Abstract
Background
This study examined the association between methylenetetrahydrofolate reductase (MTHFR) polymorphisms and survival of patients with colorectal cancer (CRC) treated with 5-fluorouracil (5-FU)-based chemotherapy in Taiwan.
Methods
We genotyped MTHFR polymorphisms C677T (rs1801133) and A1298C (rs1801131) for 498 CRC patients treated with 5-FU-based chemotherapy after receiving surgery. Survival analyses on MTHFR polymorphisms were performed using log-rank test and Kaplan–Meier curve. Cox proportional hazards models were used to calculate the hazard ratios (HRs) and 95% confidence intervals (CIs) for the association between MTHFR genotypes and survival.
Results
Overall survival (OS) was significantly longer in CRC patients with MTHFR 677 CT+TT genotypes compared with those with 677 CC genotype (HR 0.77; 95% CI 0.60–0.98). Although the MTHFR A1298C polymorphism was not associated with OS in CRC, this polymorphism was associated with significantly shorter OS in rectal cancer. Among rectal cancer patients, OS was shorter for patients with AC+CC genotypes than for those with the AA genotype (HR 1.95; 95% CI 1.35–2.83). In haplotype analysis, better OS was found for colon cancer patients carrying the MTHFR 677T-1298A haplotype (HR 0.73; 95% CI 0.55–0.97), but worse survival was linked to rectal cancer patients carrying the MTHFR 677C-1298C haplotype (HR 1.53; 95% CI 1.08–2.18).
Conclusions
Our findings suggest that MTHFR genotypes provide prognostic information for CRC patients treated with 5-FU-based chemotherapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
5-Fluorouracil (5-FU), a drug of choice for systemic chemotherapy regimens in colorectal cancer (CRC) patients, exerts its anticancer activity through inhibiting the action of thymidylate synthase (TS) [1]. A stable ternary complex is formed between a 5-FU active metabolite (fluorodeoxyuridine monophosphate), the enzyme and 5,10-methylenetetrahydrofolate (5,10-methyleneTHF) which results in TS inhibition. The optimal inhibition of TS depends on elevated cellular concentrations of 5,10-methyleneTHF.
The concentration of 5,10-methyleneTHF is controlled by the activity of methylenetetrahydrofolate reductase (MTHFR) [2], a critical enzyme that is involved in folate metabolism. This enzyme irreversibly converts 5,10-methyleneTHF to 5-methyltetrahydrofolate (5,10-methylTHF), which is essential for the methylation of homocysteine to methionine [3]. In addition, 5,10-methyleneTHF is also required for conversion of deoxyuridine monophosphate to deoxythymidine monophosphate by TS, which then affects DNA synthesis [4].
Decreased MTHFR enzyme activity results in higher concentrations of 5,10-methyleneTHF that conversely favor the formation and stability of the inhibitory ternary complex. Thus, patients with a genotype associated with decreased MTHFR enzyme activity should be more sensitive to 5-FU than patients with a genotype related to normal MTHFR enzyme expression. MTHFR has several polymorphisms on chromosome 1p [5]. Of these, the 677C>T (Ala to Val) and 1298A>C (Glu to Ala) single nucleotide polymorphisms (SNPs) are the two most commonly connected with altered enzyme activity [6,7,8]. The 677C>T substitution renders the enzyme thermolabile, and subjects with TT and CT genotypes have approximately 70 and 35% reduced enzyme activity, respectively [6]. The 1298A>C transition also leads to decreased enzyme activity, although not to the same extent as the 677T allele.
Several clinical studies have investigated these two MTHFR polymorphisms and clinical outcome in 5-FU-based chemotherapy for CRC, but the data are inconsistent and controversial. Wisotzkey et al. was the first to exam the association between the MTHFR C677T polymorphism and clinical response to 5-FU therapy in colon cancer patients, but they were unable to demonstrate an overall survival (OS) difference between patients carrying different MTHFR genotypes [9]. In addition, some studies also suggest that MTHFR polymorphisms cannot be considered as an independent predictor of clinical outcome in CRC patients receiving 5-FU-based chemotherapy [2, 10,11,12,13,14,15,16,17,18,19,20]. By contrast, many studies have reported an association between MTHFR polymorphisms and tumor response to 5-FU-based chemotherapy [21,22,23,24,25,26,27,28,29,30,31,32,33,34,35].
We conducted a retrospective study to test the hypothesis that MTHFR C677T and A1298C polymorphisms are associated with prognosis in CRC patients treated with 5-FU-based chemotherapy. We also explored whether the association differs between colon and rectal cancer.
Materials and methods
Patients
The clinicopathological characteristics and chemotherapy regimens of the patients have been clearly described in our previous studies [36, 37]. Briefly, 498 sporadic CRC patients at stages II–IV who received postoperative 5-FU-based chemotherapy as a first-line treatment were enrolled at Chang Gung Memorial Hospital (CGMH) between January 1995 and December 2001 and were followed up every 3–6 months at the outpatient clinic of the Colorectal Section at CGMH. Operative procedures were based on the clinical condition of the patients. We had no predefined criteria of resectability with regard to the tumor characteristics, such as metastatic sites and the number and size of the tumors, etc. All the patients with stages II–III had a curative resection but only 26 (17.1%) patients with stage IV cancer had a curative-intent resection. The study protocol was approved by the Institutional Review Board at China Medical University Hospital (DMR96-IRB-001) and Chang Gung Memorial Hospital (97-0424B). Informed consent was obtained from all individual participants included in the study.
The chemotherapy regimens included oral tegafur–uracil (UFUR) and weekly and monthly intravenous 5-FU plus levamisole or leucovorin (LV) (Supplementary Table S1). The oral tegafur (300–350 mg/m2/day) plus levamisole (45 mg/day) regimen was administered daily in 3 doses for 28 days for a 12-month treatment period. The weekly treatment included (1) weekly intravenous bolus injection 5-FU (450 mg/m2/day) plus LV (50 mg/day); (2) intravenous injection of 5-FU (450 mg/m2/day) weekly plus oral levamisole (50 mg/day) for 3 days every 2 weeks; or (3) a 1-day infusion of high-dose 5-FU (2600 mg/m2/day) plus weekly LV (150 mg/day). The monthly treatment included (1) a continuous 5-day infusion of 5-FU (800 mg/m2/day) plus LV (50 mg/day) administered once monthly; or (2) a continuous 5-day infusion of 5-FU (800 mg/m2/day) administered monthly and combined with oral levamisole (50 mg/day) for 3 days every 2 weeks. The weekly and monthly intravenous medications lasted 12 months for the weekly regimen and 6 months for the monthly regimen. Treatment courses were continuous unless there was evidence of progression, unacceptable toxicity, or the patient chose to discontinue treatment. Patients were allowed to undergo further chemotherapy at the physician’s discretion. The chemotherapeutic toxicity was graded according to the Common Toxicity Criteria of the National Cancer Institute. These toxic events were dichotomously classified for statistical analysis as none-to-moderate (grade 0–2) and severe (grade 3–4).
Clinical data
Clinical data included medical history, physical examination (rectal and perineal examination), and measurement of carcinoembryonic antigen (CEA) collected during the follow-up period. Liver sonographs, chest X-ray, and colonoscopy (or a barium enema) were performed annually. The criteria for establishing recurrent disease included histological confirmation, or the disease being evident on radiographic or sonographic studies with subsequent clinical progression. We not only collected recurrence and health status of the patient at the follow-up clinical visit but also identified cause of death from the death certificates. The prospective follow-up data were available for all patients until May 2008, with a median follow-up time of 48 months (range 1.5−133 months). Disease-free survival (DFS) was measured from the date of surgery to the time of tumor recurrence, metastasis, or death from any cause. OS was measured from the date of surgery to death from any cause. Patients who were alive at the last follow-up evaluation were censored at that time.
MTHFR polymorphisms
Genomic DNA was extracted from buffy coat cells using standard procedures with sodium dodecyl sulfate (VS)-proteinase K-RNase digestion and phenol–chloroform extraction. The MTHFR C677T (rs1801133) and MTHFR A1298C (rs1801131) polymorphisms were detected using polymerase chain reaction (PCR) and restriction fragment length polymorphism methods [6]. All PCR reactions were performed in a 20-µl final volume containing 5 pM of each primer, 50 ng genomic DNA, 1.5 mM MgCl2, 200 µM dNTPs and 1.0 unit of Taq DNA polymerase in the buffer provided by the manufacturer (Fermentas, Glen Burnie, MD, USA). PCR amplification was performed in a Mastercycler gradient thermocycler (Eppendorf, Hamburg, Germany).
The MTHFR C677T polymorphism was determined using the following primers: sense, 5′-TGA AGG AGA AGG TGT CTG CGG GA-3′; antisense, 5′-AGG ACG GTG CGG TGA GAG TG-3′. The PCR program was a 2-min denaturing step at 94 °C, followed by 30 cycles of 30 s at 94 °C, 30 s at 60 °C and 30 s at 72 °C. The 198 bp PCR product was digested with HinfI (New England BioLabs, Beverly, MA): the T allele was cut into 175 and 23 bp fragments; the C allele was not digested. The MTHFR A1298C polymorphism was determined using the following primers: sense, 5′-ATG TGG GGG GAG GAG CTG AC-3′; antisense, 5′-GTC TCC CAA CTT ACC CTT CTC CC-3′. The PCR program was a 5-min denaturing step at 92 °C, followed by 35 cycles of 60 s at 92 °C, 30 s at 60.5 °C and 30 s at 72 °C. The 241 bp PCR product was digested with MboII (Fermentas): the A allele was cut into 204 and 37 bp fragments; the C allele was not digested. For quality control, 10% of randomly selected samples were repeated and showed 100% concordance for both SNPs.
Statistical analysis
We compared the distributions of each categorical variable by chi-squared test and of continuous variables by Student’s t test. A univariate analysis of Kaplan–Meier estimates and log-rank test were used to compare survival curves. Cox proportional hazards regression models were used to estimate the hazard ratios (HRs) and their 95% confidence intervals (CIs) on survival according to different genotype. Covariates, such as age, gender, TNM stage and tumor subsite, were included in the multivariate Cox proportional hazards models to estimate adjusted HRs and 95% CIs. HRs for survival risk differences by tumor subsite were estimated using stratification analysis. All analyses were performed using an SAS statistical package (version 9.4 for Windows; SAS Institute, Cary, NC, USA), and two-sided P values of <0.05 were considered statistically significant. Haplotypes were constructed and analyzed using THESIAS program (JAVA version) [38].
Results
Patient characteristics
The clinicopathological characteristics of the CRC patients are summarized in Table 1. The total of 498 patients included 287 colon cancer patients and 211 rectal cancer patients with a mean age of 57.2 and 60.5 years, respectively (P = 0.003). The percentages of stage III and moderately differentiated tumors were significantly higher in rectal cancer patients (61.6 and 84.3%, respectively) than in colon cancer patients (49.5 and 74.0%, respectively). The distributions of sex, family history of cancer and CEA were not different between colon and rectal cancer patients. The genotypic distributions of the two MTHFR polymorphisms for CRC patients are also shown in Table 1. The frequencies for the MTHFR 677T and MTHFR 1298C alleles were 28.5 and 23.5%, respectively. The distributions of the two MTHFR polymorphisms were not significantly different between colon and rectal cancer patients even when stratified by tumor stage (Supplementary Table S2).
Association between MTHFR genotype and toxicity
Table 2 shows the incidence of severe toxicity (grade 3–4) by MTHFR polymorphisms and tumor site. The percentages of severe leukopenia were significantly higher in patients carrying the MTHFR 677 TT genotype (7.1%) than in those carrying the CC or CT genotype (0.8 and 0.9%, respectively) among all CRC patients (P = 0.01), as well as among colon cancer patients (P = 0.003). However, the MTHFR A1298C polymorphism was not associated with chemotherapeutic toxicity.
Association between MTHFR genotype and DFS
Survival analyses by Kaplan–Meier estimates and log-rank test found no significant association between the two MTHFR polymorphisms and DFS (data not shown). In addition, Cox proportional hazards model with an adjustment for age, gender, TNM stage, and tumor subsite showed that the MTHFR polymorphisms were not associated with DFS. Stratified analyses by tumor sites for DFS of colorectal cancer associated with MTHFR polymorphisms also showed no significant association.
Association between MTHFR genotype and OS
For all CRC patients, the MTHFR 677 TT genotype and CT+TT genotype were associated with better OS than the CC genotype (HR 0.55; 95% CI 0.31–0.98; HR 0.77; 95% CI 0.60–0.98, respectively) (Table 3). The MTHFR A1298C polymorphism was not associated with OS.
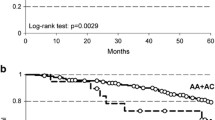
In stratification analysis for tumor subsite, the Kaplan–Meier survival curve showed a significant association between MTHFR A1298C polymorphism and OS in rectal cancer (log-rank test P = 0.01, Fig. 1d). The multivariable Cox proportional hazards model also showed that rectal cancer patients with MTHFR A1298C CC and AC genotypes had 2.08-fold (HR 2.08; 95% CI 0.98–4.43) and 1.93-fold (HR 1.93; 95% CI 1.32–2.84) poorer OS than patients with the AA genotype. Meanwhile, rectal cancer patients carrying the MTHFR A1298C C allele (AC+CC) had a shorter survival compared to those with the AA genotype (42.2 vs 53.4 months; HR 1.95, 95% CI 1.35–2.83). Interestingly, an inverse relationship between 1298C allele and survival was prominent in stage IV rectal cancer patients (HR 2.65; 95% CI 1.52–4.60) (Supplementary Table S3). However, the MTHFR C677T polymorphism was not associated with OS in rectal cancer (Fig. 1c). In addition, OS in colon cancer patients did not differ between the two MTHFR polymorphisms as shown in the survival curves (Fig. 1a, b) or in the Cox hazards models (Table 3).
Haplotype analysis
We further conducted haplotype analysis for MTHFR C677T and A1298C polymorphisms and survival in CRC patients. The univariate Kaplan–Meier survival curve showed significant differences in OS among the 4 haplotypes analyzed in all CRC patients (log-rank test P = 0.03, Fig. 2a), as well as in colon cancer patients (log-rank test P = 0.05, Fig. 2b). However, OS in rectal cancer patients did not differ between the 4 haplotypes as shown in Fig. 2c (log-rank test P = 0.08).
Table 4 shows the adjusted associations between the MTHFR haplotypes and OS in the multivariate Cox proportional hazards models. There was significantly better OS for all CRC patients with the T/A haplotype compared with the most common C/A haplotype (HR 0.77; 95% CI 0.62–0.96), as well as in colon cancer (HR 0.73; 95% CI 0.55–0.97). In contrast, the C/C haplotype was significantly associated with poor OS in rectal cancer patients (HR 1.53; 95% CI 1.08–2.18). However, the significantly better survival presented in the T/A haplotype disappeared when stratified by tumor stage (Supplementary Table S4). Surprisingly, the worse OS inherited from the C/C haplotype was enhanced in stage IV rectal cancer patients (HR 2.36; 95% CI 1.41–3.95). No association was observed between the MTHFR haplotype and DFS in CRC patients even when stratified by tumor subsite.
Discussion
We carried out an analysis of MTHFR gene polymorphisms in 498 CRC patients homogenously treated with 5-FU-based chemotherapy. The results suggested that the MTHFR 677T allele could predict better OS in CRC patients treated with 5-FU. Furthermore, the 1298C allele was associated with worse OS in rectal but not in colon cancer cases.
Although several clinical studies have correlated these two MTHFR polymorphisms with OS in CRC patients treated with 5-FU-based chemotherapy, the results are inconsistent and contradictory. Some studies have reported that C677T genetic variant is significantly associated with increased tumor response rate to 5-FU-based therapy [21,22,23, 28]. The 677T allele polymorphic genotype has also been associated with significantly increased time to survival [23, 29, 32, 34, 35], while negative effects on OS were reported [27, 31, 33, 39]. On the contrary, the 1298C allele has been correlated with an increased risk of severe adverse events or worse survival after 5-FU-based chemotherapy [22, 25, 27, 28, 34, 35]. However, several other studies do not show any correlation between MTHFR polymorphisms and clinical outcome in CRC patients treated with 5-FU-based chemotherapy [2, 9, 10, 15,16,17,18,19,20, 40,41,42,43,44,45].
Haplotype analyses have shown that MTHFR C667T and A1298C polymorphisms are in high linkage disequilibrium and this approach is also more predictive than the single polymorphism for total plasma homocysteine levels [46]. A few studies have started to investigate the relationship between MTHFR haplotype and clinical outcome in CRC patients treated with chemotherapy. Terrazzino et al. conducted a C677T/A1298C haplotype analysis in rectal cancer patients undergoing preoperative 5-FU-based chemoradiation [24], and found that the T/A haplotype predisposed to a worse response and lower tumor regression rate compared to other haplotype combinations. However, using a similar setting, Thomas et al. reported that the C/C haplotype was associated with a protective effect on the incidence of severe diarrhea or mucositis [47]. In our haplotype analysis, the T/A haplotype was significantly related to better OS in CRC patients, while the C/C haplotype was significantly associated with poor OS in rectal cancer patients.
Discrepancies across these studies might occur because of the sample size or ethnic differences in allele frequency for the MTHFR polymorphisms. The frequencies for the 677T allele and 1298C amongst the patients in our study (28.5 and 23.5%, respectively) were similar to the results from other studies conducted in Taiwan [14, 34], but appear to be lower than those reported in white American (32.7 and 30.6%) [25], Italian (48.2 and 29.5%) [12], French (36.1 and 30.4%) [22], or Spanish (39.4 and 30.9%) [10] populations. Moreover, inadequate study design such as a too-small sample size (most are <200) and/or inadequate controlling for certain confounders (such as age and tumor stage) should also be considered as contributors for the differing results.
Possible explanations for divergent findings in clinical studies should also consider the different 5-FU chemotherapy regimens used, i.e., 5-FU-monotherapy (5-FU/LV) or 5-FU-combined chemotherapy (5-FU/irinotecan or 5-FU/oxaliplatin). An ideal model to establish the relationship between MTHFR polymorphisms and survival would be the study of patients treated with a single drug, but this situation is extremely uncommon in clinical practice, where combined chemotherapy regimens are mainly used. In the latter scenario, the lack of correlation between MTHFR genotypes and survival after 5-FU treatment could result from the effects of an additional drug. Fortunately, we could only include CRC patients treated with a single antineoplastic agent because combined chemotherapy regimens were not supported by the Taiwan Nation Health Insurance before 2000. Our findings are consistent with the correlations found by several investigators in their studies of cancer patients treated with 5-FU-monotherapy [21,22,23]. Further studies are needed to confirm the role of MTHFR genotype and haplotype on survival of CRC patients receiving 5-FU chemotherapy.
Several potential limitations of our retrospective study should be discussed. First, as all patients included in this study presented at stage III, stage IV or high-risk stage II disease, it was not possible to assess genotype associated with clinical outcome in untreated patients. Second, our findings are based on a study population uniformly treated with 5-FU-based chemotherapy. Although new chemotherapy combinations, such as FOLFOX or FOLFIRI, have been shown to have superior effectiveness and are considered as the standard treatment for CRC patients, these chemotherapies with oxaliplatin or irinotecan could blur the influence of MTHFR polymorphisms on the treatment outcomes. Third, our study did not include other genetic polymorphisms in 5-FU-metabolizing enzymes that might be relevant, such as dihydropyrimidine dehydrogenase, TS, MTR, or ABCG2. We expect to extend our investigations to other relevant candidate polymorphisms. Fourth, all the CRC patients are of Chinese ethnicity. Hence, the ability to generalize the results to other racial/ethnic groups awaits further study. Nevertheless, our study which included newly diagnosed and histologically confirmed CRC patients and a relatively large sample size had sufficient power to yield important insights into the overall value of MTHFR genotypes as a prognostic marker.
To the best of our knowledge, this is one of the largest studies to show a relationship between two MTHFR polymorphisms and OS in CRC patients treated with 5-FU-based chemotherapy in a Chinese population. Our findings suggest that the MTHFR 677T allele exerts a protective role in OS in CRC patients, while the MTHFR 1298C allele may have a harmful effect on OS in rectal cancer. Further prospective studies measuring MTHFR gene expression related to its genotype are warranted to understand the role of MTHFR polymorphisms as a predictor of chemotherapeutic benefit in CRC patients.
References
Longley DB, Harkin DP, Johnston PG (2003) 5-Fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer 3:330–338
Lurje G, Zhang W, Yang D et al (2008) Thymidylate synthase haplotype is associated with tumor recurrence in stage II and stage III colon cancer. Pharmacogenet Genom 18:161–168
Scott J, Weir D (1994) Folate/vitamin B12 inter-relationships. Essays Biochem 28:63–72
Rosenblatt DS (2001) Methylenetetrahydrofolate reductase. Clin Invest Med 24:56–59
Rozen R (1996) Molecular genetics of methylenetetrahydrofolate reductase deficiency. J Inherit Metab Dis 19:589–594
Frosst P, Blom HJ, Milos R et al (1995) A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet 10:111–113
van der Put NM, Gabreels F, Stevens EM et al (1998) A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neural-tube defects? Am J Hum Genet 62:1044–1051
Weisberg I, Tran P, Christensen B et al (1998) A second genetic polymorphism in methylenetetrahydrofolate reductase (MTHFR) associated with decreased enzyme activity. Mol Genet Metab 64:169–172
Wisotzkey JD, Toman J, Bell T et al (1999) MTHFR (C677T) polymorphisms and stage III colon cancer: response to therapy. Mol Diagn 4:95–99
Marcuello E, Altes A, Menoyo A et al (2006) Methylenetetrahydrofolate reductase gene polymorphisms: genomic predictors of clinical response to fluoropyrimidine-based chemotherapy? Cancer Chemother Pharmacol 57:835–840
Suh KW, Kim JH, do Kim Y et al (2006) Which gene is a dominant predictor of response during FOLFOX chemotherapy for the treatment of metastatic colorectal cancer, the MTHFR or XRCC1 gene? Ann Surg Oncol 13:1379–1385
Ruzzo A, Graziano F, Loupakis F et al (2007) Pharmacogenetic profiling in patients with advanced colorectal cancer treated with first-line FOLFOX-4 chemotherapy. J Clin Oncol 25:1247–1254
Ruzzo A, Graziano F, Loupakis F et al (2008) Pharmacogenetic profiling in patients with advanced colorectal cancer treated with first-line FOLFIRI chemotherapy. Pharmacogenom J 8:278–288
Huang MY, Fang WY, Lee SC et al (2008) ERCC2 2251A>C genetic polymorphism was highly correlated with early relapse in high-risk stage II and stage III colorectal cancer patients: a preliminary study. BMC Cancer 8:50
Sharma R, Hoskins JM, Rivory LP et al (2008) Thymidylate synthase and methylenetetrahydrofolate reductase gene polymorphisms and toxicity to capecitabine in advanced colorectal cancer patients. Clin Cancer Res 14:817–825
Afzal S, Jensen SA, Vainer B et al (2009) MTHFR polymorphisms and 5-FU-based adjuvant chemotherapy in colorectal cancer. Ann Oncol 20:1660–1666
Gusella M, Frigo AC, Bolzonella C et al (2009) Predictors of survival and toxicity in patients on adjuvant therapy with 5-fluorouracil for colorectal cancer. Br J Cancer 100:1549–1557
Etienne-Grimaldi MC, Milano G, Maindrault-Goebel F et al (2010) Methylenetetrahydrofolate reductase (MTHFR) gene polymorphisms and FOLFOX response in colorectal cancer patients. Br J Clin Pharmacol 69:58–66
Boige V, Mendiboure J, Pignon JP et al (2010) Pharmacogenetic assessment of toxicity and outcome in patients with metastatic colorectal cancer treated with LV5FU2, FOLFOX, and FOLFIRI: FFCD 2000–05. J Clin Oncol 28:2556–2564
Joerger M, Huitema AD, Boot H et al (2015) Germline TYMS genotype is highly predictive in patients with metastatic gastrointestinal malignancies receiving capecitabine-based chemotherapy. Cancer Chemother Pharmacol 75:763–772
Cohen V, Panet-Raymond V, Sabbaghian N et al (2003) Methylenetetrahydrofolate reductase polymorphism in advanced colorectal cancer: a novel genomic predictor of clinical response to fluoropyrimidine-based chemotherapy. Clin Cancer Res 9:1611–1615
Etienne MC, Formento JL, Chazal M et al (2004) Methylenetetrahydrofolate reductase gene polymorphisms and response to fluorouracil-based treatment in advanced colorectal cancer patients. Pharmacogenetics 14:785–792
Jakobsen A, Nielsen JN, Gyldenkerne N et al (2005) Thymidylate synthase and methylenetetrahydrofolate reductase gene polymorphism in normal tissue as predictors of fluorouracil sensitivity. J Clin Oncol 23:1365–1369
Terrazzino S, Agostini M, Pucciarelli S et al (2006) A haplotype of the methylenetetrahydrofolate reductase gene predicts poor tumor response in rectal cancer patients receiving preoperative chemoradiation. Pharmacogenet Genom 16:817–824
Zhang W, Press OA, Haiman CA et al (2007) Association of methylenetetrahydrofolate reductase gene polymorphisms and sex-specific survival in patients with metastatic colon cancer. J Clin Oncol 25:3726–3731
Pare L, Salazar J, del Rio E et al (2008) Methylenetetrahydrofolate reductase gene polymorphisms: genomic predictors of clinical response to fluoropyrimidine-based chemotherapy in females. J Clin Oncol 26:3468 (author reply 3468–3469)
Capitain O, Boisdron-Celle M, Poirier AL et al (2008) The influence of fluorouracil outcome parameters on tolerance and efficacy in patients with advanced colorectal cancer. Pharmacogenom J 8:256–267
Fernandez-Peralta AM, Daimiel L, Nejda N et al (2010) Association of polymorphisms MTHFR C677T and A1298C with risk of colorectal cancer, genetic and epigenetic characteristic of tumors, and response to chemotherapy. Int J Colorect Dis 25:141–151
Castillo-Fernandez O, Santibanez M, Bauza A et al (2010) Methylenetetrahydrofolate reductase polymorphism (677 C>T) predicts long time to progression in metastatic colon cancer treated with 5-fluorouracil and folinic acid. Arch Med Res 41:430–435
Pardini B, Kumar R, Naccarati A et al (2011) 5-Fluorouracil-based chemotherapy for colorectal cancer and MTHFR/MTRR genotypes. Br J Clin Pharmacol 72:162–163
Taflin H, Wettergren Y, Odin E et al (2011) Gene polymorphisms MTHFRC677T and MTRA2756G as predictive factors in adjuvant chemotherapy for stage III colorectal cancer. Anticancer Res 31:3057–3062
Delgado-Plasencia L, Medina-Arana V, Bravo-Gutierrez A et al (2013) Impact of the MTHFR C677T polymorphism on colorectal cancer in a population with low genetic variability. Int J Colorect Dis 28:1187–1193
Zhao J, Li W, Zhu D et al (2014) Association of single nucleotide polymorphisms in MTHFR and ABCG2 with the different efficacy of first-line chemotherapy in metastatic colorectal cancer. Med Oncol 31:802
Wu NC, Su SM, Lin TJ et al (2015) Methylenetetrahydrofolate reductase C677T and A1298C polymorphisms and fluorouracil-based treatment in Taiwan colorectal cancer. Anticancer Drugs 26:888–893
Cecchin E, Perrone G, Nobili S et al (2015) MTHFR-1298 A>C (rs1801131) is a predictor of survival in two cohorts of stage II/III colorectal cancer patients treated with adjuvant fluoropyrimidine chemotherapy with or without oxaliplatin. Pharmacogenom J 15:219–225
Lai CY, Sung FC, Hsieh LL et al (2013) Associations between genetic polymorphisms of epidermal growth factor receptor (EGFR) and survival of colorectal cancer (CRC) patients treated with 5-fluorouracil-based chemotherapy. Ann Surg Oncol 20(Suppl 3):S599–S606
Lai CY, Hsieh LL, Sung FC et al (2013) Tumor site- and stage-specific associations between allelic variants of glutathione S-transferase and DNA-repair genes and overall survival in colorectal cancer patients receiving 5-fluorouracil-based chemotherapy. PLoS One 8:e69039
Tregouet DA, Garelle V (2007) A new JAVA interface implementation of THESIAS: testing haplotype effects in association studies. Bioinformatics 23:1038–1039
Ulrich CM, Rankin C, Toriola AT et al (2014) Polymorphisms in folate-metabolizing enzymes and response to 5-fluorouracil among patients with stage II or III rectal cancer (INT-0144; SWOG 9304). Cancer 120:3329–3337
Budai B, Komlosi V, Adleff V et al (2012) Impact of SHMT1 polymorphism on the clinical outcome of patients with metastatic colorectal cancer treated with first-line FOLFIRI+bevacizumab. Pharmacogenet Genom 22:69–72
Huang MY, Huang ML, Chen MJ et al (2011) Multiple genetic polymorphisms in the prediction of clinical outcome of metastatic colorectal cancer patients treated with first-line FOLFOX-4 chemotherapy. Pharmacogenet Genom 21:18–25
Kumamoto K, Ishibashi K, Okada N et al (2013) Polymorphisms of, and -3′UTR are associated with the clinical outcome of mFOLFOX6 in colorectal cancer patients. Oncol Lett 6:648–654
van Huis-Tanja LH, Gelderblom H, Punt CJ et al (2013) MTHFR polymorphisms and capecitabine-induced toxicity in patients with metastatic colorectal cancer. Pharmacogenet Genom 23:208–218
Etienne-Grimaldi MC, Bennouna J, Formento JL et al (2012) Multifactorial pharmacogenetic analysis in colorectal cancer patients receiving 5-fluorouracil-based therapy together with cetuximab-irinotecan. Br J Clin Pharmacol 73:776–785
Negandhi AA, Hyde A, Dicks E et al (2013) MTHFR Glu429Ala and ERCC5 His46His polymorphisms are associated with prognosis in colorectal cancer patients: analysis of two independent cohorts from Newfoundland. PLoS One 8:e61469
De Mattia E, Toffoli G (2009) C677T and A1298C MTHFR polymorphisms, a challenge for antifolate and fluoropyrimidine-based therapy personalisation. Eur J Cancer 45:1333–1351
Thomas F, Motsinger-Reif AA, Hoskins JM et al (2011) Methylenetetrahydrofolate reductase genetic polymorphisms and toxicity to 5-FU-based chemoradiation in rectal cancer. Br J Cancer 105:1654–1662
Acknowledgements
This study was supported by the National Sciences Council, Executive Yuan, Taiwan (NSC97-2314-B-039-020-MY3) and Taipei Medical University (TMU-NTUST-103-11, TMU-NTUST-104-01).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Dr. Ling-Ling Hsieh died on March 19, 2016.
Electronic supplementary material
Below is the link to the electronic supplementary material.
About this article
Cite this article
Yeh, CC., Lai, CY., Chang, SN. et al. Polymorphisms of MTHFR C677T and A1298C associated with survival in patients with colorectal cancer treated with 5-fluorouracil-based chemotherapy . Int J Clin Oncol 22, 484–493 (2017). https://doi.org/10.1007/s10147-016-1080-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10147-016-1080-z