Abstract
Non-small cell lung cancer (NSCLC) represents for approximately 85% of all lung cancers, which is the most common cancer worldwide. Tumor-associated macrophages (TAM) are crucial for tumor progression, which was widely believed to be mediated by long non-coding RNAs (LncRNAs). We aimed to explore the effect of one LncRNA, GNAS-AS1, in TAM-associated NSCLC progression. Relative mRNA levels were determined by qRT-PCR. Western blot and ELISA were used to detect protein levels. Proliferation in vitro was assessed by MTT and clone formation assays. Migration and invasion of cell lines were evaluated by transwell-based assays. Interaction between molecules was detected by luciferase report assay. GNAS-AS1 expression was dramatically enhanced in TAM, NSCLC cell lines, and clinical tumor tissues, and negatively correlated with overall survival of NSCLC patients. GNAS-AS1 promoted macrophage M2 polarization and NSCLC cell progression via directly inhibiting miR-4319, which could target N-terminal EF-hand calcium binding protein 3 (NECAB3) to inhibit its expression. GNAS-AS1/miR-4319/NECAB3 axis promotes tumor progression of NSCLC by altering macrophage polarization. This novel mechanism may provide potential strategy for NSCLC treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lung cancer is the most common cancer in world and is one of the most common causes of cancer-related deaths (Molina et al. 2008). According to histological characteristics, lung cancers are divided into two subtypes: small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). NSCLC represents for approximately 85% of all the lung cancer cases (Leung et al. 2016). Due to the limitation of early-stage (stages I and II) detection, most NSCLC patients were diagnosed at late stages (stage III/IV) (Kogita et al. 2014). Despite progresses in diagnosis and treatment, the 5-year overall survival of NSCLC patients is only 11% (Verdecchia et al. 2007). Therefore, further understanding of the mechanism that regulates NSCLC development and progression is urgently needed to provide more effective strategies for NSCLC treatment.
Tumor micro-environment (TME) includes cytokines, fibroblast, and various immune cells, and is commonly accepted to be essential for tumor development and progression (Whiteside 2008). In TME, macrophages undergo polarization into M1 or M2 subtypes in response to different stimulations (Martinez and Gordon 2014). Tumor-associated macrophages (TAM) display M2 phenotype and promote tumor development and progression in various cancers (Noy and Pollard 2014). In NSCLC patients, high TAM density was reported to be correlated with poor prognosis (Li et al. 2012). TAM promoted tumor metastasis via elevated SOX9/C-jun/SMAD3 pathway (Zhang et al. 2017).
Long non-coding RNAs (LncRNAs) are endogenous RNAs with the length of over 200-bp nucleotides that do not encode proteins (Quinn and Chang 2016). Accumulating evidences have demonstrated the essential role of LncRNAs in regulating numerous biological processes, such as DNA damage, miRNA silence, cell apoptosis and development, inflammation, and malignant transformation of tumor cells (Archer et al. 2015; Piccoli et al. 2015). GNAS-AS1 attracted our attention due to accumulating research into its effect on osteoarthritis and breast cancer (Lee et al. 2016, 2017; Park et al. 2017). However, the role of GNAS-AS1 in NSCLC remains unclear.
In this study, we aimed to explore the role of GNAS-AS1 in NSCLC development. We found that the expression levels of GNAS-AS1 was dramatically enhanced in TAM, NSCLC cell lines, and clinical tumor tissues. Consistently, GNAS-AS1 expression was observed to be negatively correlated with overall survival and metastasis-free survival of NSCLC patients. In addition, GNAS-AS1 was found to promote macrophage M2 polarization and NSCLC cell progression. The effect of GNAS-AS1 was achieved via directly inhibiting miR-4319, which could target N-terminal EF-hand calcium-binding protein 3 (NECAB3) to repress its expression.
Methods
Clinical NSCLC samples and cell lines
A total of 50 clinical NSCLC tissues and their adjacent normal tissues were obtained in Shanghai Pulmonary Hospital, Tongji University. All tissues were stored in freezer after being frozen in liquid nitrogen. Written consents from all patients were obtained before operation. The study was approved by the ethics committee of Shanghai Pulmonary Hospital, Tongji University.
Human NSCLC cell lines (PC9, SPCA1, H358, A549, H1299) and bronchial epithelial cells (16HBE) were purchased from Shanghai Cell Bank of China. For cell culture, RPMI medium with 10% fetal bovine serum (FBS) was used for all cell lines. The cell lines were kept in humidified incubator with 5% (v/v) CO2.
Macrophage polarization of human PBMC
Blood from healthy donors was used for isolation of peripheral blood mononuclear cells (PBMCs). Macrophage polarization from human PBMCs was reported previously (Huang et al. 2017). Briefly, PBMCs were separated by Ficoll density gradient (Axis-Shield). Then, CD14+ monocytes were sorted using anti-CD14 kit (Miltenyi) according to the manufacturer’s instructions. The sorted CD14+ monocytes were cultured in RPMI medium with 10% FBS for 7 days, followed by macrophage induction by 50 ng/ml M-CSF. M0 cells were collected after 48 h of incubation with serum-free medium; M1 macrophages were collected after overnight stimulation with 100 ng/ml lipopolysaccharides and 100 ng/ml IFN-γ; M2 macrophages were collected after overnight stimulation with 20 ng/ml IL-4.
For TAM, monocytes were first cultured for 7 days in RPMI medium containing 10% FBS with 50% culture medium from H1299 cells. The culture medium was obtained from 48 h of serum-starved culture of H1299 cells.
RNA isolation and qRT-PCR
The RNeasy kit (Qiagen, Valencia, CA, USA) was used for RNA isolation from cells and tissues according to the manufacturer’s instructions. Then, Prime-Script RT Kit (Takara, Dalian, China) was used for cDNA synthesis. The primers for qRT-PCR were as follows (5′-3′): GNAS-AS1, F: GAC GCC TTT CCT ACG G, R: TGG TAA CGC ACC TTC G; IL-10, F: TCA AGG ATG CAC ATC AAA AGG C, R: AGG CAG CAA CTT CCT CCC T; Arg-1, F: GTG GAA ACT TGC ATG GAC AAC, R: AAT CCT GGC ACA TCG GGA ATC; miR-4319, F: GCA CAG CTC CCT GAG CAA, R: CAG TGC GTG TCG TGG AGT; U6, F: TGC GGG TGC TCG CTT CGG CAG C, R: GTG CAG GGT CCG AGG T; NECAB3, F: TCT GGC AGG ATG AGG C, R: GAG GCT GGG AAG AAC AC; GAPDH, F: GGA GCG AGA TCC CTC CAA AAT, R: GGC TGT TGT CAT ACT TCT CAT GG. Relative mRNA expression levels were determined by the 2−ΔΔCt method.
THP-1-differentiated macrophage
The human monocytic THP-1 cells were purchased from ATCC. For macrophage differentiation, THP-1 cells were cultured in RPMI medium with 100 ng/ml PMA (Sigma, St. Louis, MO) for 48 h (Zhou et al. 2018).
Plasmids and cell infection
GNAS-AS1 overexpression plasmid (pSin-GNAS-AS1) was constructed using the following primers (5′-3′): F: CTA GAA TTC TAG GGG GCG CCG CGT T, R: CTA GGA TCC TTG ACA GGG TGC ATC TGG. The empty vector (pSin-VEC) was used as control. Then, the plasmids were transfected into THP-1-differentiated macrophages, respectively, using Lipofectamine 2000 (Invitrogen, Waltham, MA) according to the manufacturer’s instructions.
Enzyme-linked immunosorbent assay
The concentrations of IL-10 and Arg-1 in culture medium were detected by commercial enzyme-linked immunosorbent assay (ELISA) kits (eBioscience, San Diego, CA) following the manufacturer’s protocol.
Clone formation assay
A total of 500 cells were added into 6-well plates, followed by culturing in RPMI medium with 10% FBS for 12 days. Then, purple crystal staining was performed to visualize colony formation. Colonies containing more than 50 cells were calculated under the microscope.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was used to investigate cell viability. Briefly, 5000 cells from different groups were added into 96-well plates for 24 h of culture. Then, the MTT kit (Thermo Fisher, Waltham, MA) was used to detect cell viability according to the manufacturer’s protocol.
Migration and invasion assays
For invasion assay, 1 × 105 cells were added into matrigel-coated upper chamber with serum-free medium, while 10% FBS medium was added into lower compartment. After 48 h of culturing, the matrigel membranes were fixed and stained with crystal violet solution. Then, the invasion number was calculated. For migration assay, 2 × 105 cells were seeded into 6-well plates and allowed to grow to full confluency, then a vertical scratch wound was made on the cell monolayer using a sterile 200-μL micro pipette tip. The wound closure was monitored and calculated after 24 h.
Luciferase reporter assays
Plasmids containing pGL3-NECAB3-3′UTR (WT) and pGL3-NECAB3-3′UTR (MT) were purchased from GenePharma (Shanghai, China). A total of 3.5 × 104/well THP-1-differentiated macrophages or HEK193T cells were added into 24-well plates and cultured for 24 h. Then, plasmids containing pGL3-NECAB3-3′UTR (WT) or pGL3-NECAB3-3′UTR (MT) were co-transfected with miR-4319 or miR-NC, respectively, using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocols. Forty-eight hours later, luciferase activity was detected by Luciferase Reporter Assay Kit (Promega, Madison, WI) following the manufacturer’s protocol.
Biotin RNA pull-down assay
For biotin pull-down assay, lysates from 2 × 107 THP-1-differentiated macrophages were incubated with 3 μg in vitro–synthesized biotin-labeled GNAS-AS1 sense or anti-sense oligomers. After 3-h incubation, streptavidin-conjugated beads (Invitrogen) were used to isolate RNA-RNA complex for further detection.
Western blot
THP-1-differentiated macrophages were lysed in radioimmunoprecipitation buffer (Sigma, St. Louis, MO). Then, the lysates were electrophoresed into 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel and transferred onto membranes (Millipore, Billerica, MA). The membranes were incubated with anti-NECAB3 or anti-actin primary antibodies (Cell Signaling Technology, Danvers, MA) at 4 °C overnight after blocking with 5% milk at room temperature for 1 h. Then, the members were subjected to incubation with horseradish peroxidase (HRP)–conjugated secondary antibody for 1 h at room temperature. Finally, signals were visualized via chemiluminescence reaction with HRP substrate (Millipore).
Statistical analysis
All data were presented as mean ± standard deviation (SD) from 3 separate experiments. Student’s t test and one-way ANOVA were used to determine the statistical significance. Kaplan-Meier’s analysis was used for detecting the correlation between GNAS-AS1 expression and the overall or metastasis-free survival of NSCLC patients. In all experiments, p < 0.05 was considered significant.
Results
GNAS-AS1 expression is increased in TAM and NSCLC cell lines
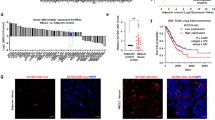
Given the crucial role of M2 macrophages and TAM in NSCLC development (Wang et al. 2011), we first determined GNAS-AS1 expression levels in various macrophages (M0, M1, M2), whose identities were confirmed by cell surface marker staining (Fig. S1). The expression levels of GNAS-AS1 were dramatically increased in M2 macrophages and TAM when compared with M0 and M1 macrophages (Fig. 1a), indicating the effect of GNAS-AS1 in promoting tumor development. To further explore the effect of GNAS-AS1 in NSCLC, we performed qRT-PCR in human bronchial epithelial cells (16HBE) and NSCLC cell lines (PC9, SPCA1, H358, A549, H1299). Higher GNAS-AS1 expressions were observed in all NSCLC cell lines than bronchial epithelial cells (Fig. 1b). In clinical NSCLC samples, the expression level of GNAS-AS1 was significantly increased in tumor tissues when compared with adjacent normal tissues (Fig. 1c). The correlation of GNAS-AS1 expression with clinicopathological features in 50 NSCLC patients is shown in Table 1. Tumor tissues from patients with lymph node metastasis displayed higher GNAS-AS1 expression than non-metastasis (Fig. 1d). Moreover, high GNAS-AS1 expression in NSCLC patients negatively correlated with overall survival and metastasis-free survival (Fig. 1e, f). These results demonstrated the effect of GNAS-AS1 in promoting NSCLC development.
The expression levels of GNAS-AS1 in different types of macrophage and NSCLC. a The expression levels of GNAS-AS1 in M0, M1, M2, and TAMs were detected by qRT-PCR. b The expression levels of GNAS-AS1 in NSCLC cells (PC9, SPCA1, H358, A549, H1299) and human bronchial epithelial cells (16HBE) were detected by qRT-PCR. c qRT-PCR analysis of GNAS-AS1 expression in 50 NSCLC tissues and 50 adjacent normal tissues (shown as 2−ΔΔCt). d Relative expression levels of GNAS-AS1 in groups of NSCLC tissues classified based on the occurrence of lymph node metastasis (metastatic, M or non-metastatic, N-M). e and f Kaplan-Meier’s analysis of the correlation between GNAS-AS1 expression and the overall survival or metastasis-free survival of NSCLC patients. *P < 0.05; **P < 0.01; *** P < 0.001
GNAS-AS1 promotes M2 polarization and NSCLC cell proliferation
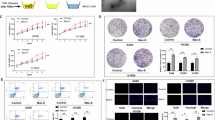
To investigate the biological effect of GNAS-AS1 in macrophages, THP-1 monocytic cell line was induced to differentiate into macrophages (THP-1-differentiated macrophage). GNAS-AS1 over-expressing THP-1-differentiated macrophage was established by transfection with GNAS-AS1 plasmid (Fig. 2a). Interleukin 4 (IL-4) administration greatly elevated mRNA and protein levels of M2 polarization markers IL-10 and Arg-1. In addition, IL-10 and Arg-1 levels were much higher in GNAS-AS1 overexpressing THP-1-differentiated macrophages after IL-4 stimulation (Fig. 2b, c), indicating that GNAS-AS1 promoted M2 polarization.
GNAS-AS1 promoted M2 polarization and NSCLC cell proliferation. a RNA level of GNAS-AS1 was determined by qRT-PCR in THP-1-differentiated macrophages transfected with GNAS-AS1 plasmid (pSin-GNAS-AS1) or empty vector (pSin-VEC). THP-1-differentiated macrophages were divided into four groups: control, exposed to IL-4 (20 ng/ml, 24 h), exposed to IL-4 and transfected with empty vector (pSin-VEC), exposed to IL-4 and transfected with GNAS-AS1 plasmid (pSin-GNAS-AS1). b The mRNA levels of IL-10 and Arg-1 (the marker of M2 polarization) were detected by qRT-PCR in these THP-1 macrophages. c The protein levels of IL-10 and Arg-1 (the marker of M2 polarization) in the cultured media of these THP-1 macrophages were detected by ELISA. d and e Cell proliferation of A549 and H1299 cells treated with the conditioned media of these THP-1 macrophages was determined by MTT and colony formation assays. The data represent the mean ± SD from three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001
A549 and H1299 NSCLC cells were cultured with conditional media from 4 groups of THP-1-differentiated macrophages (control, IL-4, IL-4 + pSin-VEC, IL-4 + pSin-GNAS-AS1), as shown in Fig. 2b, c. Similarly, IL-4-conditional media significantly enhanced cell viability (Fig. 2d) and proliferation (Fig. 2e). IL-4 plus GNAS-AS1 overexpression-conditional media further enhanced cell viability and proliferation of A549 and H1299 (Fig. 2d, e). These results indicated that GNAS-AS1 overexpression-mediated M2 polarization promoted malignant transformation of NSCLC cells.
GNAS-AS1 enhances migration and invasion of NSCLC cells
We further detected the role of GNAS-AS1 on NSCLC cell migration and invasion. The migration of A549 and H1299 cells was determined by scratch wound healing assay. As shown in Fig. 3a, b, in both A549 and H1299 cells, IL-4-conditional media significantly enhanced the percentage of wound closure, which was further enhanced by IL-4 plus GNAS-AS1 overexpression-conditional media. Cell invasion was further determined by transwell system. IL-4-conditional media enhanced A549 cell migration and H1299 cell invasion, which were further enhanced by IL-4 plus GNAS-AS1 overexpression-conditional media (Fig. 3c, d). To rule out the possibility that conditional media from NSCLC cell lines overexpressing GNAS-AS1 also promoted macrophage M2 polarization, we detected mRNA and protein levels of IL-10 and Arg-1, and found conditional media from NSCLC cell lines overexpressing GNAS-AS1 had no significant effects on macrophage M2 polarization (Fig. S2). These results indicated that GNAS-AS1 overexpression-mediated M2 polarization promoted NSCLC migration and invasion. Of note, these observed enhancing effect of GNAS-AS1 overexpression on NSCLC migration and invasion was independent of epithelial-mesenchymal transition (EMT), as EMT markers, such as N-cadherin, Twist, and Slug, were unaffected (Fig. S3a and S3b).
The conditioned medium from M2 macrophages overexpressing GNAS-AS1 enhanced migration and invasion of NSCLC cells. THP-1-differentiated macrophages were divided into four groups: control, exposed to IL-4 (20 ng/ml, 24 h), exposed to IL-4 and transfected with empty vector (pSin-VEC), exposed to IL-4 and transfected with GNAS-AS1 plasmid (pSin-GNAS-AS1). a and b The migration of A549 and H1299 cells treated with the conditioned media of these THP-1 macrophages was determined by scratch wound healing assay. c and d The migratory and invaded ability of A549 and H1299 cells treated with the conditioned media of these THP-1 macrophages was determined by transwell assays. The data represent the mean ± SD from three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001
GNAS-AS1 knockdown inhibits NSCLC cell progression
We further knocked down GNAS-AS1 expression to confirm the biological role of GNAS-AS1 in NSCLC cells. We first demonstrated the successful down-regulation of GNAS-AS1 detected by qRT-PCR (Fig. 4a). GNAS-AS1 downregulation (sh-GNAS-AS1-1, sh-GNAS-AS1-2) significantly inhibited cell proliferation (Fig. 4b), clone formation (Fig. 4c), and migration (Fig. 4d) of both A549 and H1299 cells. Moreover, transwell assay showed that GNAS-AS1 downregulation dramatically suppressed the migrasive and invasive abilities of A549 and H1299 cells (Fig. 4e, f). These results indicated that GNAS-AS1 knockdown inhibited NSCLC cell progression.
Knockdown of GNAS-AS1 inhibited NSCLC cells progression. a The relative expression levels of GNAS-AS1 in A549 and H1299 cells transfected with GNAS-AS1 shRNAs (sh-GNAS-AS1-1, sh-GNAS-AS1-2) or empty vector (sh-CTR) were detected by qRT-PCR. b and c Cell proliferation of A549 and H1299 cells transfected with GNAS-AS1 shRNAs (sh-GNAS-AS1-1, sh-GNAS-AS1-2) or empty vector (sh-CTR) was determined by MTT and colony formation assays. d The migration of A549 and H1299 cells transfected with GNAS-AS1 shRNAs (sh-GNAS-AS1-1, sh-GNAS-AS1-2) or empty vector (sh-CTR) was determined by scratch wound healing assay. e and f The migratory and invaded ability of A549 and H1299 cells transfected with GNAS-AS1 shRNAs (sh-GNAS-AS1-1, sh-GNAS-AS1-2) or empty vector (sh-CTR) was determined by transwell assays. The data represent the mean ± SD from three independent experiments. *P < 0.05; **P < 0.01. Student’s t test
GNAS-AS1 directly inhibits miR-4319 expression
We next investigated the mechanism underlying GNAS-AS1 function in NSCLC. LncRNAs exert their biological roles via competitively binding endogenous RNAs (Quinn and Chang 2016). Accordingly, microRNA (miR)-4319 was predicted to be the potential binding microRNA of GNAS-AS1 through miRcode (http://www.mircode.org/mircode/). The binding sites between miR-4319 and GNAS-AS1 were predicted by RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid). Mutations in the binding sites were carried out for luciferase reporter assay (Fig. 5a). Luciferase activity was significantly decreased in THP-1-differentiated macrophages and HEK293T cells co-transfected with miR-4319 and luciferase reporters containing GNAS-AS1 wt, when compared with negative control (miR-NC). However, no significant difference of luciferase activity in both cells was observed between miR-4319 and miR-NC, when co-transfected with luciferase reporters containing GNAS-AS1 mt (Fig. 5b, c), indicating that miR-4319 directly targeted GNAS-AS1.
GNAS-AS1 could negatively regulate miR-4319 by a direct interaction. a The prediction for miR-4319 binding sites on GNAS-AS1 transcript and schematic of luciferase reporter vector constructs GNAS-AS1 wild-type (GNAS-AS1 wt) and the miR-4319 binding site mutated (GNAS-AS1 mt) one. b and c The luciferase activities in THP-1-differentiated macrophages and HEK293T cells co-transfected with miR-4319 or miR-NC mimic and luciferase reporters containing GNAS-AS1 wt or GNAS-AS1 mt. Data are presented as the relative ratio of hRluc luciferase activity to hluc+ luciferase activity. d The relative expression levels of GNAS-AS1 and miR-4319 in THP-1-differentiated macrophages transfected with GNAS-AS1 shRNAs (sh-GNAS-AS1-1, sh-GNAS-AS1-2) or empty vector (sh-CTR) were detected by qRT-PCR. e The relative expression levels of GNAS-AS1 and miR-4319 in THP-1-differentiated macrophages transfected with GNAS-AS1 plasmid (pSin-GNAS-AS1) or empty vector (pSin-VEC) were detected by qRT-PCR. f Lysates from THP-1-differentiated macrophages were incubated with in vitro–synthesized biotin-labeled sense or antisense DNA probes against GNAS-AS1 for biotin pull-down assay, followed by the real-time RT-PCR analysis to examine GNAS-AS1 and miR-4319 levels. g Lysates from THP-1-differentiated macrophages were incubated with in vitro–synthesized biotin-labeled GNAS-AS1 and antisense RNA for biotin pull-down assay, followed by the real-time RT-PCR analysis to examine miR-4319 level. The data represent the mean ± SD from three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001
GNAS-AS1 downregulation in THP-1-differentiated macrophages by shRNAs dramatically promoted miR-4319 expression levels (Fig. 5d). Consistently, GNAS-AS1 overexpression significantly inhibited miR-4319 expression level (Fig. 5e). In a pull-down assay, GNAS-AS1 anti-sense probe-enriched complex from THP-1-differentiated macrophages showed elevated expression levels of GNAS-AS1 and miR-4319, compared with sense probe (Fig. 5f), indicating GNAS-AS1 indeed targeted miR-4319. Moreover, miR-4319 expression level was significantly elevated in complex enriched with biotin-labeled GNAS-AS1 compared with anti-sense RNA (Fig. 5g). These results demonstrated that GNAS-AS1 directly inhibited miR-4319 expression.
NECAB3 is the direct target of miR-4319
We further explored the biological target of miR-4319. NECAB3 was predicted to be the potential target of miR-4319 using Targetscan (http://www.targetscan.org/vert_71/). The binding sites between 3′UTR of NECAB3 and miR-4319 (both wild type and mutant) were illustrated in Fig. 6a. Luciferase activity in THP-1-differentiated macrophages and HEK293T cells co-transfected with miR-4319 and NECAB3 3′UTR (WT) was dramatically inhibited, when compared with miR-NC. However, no difference in luciferase activity was observed when the cells were transfected with NECAB3 3′UTR (MT) (Fig. 6b, c), indicating that NECAB3 was the direct target of miR-4319. Pull-down assay in THP-1-differentiated macrophages showed that NECAB3 mRNA level was much higher in biotinylated miR-4319 (bio-miR-4319)-mediated complex than biotinylated control random RNA (bio-miR-NC) (Fig. 6d), suggesting the biological combination between miR-4319 and NECAB3.
MiR-4319 targeted NECAB3 to regulate its expression. a The sequences of miR-4319 binding site within NECAB3 3′UTR, including wild-type (NECAB3 3′UTR (WT)) and mutant (NECAB3 3′UTR (MT)). b and c Luciferase reporter assay was performed in THP-1-differentiated macrophages and HEK293T cells co-transfected with miR-4319 or miR-NC mimics and plasmid containing NECAB3 3′UTR (WT) or NECAB3 3′UTR (MT). d Detection of NECAB3 mRNAs in biotinylated miRNA/target mRNA complex by qRT-PCR. The relative level of NECAB3 mRNA in the complex pull-down by using biotinylated miR-4319 (bio-miR-4319) was compared with that of the complex pull-down by using the biotinylated control random RNA (bio-miR-NC). e–g The relative expression levels of NECAB3 in THP-1-differentiated macrophages transfected with indicated microRNA mimics and microRNA inhibitors or their respective negative controls were detected by qRT-PCR and Western blot. h–j The relative expression levels of NECAB3 in THP-1-differentiated macrophages transfected GNAS-AS1 shRNAs (sh-GNAS-AS1-1, sh-GNAS-AS1-2) or empty vector (sh-CTR) and GNAS-AS1 plasmid (pSin-GNAS-AS1) or empty vector (pSin-VEC) were detected by qRT-PCR and Western blot. Data are representative of three or more independent experiments. Data are the mean ± SD. *P < 0.05, **P < 0.01, ns = not significant
NECAB3 expression level was attenuated in miR-4319-transfected THP-1-differentiated macrophages (Fig. 6e), which was elevated upon treatment with miR-4319 inhibitor (Fig. 6f). Consistently, NECAB3 protein level in THP-1-differentiated macrophages showed similar trend after different treatment (Fig. 6g), further demonstrating that NECAB3 was the target of miR-4319. GNAS-AS1 downregulation inhibited NECAB3 mRNA expression (Fig. 6h). Meanwhile, GNAS-AS1 upregulation enhanced NECAB3 mRNA expression in THP-1-differentiated macrophages (Fig. 6i). This similar trend in NECAB3 protein level was observed after different treatment (Fig. 6j). These results indicated that GNAS-AS1 and NECAB3 could competitively bind with miR-4319.
GNAS-AS1/miR-4319/NECAB3 axis regulates M2 polarization and NSCLC cell progression
Given the competitive combination of GNAS-AS1 and NECAB3 with miR-4319, we next determined its biological effect in M2 polarization and NSCLC cell progression. NECAB3 mRNA and protein levels were inhibited after GNAS-AS1 downregulation, which could be recovered after miR-4319 inhibitor administration (Fig. 7a, b). In IL-4-induced M2 macrophages, GNAS-AS1 downregulation inhibited the mRNA and protein levels of IL-10 and Arg-1, which was rescued by miR-4319 inhibitor administration (Fig. 7c, d).
GNAS-AS1/miR-4319/NECAB3 axis regulated M2 polarization and NSCLC cell progression. a and b Expression levels of NECAB3 was determined by qRT-PCR and Western blot in THP-1-differentiated macrophages co-transfected with empty vector and inhibitor negative control (pSin-VEC + NC-inhibitor) or GNAS-AS1 plasmid and inhibitor negative control (pSin-GNAS-AS1+NC-inhibitor) or GNAS-AS1 plasmid and miR-4319 inhibitor (pSin-GNAS-AS1+miR-4319-inhibitor). c–h THP-1-differentiated macrophages were divided into three groups: exposed to IL-4 (20 ng/ml, 24 h) and co-transfection with empty vector and inhibitor negative control (IL-4+pSin-VEC+NC-inhibitor), exposed to IL-4 and co-transfection with GNAS-AS1 plasmid and inhibitor negative control (IL-4+pSin-GNAS-AS1+NC-inhibitor), exposed to IL-4 and co-transfection with GNAS-AS1 plasmid and miR-4319 inhibitor (IL-4+pSin-GNAS-AS1+miR-4319-inhibitor). c The mRNA levels of IL-10 and Arg-1 (the marker of M2 polarization) were detected by qRT-PCR in these THP-1 macrophages. d The protein levels of IL-10 and Arg-1 (the marker of M2 polarization) in the cultured media of these THP-1 macrophages were detected by ELISA. e and f Cell proliferation of A549 cells treated with the conditioned media of these THP-1 macrophages was determined by MTT and colony formation assays. g The migration of A549 cells treated with the conditioned media of these THP-1 macrophages was determined by scratch wound healing assay. h The migratory and invaded ability of A549 cells treated with the conditioned media of these THP-1 macrophages was determined by transwell assays. The data represent the mean ± SD from three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001
A549 cells were treated with different conditional media as shown in Fig. 7c, d. Conditional media from GNAS-AS1 downregulated THP-1-differentiated macrophages suppressed cell viability, clone formation, migration, and invasion of A549 cells, whereas further miR-4319 inhibitor administration exhibited opposite effects (Fig. 7e–h). These results demonstrated that GNAS-AS1 promoted NECAB3 via inhibiting miR-4319, and GNAS-AS1/miR-4319/NECAB3 axis regulated M2 polarization and NSCLC cell progression.
Discussion
NSCLC constitutes the majority of lung cancers, which is also the most common type of cancer in the world (Siegel et al. 2018). Strategies for NSCLC treatment are limited due to high recurrence and metastasis rates (Temel et al. 2010). Therefore, NSCLC has been the acute challenge to public health worldwide. TAM was widely believed to be important in promoting NSCLC development (Mei et al. 2016). Moreover, accumulating studies demonstrated the novel role of LncRNAs in various biological progresses including tumor development (Gutschner and Diederichs 2012; Mercer et al. 2009). Accordingly, we explored the effect of LncRNA-mediated TAM in NSCLC. Macrophage polarization in dynamic TME is crucial for immune response and further tumor development (Mantovani et al. 2002; Sica and Bronte 2007). Macrophage activation has been proposed based on dichotomy: M1 (classic) and M2 (alternative) (Martinez and Gordon 2014). M1 macrophages induce inflammatory responses, while M2 macrophages antagonizes inflammatory responses (Martinez and Gordon 2014). M2 macrophages, such as TAM, promote several types of tumor development (Mantovani et al. 2002; Sica et al. 2006; Solinas et al. 2009). As TAM represents for the crucial orchestrators for immune escape, therapies targeting TAM are attractive for anti-tumor treatment (Noy and Pollard 2014). Macrophage colony-stimulating factor 1 (CSF-1) is the key survival factor for TAM. Monoclonal antibody that suppresses activation of CSF-1 receptor leads to cell death of TAM in vitro. CSF-1 receptor inhibition significantly eliminated TAM and promoted ratio of CD8+/CD4+ T cells in animal models. More importantly, patients administrated with CSF-1 receptor exhibited dramatically decreased CSF-1R+CD163+ TAM (Ries et al. 2014). Reduction of TAM in tumor stromal significantly switched TME and therefore led to dramatical tumor suppression (Luo et al. 2006). We first successfully induced macrophage differentiation from THP-1 cells by IL-4 stimulation. In GNAS-AS1-overexpressing THP-1-differentiated macrophages, TAM markers IL-10 and Arg-1 were markedly enhanced. Conditional media from GNAS-AS1-overexpressing THP-1-differentiated macrophages (with higher TAM expression) promoted cell viability, migration, and invasion of NSCLC cells A549 and H1299, demonstrating the role of TAM in promoting NSCLC malignant transformation.
The effects of LncRNAs were achieved by competitively binding with endogenous RNAs. In pseudohypoparathyroidism patients, GNAS-AS1 was confirmed to be the most significant differentially methylated regions (Rochtus et al. 2016). MiR-4319 was found to be directly suppressed by GNAS-AS1. Indeed, no significant difference in luciferase activity was observed when binding sites between GNAS-AS1 and miR-4319 were mutated. GNAS-AS1 downregulation increased miR-4319 expression levels. Furthermore, the effect of GNAS-AS1 downregulation in M2 macrophage polarization and NSCLC cell progression was rescued by miR-4319 inhibitor. To the best of our knowledge, this is the first report that demonstrates direct binding between GNAS-AS1 and miR-4319.
NECAB3 promotes hypoxia-inducible factor-1 activation. Downregulation of NECAB3 in tumor cells results in suppressed tumorigenicity (Nakaoka et al. 2016), indicating the crucial role of NECAB3 in promoting cancer development. However, the role of NECAB3 in macrophages remains unclear. In this study, we first showed that NECAB3 expression was enhanced in THP-1-differentiated macrophages. Moreover, NECAB3 was the direct target of miR-4319. MiR-4319 inhibition elevated NECAB3 expression levels, and promoted NSCLC cell malignant transformation which was suppressed by GNAS-AS1 downregulation.
In summary, we report that GNAS-AS1 expression levels are markedly increased in TAM, NSCLC cell lines, and NSCLC patient tumor tissues. NSCLC patients with higher GNAS-AS1 expression levels showed poor overall survival and metastasis-free survival. GNAS-AS1 promotes macrophage M2 polarization and NSCLC cell progression via directly inhibiting miR-4319. Furthermore, NECAB3 is the direct target of miR-4319 in GNAS-AS1-promoted M2 polarization and NSCLC cell progression. This research provides a potential mechanical basis for more effective NSCLC treatment.
Conclusion
GNAS-AS1/miR-4319/NECAB3 axis promotes tumor progression of non-small cell lung cancer (NSCLC) by altering macrophage polarization. This research provides a potential mechanical basis for more effective NSCLC treatment.
References
Archer K, Broskova Z, Bayoumi AS, Teoh JP, Davila A, Tang Y, Su H, Kim IM (2015) Long non-coding RNAs as master regulators in cardiovascular diseases. Int J Mol Sci 16:23651–23667. https://doi.org/10.3390/ijms161023651
Gutschner T, Diederichs S (2012) The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol 9:703–719. https://doi.org/10.4161/rna.20481
Huang JK, Ma L, Song WH, Lu BY, Huang YB, Dong HM, Ma XK, Zhu ZZ, Zhou R (2017) LncRNA-MALAT1 promotes angiogenesis of thyroid cancer by modulating tumor-associated macrophage FGF2 protein secretion. J Cell Biochem 118:4821–4830. https://doi.org/10.1002/jcb.26153
Kogita A, Togashi Y, Hayashi H, Sogabe S, Terashima M, De Velasco MA, Sakai K, Fujita Y, Tomida S, Takeyama Y, Okuno K, Nakagawa K, Nishio K (2014) Hypoxia induces resistance to ALK inhibitors in the H3122 non-small cell lung cancer cell line with an ALK rearrangement via epithelial-mesenchymal transition. Int J Oncol 45:1430–1436. https://doi.org/10.3892/ijo.2014.2574
Lee J, Jung JH, Chae YS, Park HY, Kim WW, Lee SJ, Jeong JH, Kang SH (2016) Long noncoding RNA snaR regulates proliferation, migration and invasion of triple-negative breast cancer cells. Anticancer Res 36:6289–6295. https://doi.org/10.21873/anticanres.11224
Lee J, Park HY, Kim WW, Lee SJ, Jeong JH, Kang SH, Jung JH, Chae YS (2017) Biological function of long noncoding RNA snaR in HER2-positive breast cancer cells. Tumour Biol 39. https://doi.org/10.1177/1010428317707374
Leung AW, de Silva T, Bally MB, Lockwood WW (2016) Synthetic lethality in lung cancer and translation to clinical therapies. Mol Cancer 15:61. https://doi.org/10.1186/s12943-016-0546-y
Li CG, Huang XE, Xu L, Li Y, Lu YY (2012) Clinical application of serum tumor associated material (TAM) from non-small cell lung cancer patients. Asian Pac J Cancer Prev 13:301–304
Luo Y, Zhou H, Krueger J, Kaplan C, Lee SH, Dolman C, Markowitz D, Wu W, Liu C, Reisfeld RA, Xiang R (2006) Targeting tumor-associated macrophages as a novel strategy against breast cancer. J Clin Invest 116:2132–2141. https://doi.org/10.1172/jci27648
Mantovani A, Sozzani S, Locati M, Allavena P, Sica A (2002) Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol 23:549–555
Martinez FO, Gordon S (2014) The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep 6:13. https://doi.org/10.12703/P6-13
Mei J, Xiao Z, Guo C, Pu Q, Ma L, Liu C, Lin F, Liao H, You Z, Liu L (2016) Prognostic impact of tumor-associated macrophage infiltration in non-small cell lung cancer: a systemic review and meta-analysis. Oncotarget 7:34217–34228. https://doi.org/10.18632/oncotarget.9079
Mercer TR, Dinger ME, Mattick JS (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet 10:155–159. https://doi.org/10.1038/nrg2521
Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA (2008) Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc 83:584–594. https://doi.org/10.4065/83.5.584
Nakaoka HJ, Hara T, Yoshino S, Kanamori A, Matsui Y, Shimamura T, Sato H, Murakami Y, Seiki M, Sakamoto T (2016) NECAB3 promotes activation of hypoxia-inducible factor-1 during normoxia and enhances tumourigenicity of cancer cells. Sci Rep 6:22784. https://doi.org/10.1038/srep22784
Noy R, Pollard JW (2014) Tumor-associated macrophages: from mechanisms to therapy. Immunity 41:49–61. https://doi.org/10.1016/j.immuni.2014.06.010
Park S, Lee M, Chun CH, Jin EJ (2017) The lncRNA, Nespas, is associated with osteoarthritis progression and serves as a potential new prognostic biomarker. Cartilage:148–156. https://doi.org/10.1177/1947603517725566
Piccoli MT, Gupta SK, Thum T (2015) Noncoding RNAs as regulators of cardiomyocyte proliferation and death. J Mol Cell Cardiol 89:59–67. https://doi.org/10.1016/j.yjmcc.2015.02.002
Quinn JJ, Chang HY (2016) Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 17:47–62. https://doi.org/10.1038/nrg.2015.10
Ries CH, Cannarile MA, Hoves S, Benz J, Wartha K, Runza V, Rey-Giraud F, Pradel LP, Feuerhake F, Klaman I, Jones T, Jucknischke U, Scheiblich S, Kaluza K, Gorr IH, Walz A, Abiraj K, Cassier PA, Sica A, Gomez-Roca C, de Visser KE, Italiano A, Le Tourneau C, Delord JP, Levitsky H, Blay JY, Ruttinger D (2014) Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy. Cancer Cell 25:846–859. https://doi.org/10.1016/j.ccr.2014.05.016
Rochtus A, Martin-Trujillo A, Izzi B, Elli F, Garin I, Linglart A, Mantovani G, Perez de Nanclares G, Thiele S, Decallonne B, Van Geet C, Monk D, Freson K (2016) Genome-wide DNA methylation analysis of pseudohypoparathyroidism patients with GNAS imprinting defects. Clin Epigenetics 8:10. https://doi.org/10.1186/s13148-016-0175-8
Sica A, Bronte V (2007) Altered macrophage differentiation and immune dysfunction in tumor development. J Clin Invest 117:1155–1166. https://doi.org/10.1172/jci31422
Sica A, Schioppa T, Mantovani A, Allavena P (2006) Tumour-associated macrophages are a distinct M2 polarised population promoting tumour progression: potential targets of anti-cancer therapy. Eur J Cancer 42:717–727. https://doi.org/10.1016/j.ejca.2006.01.003
Siegel RL, Miller KD, Jemal A (2018) Cancer statistics, 2018. CA Cancer J Clin 68:7–30. https://doi.org/10.3322/caac.21442
Solinas G, Germano G, Mantovani A, Allavena P (2009) Tumor-associated macrophages (TAM) as major players of the cancer-related inflammation. J Leukoc Biol 86:1065–1073. https://doi.org/10.1189/jlb.0609385
Temel JS, Greer JA, Muzikansky A, Gallagher ER, Admane S, Jackson VA, Dahlin CM, Blinderman CD, Jacobsen J, Pirl WF, Billings JA, Lynch TJ (2010) Early palliative care for patients with metastatic non-small-cell lung cancer. N Engl J Med 363:733–742. https://doi.org/10.1056/NEJMoa1000678
Verdecchia A, Francisci S, Brenner H, Gatta G, Micheli A, Mangone L, Kunkler I (2007) Recent cancer survival in Europe: a 2000-02 period analysis of EUROCARE-4 data. Lancet Oncol 8:784–796. https://doi.org/10.1016/s1470-2045(07)70246-2
Wang R, Zhang J, Chen S, Lu M, Luo X, Yao S, Liu S, Qin Y, Chen H (2011) Tumor-associated macrophages provide a suitable microenvironment for non-small lung cancer invasion and progression. Lung Cancer 74:188–196. https://doi.org/10.1016/j.lungcan.2011.04.009
Whiteside TL (2008) The tumor microenvironment and its role in promoting tumor growth. Oncogene 27:5904–5912. https://doi.org/10.1038/onc.2008.271
Zhang S, Che D, Yang F, Chi C, Meng H, Shen J, Qi L, Liu F, Lv L, Li Y, Meng Q, Liu J, Shang L, Yu Y (2017) Tumor-associated macrophages promote tumor metastasis via the TGF-beta/SOX9 axis in non-small cell lung cancer. Oncotarget 8:99801–99815. https://doi.org/10.18632/oncotarget.21068
Zhou YX, Zhao W, Mao LW, Wang YL, Xia LQ, Cao M, Shen J, Chen J (2018) Long non-coding RNA NIFK-AS1 inhibits M2 polarization of macrophages in endometrial cancer through targeting miR-146a. Int J Biochem Cell Biol 104:25–33. https://doi.org/10.1016/j.biocel.2018.08.017
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOC 2365 kb)
Rights and permissions
About this article
Cite this article
Li, Z., Feng, C., Guo, J. et al. GNAS-AS1/miR-4319/NECAB3 axis promotes migration and invasion of non-small cell lung cancer cells by altering macrophage polarization. Funct Integr Genomics 20, 17–28 (2020). https://doi.org/10.1007/s10142-019-00696-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-019-00696-x