via the miR-134-5p/MBTD1 axis. Aging (Albany NY) 11(19):8374–8385. https://doi.org/10.18632/aging.102325 (Epub 2019 Oct 10)
Huang Q, Shi SY, Ji HB et al (2019) LncRNA BE503655 inhibits osteosarcoma cell proliferation, invasion/migration via Wnt/β-catenin pathway. Biosci Rep 39(7):BSR20182200. https://doi.org/10.1042/BSR20182200
Fu X, Wang D, Shu T et al (2020) LncRNA NR2F2-AS1 positively regulates CDK4 to promote cancer cell proliferation in prostate carcinoma. Aging Male 23(5):1073–1079. https://doi.org/10.1080/13685538.2019.1670157 (Epub 2019 Sep 28)
Liu D, Huang K, Wang T et al (2020) NR2F2-AS1 accelerates cell proliferation through regulating miR-4429/MBD1 axis in cervical cancer. Biosci Rep 40(6):BSR20194282
Mishra S, Yadav T, Rani V (2016) Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit Rev Oncol Hematol 98:12–23. https://doi.org/10.1016/j.critrevonc.2015.10.003 (Epub 2015 Oct 8)
Cai WT, Guan P, Lin MX et al. (2020) MiRNA-206 suppresses the metastasis of osteosarcoma via targeting Notch3. J Biol Regul Homeost Agents 34(3):775–783. https://doi.org/10.23812/20-72-A-26
Sun Y, Cao L, Lin JT et al (2019) Upregulated miRNA-1236-3p in osteosarcoma inhibits cell proliferation and induces apoptosis via targeting KLF8. Eur Rev Med Pharmacol Sci 23(14):6053–6061. https://doi.org/10.26355/eurrev_201907_18418
Lu Y, Wu X, Wang J (2019) Correlation of miR-425-5p and IL-23 with pancreatic cancer. Oncol Lett 17(5):4595–4599. https://doi.org/10.3892/ol.2019.10099 (Epub 2019 Mar 4)
Xiao S, Zhu H, Luo J et al (2019) miR-425-5p is associated with poor prognosis in patients with breast cancer and promotes cancer cell progression by targeting PTEN. Oncol Rep 42(6):2550–2560
Yang G, Zhang C, Wang N et al (2019) miR-425-5p decreases LncRNA MALAT1 and TUG1 expressions and suppresses tumorigenesis in osteosarcoma via Wnt/β-catenin signaling pathway. Int J Biochem Cell Biol 111:42–51
Li H, Zhang K, Liu LH et al (2015) MicroRNA screening identifies circulating microRNAs as potential biomarkers for osteosarcoma. Oncol Lett 10(3):1662–1668
Fu D, Li J, Wei J et al (2018) HMGB2 is associated with malignancy and regulates Warburg effect by targeting LDHB and FBP1 in breast cancer. Cell Commun Signal 16(1):8
Fang J, Ge X, Xu W et al (2020) Bioinformatics analysis of the prognosis and biological significance of HMGB1, HMGB2, and HMGB3 in gastric cancer. J Cell Physiol 235(4):3438–3446. https://doi.org/10.1002/jcp.29233 (Epub 2019 Oct 16)
Cui G, Cai F, Ding Z et al (2019) HMGB2 promotes the malignancy of human gastric cancer and indicates poor survival outcome. Hum Pathol 84:133–141. https://doi.org/10.1016/j.humpath.2018.09.017 (Epub 2018 Oct 5)
Mo Y, Fang RH, Wu J et al (2019) MicroRNA-329 upregulation impairs the HMGB2/β-catenin pathway and regulates cell biological behaviors in melanoma. J Cell Physiol 234(12):23518–23527
Suzuki S, Kato H, Fuji S et al (2018) Early detection of prostate carcinogens by immunohistochemistry of HMGB2. J Toxicol Sci 43(6):359–367
Zhang P, Lu Y, Gao S (2019) High-mobility group box 2 promoted proliferation of cervical cancer cells by activating AKT signaling pathway. J Cell Biochem 120(10):17345–17353. https://doi.org/10.1002/jcb.28998 (Epub 2019 Jun 18)
Yang S, Ye Z, Wang Z et al (2020) High mobility group box 2 modulates the progression of osteosarcoma and is related with poor prognosis. Ann Transl Med 8(17):1082. https://doi.org/10.21037/atm-20-4801
Yang XR, Xiong Y, Duan H et al (2015) Identification of genes associated with methotrexate resistance in methotrexate-resistant osteosarcoma cell lines. J Orthop Surg Res 4(10):136. https://doi.org/10.1186/s13018-015-0275-8
Tang C, Yang Z, Chen D et al (2017) Downregulation of miR-130a promotes cell growth and epithelial to mesenchymal transition by activating HMGB2 in glioma. Int J Biochem Cell Biol 93:25–31
Tang X, Yu L, Bao J et al (2019) Function of long noncoding RNA UCA1 on gastric cancer cells and its clinicopathological significance in plasma. Clin Lab. https://doi.org/10.7754/Clin.Lab.2019.181233
Luan W, Ding Y, Ma S et al (2019) Long noncoding RNA LINC00518 acts as a competing endogenous RNA to promote the metastasis of malignant melanoma via miR-204-5p/AP1S2 axis. Cell Death Dis 10(11):855
Liu Y, Guo C, Li F et al (2020) LncRNA LOXL1-AS1/miR-28-5p/SEMA7A axis facilitates pancreatic cancer progression. Cell Biochem Funct 38(1):58–65. https://doi.org/10.1002/cbf.3449 (Epub 2019 Nov 15)
Zheng S, Jiang F, Ge D et al (2019) LncRNA SNHG3/miRNA-151a-3p/RAB22A axis regulates invasion and migration of osteosarcoma. Biomed Pharmacother 112:108695. https://doi.org/10.1016/j.biopha.2019.108695 (Epub 2019 Feb 20)
Shi D, Wu F, Mu S et al (2019) LncRNA AFAP1-AS1 promotes tumorigenesis and epithelial-mesenchymal transition of osteosarcoma through RhoC/ROCK1/p38MAPK/Twist1 signaling pathway. J Exp Clin Cancer Res 38(1):375. https://doi.org/10.1186/s13046-019-1363-0
Xu R, Feng F, Yu X et al (2018) LncRNA SNHG4 promotes tumour growth by sponging miR-224-3p and predicts poor survival and recurrence in human osteosarcoma. Cell Prolif 51(6):e12515
Sheng K, Li Y (2019) LncRNA TUG1 promotes the development of osteosarcoma through RUNX2. Exp Ther Med 18(4):3002–3008
Zhang S, Zhang X, Sun Q et al (2019) LncRNA NR2F2-AS1 promotes tumourigenesis through modulating BMI1 expression by targeting miR-320b in non-small cell lung cancer. J Cell Mol Med 23(3):2001–2011. https://doi.org/10.1111/jcmm.14102 (Epub 2018 Dec)
Qin H, Qin C (2020) Downregulation of long non-coding RNA NR2F2-AS1 inhibits proliferation and induces apoptosis of nasopharyngeal carcinoma cells by up-regulating the expression of PTEN. Oncol Lett 19(2):1145–1150. https://doi.org/10.3892/ol.2019.11211 (Epub 2019 Dec)
Li F, Jiang Z, Shao X et al (2020) Downregulation of lncRNA NR2F2 antisense RNA 1 induces G1 arrest of colorectal cancer cells by downregulating cyclin-dependent kinase 6. Dig Dis Sci 65(2):464–469
Hui Z, Yiling C, Wenting Y et al (2015) miR-491-5p functions as a tumor suppressor by targeting JMJD2B in ERα-positive breast cancer. FEBS Lett 589(7):812–821
Cai H, Lin H, Cao W et al (2020) Downregulation of miR-519a predicts poor prognosis and contributes to tumor progression in gastric cancer. Oncol Res Treat 43(1–2):19–26
Shi X, Guan X (2019) MicroRNA-99b predicts clinical outcome of osteosarcoma and suppresses tumor cell proliferation, migration and invasion. Diagn Pathol 14(1):117
Wang L, Hu K, Chao Y et al (2020) MicroRNA-1296-5p suppresses the proliferation, migration, and invasion of human osteosarcoma cells by targeting NOTCH2. J Cell Biochem 121(2):2038–2046
Wang L, En H, Yang L et al (2019) miR-596 suppresses the expression of survivin and enhances the sensitivity of osteosarcoma cells to the molecular targeting agent anlotinib. Onco Targets Ther 12:6825–6838
Jiang Z, Zhang Y, Chen X et al (2020) Long noncoding RNA RBMS3-AS3 acts as a microRNA-4534 sponge to inhibit the progression of prostate cancer by up-regulating VASH1. Gene Ther 27(3–4):143–156
Zheng JF, Guo NH, Zi FM et al (2020) Long noncoding RNA H19 promotes tumorigenesis of multiple myeloma by activating BRD4 signaling by targeting microRNA 152–3p. Mol Cell Biol 40(3):e00382-e419
Fan Z, Beresford PJ, Zhang D et al (2002) HMG2 interacts with the nucleosome assembly protein SET and is a target of the cytotoxic T-lymphocyte protease granzyme A. Mol Cell Biol 22(8):2810–2820
Ugrinova I, Pashev IG, Pasheva EA (2009) Nucleosome binding properties and co-remodeling activities of native and in vivo acetylated HMGB-1 and HMGB-2 proteins. Biochemistry 48(27):6502–6507
Meng Q, Zhao J, Liu H et al (2014) HMGB1 promotes cellular proliferation and invasion, suppresses cellular apoptosis in osteosarcoma. Tumour Biol 35(12):12265–12274
Huang J, Ni J, Liu K et al (2012) HMGB1 promotes drug resistance in osteosarcoma. Cancer Res 72(1):230–238
Funding
This research was funded by Natural Science Foundation of Hubei Province (No. 2020CFB659).
Author information
Authors and Affiliations
Contributions
ZJ: conceived and designed the experiments; JY, HH: performed the experiments; SC, YR, WG: statistical analysis; JY, HH, SC, YR, ZJ: wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This research was authorized by the research ethics committee of Renmin Hospital of Wuhan University.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
About this article
Cite this article
Ye, J., He, H., Chen, S. et al. Long non-coding RNA NR2F2-AS1 regulates human osteosarcoma growth and metastasis through miR-425-5p-mediated HMGB2. Int J Clin Oncol 27, 1891–1903 (2022). https://doi.org/10.1007/s10147-022-02245-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10147-022-02245-2