Abstract
Terminal heat stress has detrimental effect on the growth and yield of wheat. Very limited information is available on heat stress-associated active proteins (SAAPs) in wheat. Here, we have identified 159 protein groups with 4271 SAAPs in control (22 ± 3 °C) and HS-treated (38 °C, 2 h) wheat cvs. HD2985 and HD2329 using iTRAQ. We identified 3600 proteins to be upregulated and 5825 proteins to be downregulated in both the wheat cvs. under HS. We observed 60.3% of the common SAAPs showing upregulation in HD2985 (thermotolerant) and downregulation in HD2329 (thermosusceptible) under HS. GO analysis showed proton transport (molecular), photosynthesis (biological), and ATP binding (cellular) to be most altered under HS. Most of the SAAPs identified were observed to be chloroplast localized and involved in photosynthesis. Carboxylase enzyme was observed most abundant active enzymes in wheat under HS. An increase in the degradative isoenzymes (α/β-amylases) was observed, as compared to biosynthesis enzymes (ADP-glucophosphorylase, soluble starch synthase, etc.) under HS. Transcript profiling showed very high relative fold expression of HSP17, CDPK, Cu/Zn SOD, whereas downregulation of AGPase, SSS under HS. The identified SAAPs can be used for targeted protein-based precision wheat-breeding program for the development of ‘climate-smart’ wheat.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Abiotic stresses severely limit the plant growth, development, and yield (Kumar and Rai 2014). Global climate change has large impact on the production of agriculturally important crops, as evident from the drastic reduction in the total world food grain production. Among abiotic stresses, heat stress is one of the most detrimental factors reducing the quality and quantity of the crops (Prasad et al. 2008; Kumar et al. 2015). Elevation in environmental temperature, especially during the pollination and grain-filling, causes disintegration of photosystem-II, and chlorophyll pigment, rupturing of pollens, drying of stigmatic surface, denaturation/aggregation of key pathways associated enzymes, small and disintegrated starch granules synthesis, shriveled seeds, etc. (Feller and Vaseva 2014; Kumar et al. 2013). Heat stress reduces the photosynthetic capacity of the plant through limiting the activity of the enzymes and metabolites and inflicting the oxidative damage to the different cellular organelles (Farooq et al. 2011).
Wheat, being the staple food grain crop, provides major share of carbohydrate and calorie in the diet, worldwide. Heat stress has been predicted to reduce the yield of the wheat by 15% in years to come (Qin et al. 2008). Wheat has inherent defense mechanism to cope up with the vagaries of the nature by activating the defense networks associated with molecular and biochemical changes. At molecular level, plant enhances the expression of stress-associated genes (SAGs) like HSPs, antioxidant enzyme genes, signaling molecules, etc. (Kumar et al. 2012). Biochemically, it increases the accumulation of stress-associated active proteins (SAAPs) and metabolites. In case of wheat, the information regarding the SAGs and SAAPs is scarce and very limited information is available on the public domain (Kumar et al. 2014). Heat stress is one of the major problems in agriculturally important cereal grain crops, especially wheat (Triticum aestivum), which is highly heat-sensitive. A slight elevation in temperature during pollination and grain-filling stages in wheat causes defunct pollen, drying of stigmatic surface, empty pockets in endospermic tissues, defragmented starch granules, etc. (Kumar et al. 2015). Even key regulatory enzymes associated with different pathways showed denaturation/aggregation under HS; impact is quite visible on the quality of the grain as such. The mechanism underlying thermotolerance in wheat has not been deciphered till date; reason being the partial genome sequence available, limited information about the SAGs and SAAPs are available on the public domain. There is a need to identify the candidate proteins possibly playing diverse role in different pathways operated inside the wheat under HS, so that the information generated can be used for designing ‘climate-smart’ wheat.
Plant undergoes either the avoidance or the tolerance mechanism to protect itself from the aberrant climatic conditions. Both the approaches are mediated via changes in the gene expression which causes changes in plant transcriptome, proteome, and metabolome (Rodziewicz et al. 2014). Plant proteome has become powerful tool nowadays to characterize various mechanisms of tolerance and acclimation. Proteins are directly involved in modulating the tolerance mechanism of plants under stress (Kosová et al. 2011). Fourth generation proteomic tools have paved the way for the identification of novel/ conserved SAAPs as well as elucidating their networked roles and interaction. With the advent of new technologies like mass spectrometry, ICPs, etc. proteome has opened totally new perspectives to explore the pathways in different crop species at different levels (Canovas et al. 2004).
The expression of gene at transcript level does not correlate with the changes at protein level. Hence, study on changes in plant proteome is very important since proteins, unlike transcripts, are direct effectors of plant stress response (Jeandroz and Lamotte 2017). Proteins include enzymes, components of transcription, and translation machinery, etc. that regulate plant stress response at transcript and protein levels. Studies of plant response to different stress conditions at protein level can significantly contribute to our understanding of biochemical and physiological mechanisms underlying plant stress tolerance (Kumar et al. 2015, 2016). Proteomics studies lead to identification of potential protein markers whose changes in abundance can be associated with quantitative changes in some physiological and biochemical parameters used for a description of genotype’s level of stress tolerance (Finnie 2007).
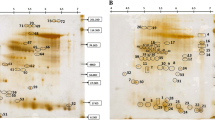
Gel-based proteomics is basically used to compare different samples exposed to differential treatment in order to observe the variations in the expression of the proteins (Chevalier 2010). Gel-based methods are mostly dominated by 2-DE followed by identification of protein spots through mass spectroscopy (MS) technique (Patterson et al. 2007; Budak et al. 2013). Fourth generation proteomic tools have been exhaustively used for the identification of novel SAAPs involved in different pathways in crops like rice, maize, Arabidopsis, etc. (Nanjo et al. 2013). Differential-proteomics approach is used for the identification and analysis of proteomes differing both in the quality and quantity of proteins (Zhang et al. 2006; Kumar et al. 2017).
Isobaric tag for relative and absolute quantitation of proteins (iTRAQ) is a novel and MS-based approach for the relative quantification of proteins (Wiese et al. 2007). It relies on the derivatization of primary amino groups in intact proteins using isobaric tag for relative and absolute quantitation. Due to the isobaric mass design of the iTRAQ reagents, differentially labeled proteins do not differ in mass; accordingly, their corresponding proteolytic peptides appear as single peaks in MS scans. The fragmentation behavior of ESI and MALDI ions of peptides generated from iTRAQ-labeled proteins is analyzed using a TOF/TOF and/or a QTOF instrument (Wiese et al. 2007). Though different approaches have been used in Triticum to identify and report novel and conserved proteins associated with metabolic and stress-related pathways, none of them provides the identification and quantitation of the proteins in a reliable manner with high accuracy in crop like wheat. The availability of limited wheat protein database and partial information about differentially expressed proteins restrict the use of most of the available bioinformatics tools, and researchers have to rely on the information available in closely related species in order to get conclusive results.
Here, we have attempted to use the iTRAQ for the identification of heat tolerance-associated active proteins (SAAPs) in pooled samples (leaves, stem, and spike) of wheat cvs. HD2985 (thermotolerant) and HD2329 (thermosusceptible) under HS. We have identified a list of SAAPs under HS, and further, they were functionally cataloged and characterized for their role in different metabolic pathways and defense-related mechanism.
Materials and methods
Plant material and stress treatment
Wheat cvs. HD2985 (thermotolerant) and HD2329 (thermosusceptible) were selected for the present investigation. The seeds were procured from Division of Genetics, Indian Agricultural Research Institute, New Delhi. Pretreated seeds (Bavistin@0.25%) of both the cvs. were sown in 24 pots (12 pots for each cultivar) and divided into two groups for the pollination and grain-filling stages. Equal quantities of perlite and FYM mixtures were used in the pots and were kept inside the regulated chamber (22 ± 3 °C, relative humidity of 75% and 8 h light with intensity of 250 μmol m−1 s−1 PAR) in the National Phytotron Facility at Indian Agricultural Research Institute, New Delhi. Irrigation was done at regular intervals and plants at the pollination and grain-filling stages (6 pots) in triplicates for each cultivar were exposed to the heat stress (38 °C for 2 h). Pots kept inside the regulated chamber at 22 ± 3 °C were used as control. The heat stress was given in a sinusoidal mode using microprocessor-regulated controller with an increase of 1 °C/10 min till the temperature reaches 38 °C, and further, it was maintained for 2 h. The temperature was decreased to optimum (22 °C ± 3) in the same fashion. For iTRAQ analysis, we used pooled samples (leaf, stem, and spike) collected from five plants (as one biological replicate); every treatment had two biological replicates. For other biochemical analysis, we collected different tissues in triplicate from 5 days after flowering (pollination) and 25 days after flowering (grain-filling) old plants for further downstream analysis. Feekes scale was strictly followed for the collection of samples (Large 1954). Validations of identified stress-associated active proteins (SAAPs) were carried out in the same contrasting wheat cultivars, i.e., HD2985 and HD2329 during grain-filling stage. Seeds were sown in nine pots for each cultivar (three pots each for control: 22 ± 3 °C, T1: 30 °C, 2 h, T2: 38 °C, 2 h) inside the growth chamber under the regulated conditions as mentioned above. Samples (root, stem, leaf, and spike) were collected in triplicate after the HS treatment and stored at − 80 °C for further down-processing. We used three biological and three technical replicates for the expression analysis.
Protein extraction and purification
The samples (leaves, stems, and spikes) collected from the control (22 ± 3 °C) and HS-treated (38 °C, 2 h) wheat cvs. HD2985 (thermotolerant) and HD2329 (thermosusceptible) were frozen on dry ice and ground to a powder in a pestle. Total protein was extracted from each tissue using the Total Protein Isolation kit (ToPI-P; ITSI- Biosciences, Pennsylvania) following the instructions as given by the manufacturers. The protease inhibitor cocktail was added to the extracted protein during the isolation to inhibit the protease activity. The protein lysates were assayed for the protein concentration using the Total Protein Assay kit (ITSI-Biosciences, Pennsylvania). Equal protein concentration from different tissue (leaf, stem, and spike) under control and HS-treated conditions was pooled separately (in duplicate) for the iTRAQ. The quality of the proteins was analyzed using the Bio Analyzer chip (Agilent, USA) and through electrophoretic separation on SDS-PAGE gel (Laemmli 1970).
iTRAQ labeling and mass spectrometry analysis
iTRAQ labeling and mass spectrometry (MS) were performed as per the protocol standardized for this experiment by the ITSI-Biosciences (Johnstown PA 15901, USA). In brief, pooled protein samples (isolated from leaf, stem, and spike) were treated with 10 mM dithiothreitol (DTT) at 55 °C for 1 h followed by alkylation in 55 mM iodoacetamide for 45 min. The supernatant from each pool was precipitated and re-suspended in a digestion and iTRAQ compatible buffer. One hundred micrograms of each pooled protein solution was digested with sequencing grade trypsin (Promega, USA) overnight. The digested samples were labeled with iTRAQ 8-plex kits (Applied Biosystems, USA) according to the manufacturer’s protocol. Wheat cv. HD2985 (control) sample was labeled with reagents 114, HD2985 (HS-treated) with 116, HD2329 (control) with 118, and HD2329 (HS-treated) with 121. The iTRAQ-labeled peptides were mixed and cleaned by strong cation exchange (SCX) chromatography on Ultremex SCX column using Shimadzu LC-20AB HPLC. The peptides were further eluted from the SCX column using 250–450 mM ammonium acetate and were dried and re-dissolved in 2% acetonitrile in 0.1% trifluoroacetic acid. The fractionated samples were loaded onto a 100 μm × 20 mm Magic C18 100 Å 5 U reverse phase trap where they were desalted online before being separated using a 75 μm × 150 mm Magic C18 200 Å 3 U reverse phase column. LC-MS/MS analysis was performed using a standard top 15 method on Thermo Scientific Q-Exactive orbitrap mass spectrometer in conjunction with a Proxeon Easy-nLC II HPLC (Thermo Scientific) and Proxeon nanospray source. Peptides were eluted using a flow rate of 300 nL/min and a gradient of 0.1% formic acid (A) and 100% acetonitrile (B). A 180 min gradient was ran with 5% to 35% B over 155 min, 35% to 80% B over 10 min, 80% B for 2 min, 80% to 5% B over 3 min, and finally held at 5% B for 10 min. Data-dependent MS/MS data was collected using normalized higher energy collision dissociation (HCD) of 30. MS1 AGC was set to 1e6 with an ion trap time of 60 ms, and MS2 AGC was set at 5e4 with 250 ms ion trap time. Unassigned charge states and charge states of + 1 and >+ 6 were excluded for MS/MS selection and dynamic exclusion was set to 15 s.
Database search and quantification
Raw data files were searched using Proteome Discoverer™ 1.4 software (Thermo Scientific, USA) and the SEQUEST algorithm (University of Washington) against the most recent database for the Triticum aestivum with ~ 19,000 proteins downloaded from National Center for Biotechnology Information (NCBI) on 18th January, 2018 with the following search parameters:
-
Trypsin was the selected enzyme allowing for up to three missed cleavages per peptide; methylthio-cysteine, N-terminal iTRAQ 8-plex, and lysine iTRAQ 8-plex were used as a static modifications; oxidation of methionine was used as variable modification.
-
The false discovery rate (FDR) at the peptide level and protein level were accepted to 5% based on the additional identification confidence. Only peptides that were identified at a high confidence level were included in the protein identification.
-
Proteins with iTRAQ ratio more than 2-fold were considered as upregulated and less than 0.67-fold was considered as downregulated. iTRAQ ratio between 2.0 and 0.67-fold in successive comparisons was considered as moderate or no changes. Other ratios were also included for the reference.
iTRAQ 8-plex was chosen for the quantification during the search simultaneously. The search results were passed through additional filters before exporting the data. For protein identification, the filters were set as follows: significance threshold P < 0.05 (with 95% confidence) and ion score or expected cutoff less than 0.05 (with 95% confidence).
Function assignment
The identified proteins were annotated using BLAST2GO (https://www.blast2go.com/) for gene ontology (GO) annotation and enrichment analysis. The sequences were blasted against non-redundant database. The dataset containing all of the EuKaryotic Orthologous Groups (KOG) numbers was then used to conduct protein–protein interaction (PPI) analysis by STRING using Cytoscape 3 v 1.3.0 software (http://apps.cytoscape.org/apps/stringapp). Heat map of the differentially abundant proteins was built using Multiple Experiment Viewer (Mev) tool (http://mev.tm4.org/). We also used the DAVID Bioinformatics Database v 6.7 developed by the National Institutes of Health (NIH) for the GO analysis (https://david.ncifcrf.gov/).
Expression analysis of identified SAAPs through quantitative real-time PCR
Designing of gene-specific primers
Twelve proteins randomly selected from the identified SAAPs in present investigation were used for the complementation at transcript level in wheat cvs. HD2985 and HD2329 under differential HS. The peptide sequence of identified proteins [HSP70, HSP17, Cu/Zn SOD, calcium-dependent protein kinase (CDPK), alpha amylase, ADP glucophosphorylase (AGPase), soluble starch synthase (SSS), starch branching enzyme (SBE), RuBisCo (LSU), RuBisCo activase (Rca), sucrose synthase (Suc Syn), and oxygen evolving enhancer protein (OEEP)] were subjected to EMBOSS Backtranseq (EMBL-EBI; https://www.ebi.ac.uk/Tools/st/emboss_backtranseq) for the retrieval of nucleotide sequence which was further used for the homology search based gene identification using BLASTn (NCBI). Nucleotide sequence of randomly selected SAAPs was mined from the GenBank database of NCBI [HSP70 (AF005993.1), HSP17 (KY075943.1), Cu/Zn SOD (U69632.1), CDPK (JX878360.1), alpha amylase (X05809.1), AGPase (KC347594.1), SSS (KJ854903.1), SBE (Y12320.1), Rub-LSU (KF801504.1), Rca (KU291395.1), SPS (AF310160.1), OEEP (KY014425.1), and β-actin (AB181991.1)] and was used for the oligo designing using Genefisher2 primer designing software (http://bibiserv.techfak.uni-bielefeld.de/genefisher2). The quality of the primers was checked using OligoAnalyzer v 3.1 (IDT, USA; https://eu.idtdna.com/calc/analyzer). The list of primers used for the quantitative expression analysis has been presented as supplementary file (Online Resource 1).
Quantitative real-time PCR
Here, we have used different tissues (spike, leaf, stem, and root) of contrasting wheat cultivars, i.e., HD2985 (thermotolerant) and HD2329 (thermosusceptible) for the expression analysis using qRT-PCR. Total RNA was isolated from the control (C: 22 ± 3 °C) and HS-treated (T1: 30 °C, 2 h; T2: 38 °C, 2 h) samples using Trizol method (Invitrogen, UK). The quality of the isolated total RNA was checked on Bio-analyzer (Agilent, USA). We observed two prominent peaks in all the samples with OD 260/280 ratio of > 2.0. First-strand cDNA synthesis was carried out using RevertAid™ H minus First Strand cDNA synthesis kit (Thermo Fisher Scientific, USA). The cDNA products were then diluted 50-fold with deionized water before using as a template in qRT-PCR. Quantitative real-time PCR was performed in triplicate reactions using the CFX96 platform (Bio Rad, UK) as mentioned in our earlier literature (Kumar et al. 2018). The reactions were conducted in 20-μL volume containing 2 μL diluted cDNA, 0.4 μL of each forward and reverse primer (10 μM), 10 μL of the KAPA SYBR® FAST universal 2×PCR Master Mix (Kappa Biosystems, Switzerland), and 7.2 μL of nuclease free water under the following cycling conditions: 95 °C for 2 min, 40 cycles at 95 °C for 20 s, 58 °C for 30 s, and 72 °C for 5 s followed by plate read. A thermal denaturing cycle at 95 °C for 15 s, 65 °C for 15 s, and 95 °C for 15 s was also added to determine the dissociation curves and the specificity of the amplifications. All the expression levels were normalized to the arithmetic mean of the selected β-actin gene (accession no AB181991.1). The relative expression was calculated using comparative Ct method (Pfaffl 2001).
In-planta validation of signaling molecule and catalytic chaperone through immunoblotting
The control (C: 22 ± 3 °C) and HS-treated (T1: 30 °C, 2 h; T2: 38 °C, 2 h) samples collected from wheat cvs. HD2985 and HD2329 during the pollination and grain-filling stages were subjected to immunoblotting as described earlier (Kumar et al. 2015, 2017). The antibodies of randomly selected signaling molecule (CDPK) and catalytic chaperone (Rca) were used for the hybridization. Monoclonal antibody of CDPK was procured commercially (Sigma-Aldrich, UK), and antibody of Rca was synthesized in rabbit from the purified wheat Rca protein available in our Stress Genomics lab (IARI, New Delhi). The dilution used for the incubation of membrane in primary monoclonal antibody and secondary antibody (peroxidase-conjugated) was 1:1000 and 1:4000. Band of desired intensity was visible after 2–5-min incubation, and the membrane was washed immediately in water and Tris-buffered saline (TBS). The dried membrane was used for photography with Gel Doc Easy (Bio Rad, UK).
Activity assay of enzymes involved in starch biosynthesis
AGPase activity was assayed as per the modified method of Kleczkowski et al. (1993). The reaction mixture of 1 mL was prepared by adding 50 mM MOPS (pH 7.4), 7.5 μM MgSO4·7H2O, enzyme extract, 3 μM 3-PGA, 0.5 μM NADP+, 0.5 μM ADP-glucose, 2 U phosphoglucomutase, and 2 U glucose-6-phosphate dehydrogenase. The reaction was started by the addition of 200 μL of sodium pyrophosphate (2.5 μM). The pyrophosphorolytic activity of AGPase was assayed spectrophotometrically by monitoring the increase in absorbance due to conversion of NADP to NADPH at 340 nm.
Soluble starch synthase (SSS) activity was assayed in the endospermic tissue as per the protocol of Nakamura et al. (1989).
Activity assay of enzymes involved in antioxidant defense network
Superoxide dismutase activity assay
Leaf (1 mg) was ground in 6 mL of ice cold 50 mM potassium phosphate buffer (pH 7.0) containing 2 mM sodium EDTA and 1% (w/v) polyvinylpolypyrrolidone (PVP). The homogenates were centrifuged at 10,000×g (4 °C) for 10 min. The tissue extracts were either stored at − 80 °C or immediately used for enzyme assay. For the quantification of soluble protein content, coomassie blue dye binding assay was used (Bradford 1976). Bovine serum albumin (BSA) was used for the preparation of the standard curve. Superoxide dismutase activity was determined by measuring its ability to inhibit the photochemical reduction of nitro-blue tetrazolium (NBT) in the presence of riboflavin in light (Giannopolitis and Ries 1977). One unit of enzyme activity was determined as the amount of the enzyme needed for the inhibition of 50% NBT reduction rate by monitoring absorbance at 560 nm with spectrophotometer.
Catalase activity assay
The activity of catalase enzyme was measured as described by Chance and Maehly (1955). Fresh leaf material (1 g) was crushed in 5 mL of ice-cold 50 mM potassium phosphate buffer (pH 7.0) containing 2 mM sodium-EDTA and 1% (w/v) polyvinylpyrrolidone (PVP). The homogenates were centrifuged at 10,000g (4 °C) for 10 min. The supernatant was used for the quantification of soluble protein content by using Bradford method (Bradford 1976). For assaying CAT activity, the decomposition of H2O2 was followed by decline in the absorbance at 240 nm. CAT activity was determined by following the consumption of H2O2 (extinction coefficient: 39.4 mmol−1 cm−1) at 240 nm over a 3-min interval.
Statistical analysis
The analysis of variance of different biochemical parameters was calculated using the SPSS v16.0 (SPSS, Inc., USA) for statistically significant differences (P ≤ 0.05) based on two-way variance analysis (ANOVA).
Results
Proteomic characterization of wheat under HS
The proteome of pooled samples (leaf, stem and spikes) of control and HS-treated wheat cvs. HD2985 and HD2329 were quantified and characterized using the iTRAQ method. High quality proteins were extracted from the different tissues, digested and iTRAQ labeled peptides were quantified using Thermo Scientific Q-Exactive orbitrap mass spectrometer in conjunction with a Proxeon Easy-nLC II HPLC (Thermo Scientific) and Proxeon nanospray source. Proteome Discoverer 1.4 (Thermo Scientific, USA) and the SEQUEST algorithm were used for the peptide identification along with Maxquant software against the recent protein database of wheat and other closely related species. We identified ~ 159 protein groups. Further, BLAST2GO analysis resulted in the identification of ~ 51,317 homologous proteins. Combining the dataset of both the cultivars, 4271 stress-associated active proteins (SAAPs) were annotated using different databases as presented in supplementary file (Online Resource 2).
Differentially abundant stress-associated active proteins
We identified 144 protein groups (2800 upregulated and 2225 downregulated SAAPs) in HS-treated wheat cv. HD2985. Similarly, HD2329 showed 147 protein groups (800 proteins upregulated and 3600 proteins downregulated) under HS. Under ambient conditions, we observed 146 differentially abundant protein groups (3225 active proteins to be upregulated and 1600 to be downregulated) by comparing thermotolerant (HD2985) with thermosusceptible (HD2329) as shown in supplementary file (Online Resource 3). Some of the identified important differentially abundant SAAPs have been shown in Table 1.
A Venn diagram was constructed showing the distribution of differentially abundant SAAPs (upregulated and downregulated) in response to HS in control and HS-treated wheat cvs. HD2985 and HD2329 (Fig. 1). We observed 60.3% of the identified differentially abundant SAAPs showing upregulation in HD2985 (thermotolerant) and downregulation in HD2329 (thermosusceptible), whereas 16.2% of differentially abundant SAAPs were observed in upregulated HD2985 and HD2329 cvs. Similarly, 19.1% of the SAAPs were observed common in downregulated HD2985 and HD2329. A list of differentially abundant SAAPs identified in HD2985 and HD2329 with their mean fold expression (Fc) has been presented in supplementary file (Online Resource 4).
Venn diagram depicting the distribution of upregulated and downregulated SAAPs identified in wheat cvs. HD2985 and HD2329 under HS; Up_reg_HD2985: upregulated SAAPs in HD2985, Down_reg_HD2985: downregulated SAAPs in HD2985, Up_reg_HD2329: upregulated SAAPs in HD2329, Down_reg_HD2329: downregulated SAAPs in HD2329; maximum SAAPs showed upregulation in HD2985 and downregulation in HD2329 under HS
A heat map of the identified differentially abundant SAAPs was generated in different combinations—HS-treated HD2985 (116/114), controls of HD2985 vs HD2329 (118/114), HS-treated HD2329 vs control of HD2985 (121/114), controls of HD2329 vs HD2985 (118/116), HS-treated HD2329 vs HD2985 (121/116), and HS-treated HD2329 (121/118) (Fig. 2). We observed more than 1.6-fold expressions in most of the SAAPs identified in HS-treated HD2985, as compared to HS-treated HD2329 where mostly downregulation was observed. Similar variations in the expression were observed in the control samples of wheat cv. HD2985 and HD2329. Some of the SAAPs identified in HS-treated HD2329 like oxygen evolving enhancer protein, PS-I, 1,4-beta-D xylan xylanohydrolase, alpha-amylase inhibitor, etc. showed very high fold expression, as compared to control of HD2985. We identified few unique differentially abundant proteins based on digital fold expression showing upregulation in HS-treated wheat cv. HD2985 (thermotolerant) and downregulation in HS-treated wheat cv. HD2329 (thermosusceptible; Fig. 3; Online Resource 5)). For example, Cu/Zn-SOD showed 4.1-fold upregulation in HS-treated HD2985 and 1.2-fold downregulation in HS-treated HD2329. Similarly, gamma gliadin (seed storage protein) showed 3.7-fold increase in expression in HD2985 and 2.3-fold downregulation in HD2329 under HS. The variations in the expression of these SAAPs may be the reason behind the diversity in the HS-tolerance capacity of different wheat germplasm.
Heat map analysis of differentially abundant stress-associated active proteins (SAAPs) in contrasting wheat cultivars under HS using isobaric tag for relative and absolute quantitation of proteins (iTRAQ); upregulated and downregulated proteins observed in HS-treated HD2985 (116/114), HS-treated HD2329 (121/118), and control samples (HD2985 versus HD2329) (116/121); C: 22 ± 3 °C, HS: 38 °C, 2 h)
Unique differentially abundant stress-associated active proteins (SAAPs) identified using iTRAQ showing upregulation in HS-treated HD2985 (thermotolerant) and downregulation in HS-treated HD2329 (thermosusceptible); blue and red bars represent the numbers of upregulated and downregulated proteins, respectively
Gene ontology analysis of SAAPs
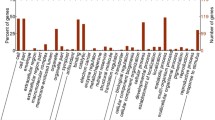
Identified proteins in wheat cvs. HD2985 and HD2329 and their predicted homologs were further characterized using gene ontology (GO) analysis tool. Gene ontology analysis of SAAPs showed 51%, 39%, and 10% to have established molecular functions, biological processes, and cellular component.
Based on the molecular function, the identified proteins were classified into 58 groups with the involvement of 6870 proteins. We observed proton transport (36%) to be most altered under HS followed by membrane-related protein, ion-binding protein, ATP hydrolysis and metabolic related proteins, phosphatase protein, aldolase, Dikinase, ATP synthase, hydrolase, and RuBisCo (Fig. 4). Similarly, 69 biological processes consisting of 5145 proteins were observed to be significantly altered under HS. Photosynthesis (35%) was observed to be most affected under HS followed by carbohydrate metabolism, binding protein, enzymatic activity, etc. Some of the photosynthesis-related processes observed to be highly sensitive to HS were carbon fixation, PSII, electron transport chain (ETC), and oxygen evolving complex. Cellular component analysis showed 44 groups with more than 2360 proteins to be altered under HS. Some of the cellular-related activities identified were protein heterodimerization activity, protein folding, proteolysis, protein peptidyl-prolyl isomerization, protein kinase activity, and protein phosphorylation. Maximum proteins were observed to be associated with ATP binding (27%) followed by ETC proteins (Fig. 4).
Localization of identified SAAPs in wheat under HS
The SAAPs identified in wheat cvs. HD2985 and HD2329 under HS-treated condition were analyzed for their localization inside the cells (Fig. 5). We observed ~ 950 proteins to be chloroplast localized followed by thylakoid membrane (425 proteins), cytoplasm (280 proteins), and nucleus (250 proteins). The localization of identified SAAPs was observed all across the different organelles like plasma membrane, mitochondria, and ribosome. The localization of maximum SAAPs in chloroplast makes us to conclude that chloroplast is most affected under HS followed by other important organelles like mitochondria and ribosomes. These SAAPs especially signaling molecules (MAPKs and CDPKs), HSPs (HSP17, HSP20, HSP26, and HSC70) and antioxidant enzymes (SOD, CAT, and APX) might be playing role in imparting tolerance to the organelles in wheat under HS.
Functional cataloging of identified SAAPs on the basis of pathway involvement
Based on the information available on public domain, the SAAPs identified in wheat through iTRAQ were characterized for their involvement in different pathways (Fig. 6a). Most of the SAAPs were observed to be involved in antibiotic biosynthesis followed by photosynthesis, glyoxylate and decarboxylate pathways, glycolysis and gluconeogenesis pathways. Some of the other pathways showing involvement of these SAAPs are purine metabolism, starch and sucrose metabolism, nitrogen metabolism, and oxidative phosphorylation. We observed ~ 20 uncharacterized SAAPs to be associated with some novel pathways showing differential expression in response to HS. The present findings make us to conclude that carbon assimilatory pathway (source) is most altered under HS followed by starch biosynthesis pathway (sink) in wheat. A list of pathways associated SAAPs identified in control and HS-treated wheat cvs. HD2985 and HD2329 have been shown in supplementary file (Online Resource 6).
Functional cataloging of SAAPs on the basis of enzymatic function
The identified SAAPs were mapped on to different databases for tracing the enzymatic functions executed inside plant system (Fig. 6b). We observed most of the identified SAAPs showing the function of carboxylase enzyme (280 proteins) followed by phosphatase, dehydrogenase, aldolase, and kinase activities. Some of the other enzymatic function observed in HS-treated wheat cvs. HD2985 and HD2329 are invertase, isomerase, oxidase, and reductase. The abundance of carboxylase enzyme showed abundance of carboxylation reaction in wheat under HS. This may be the reason behind the turmoil in the ratio of source to sink as reflected in terms of quality and quantity of wheat grains.
List of SAAPs associated with carbohydrate metabolism
We identified ~ 951 SAAPs to be involved in carbohydrate metabolism in wheat cvs. HD2985 and HD2329 under control and HS-treated conditions. Some of the SAAPs identified are α/β-amylases, fructose 1, 6-bisphosphate 1-phosphatase, phosphoric ester hydrolase, chitinase, hydrolase, β-mannosidase, RuBisCo, and RuBisCo activase. The list of SAAPs associated with carbohydrate metabolism has been presented in supplementary file online (Online Resource 7).
We identified ~ 712 SAAPs to be involved in photosynthesis in both the cvs. under HS as shown in supplementary file online (Online Resource 8). The expression of some of the identified SAAPs involved in photosynthesis has been presented in the form of heat map (Fig. 7). Some of the identified active proteins involved in photosynthesis are photosystem-I family, photosystem antenna protein-like family, photosystem II CP47 reaction center protein, chlorophyll a/b binding protein, PS-II, OEEP, PsbO, Cyt b6f, and RuBisCo. We also identified ~ 124 SAAPs to be thylakoid localized especially dominated with photosystem I and cytochrome f families as presented in supplementary file online (Online Resource 9).
Heat map analysis of differentially abundant SAAPs identified in wheat for depicting the expression pattern under HS; expression analysis of SAAPs associated with photosynthesis, defense/stress, signaling and transport, storage, and metabolic processes has been shown through heat map; green color showing upregulation and red color showing downregulation of proteins
Expression of SAAPs involved in defense and stress tolerance
The SAAPs identified were further characterized for their functional role in defense and stress-related activities. We observed very high fold increase in the expression of Ribonuclease TUDOR-1, HSP90, HSP20, Peroxidase, and HSC70 in thermotolerant wheat cv. HD2985, whereas downregulation was observed in thermosusceptible (wheat cv. HD2329) under HS (Fig. 7). The expression of HSPs was observed most altered in both the cultivars under HS. We observed HSPs as most abundant SAAPs involved in protecting the nascent proteins from denaturation under HS, other than playing important role in protein folding and thermotolerance. The iTRAQ data was mined for the identification of different HSPs and the result is presented in the supplementary file online (Online Resource 10).
Expression of SAAPs involved in signaling, transport, and extracellular activities
We observed very high fold increase in the expression of signaling and transport proteins like protein kinase, transmembrane protein, and calmodulin in HS-treated wheat cv. HD2985 and downregulation in HS-treated wheat cv. HD2329 (Fig. 7). Under ambient temperature, we observed upregulation of basic 7s globulin 2, protein folding related proteins, etc., whereas downregulation of HSC70, peroxidase, cell redox homeostasis proteins was observed in both the cultivars. Serpin protein was observed predominantly localized in extracellular space and reported to be involved in protease inhibition and signaling especially in wheat. The iTRAQ data generated in present investigation was mined for the identification of SAAPs associated with signaling, transport, and extracellular activities and the list has been presented in supplementary file online (Online Resource 11).
Expression of SAAPs involved in storage and metabolic processes
We observed upregulation of identified storage proteins like oleosin, Globulin 3A, 3B, and gamma gliadin in wheat cv. HD2985 (thermotolerant) and downregulation in wheat cv. HD2329 (thermosusceptible) under HS. Some of the proteins like uncharacterized protein, vicilin like protein, and Globulin 1S showed upregulation in thermosusceptible wheat cv. HD2329. The expression pattern of identified SAAPs associated with storage has been depicted in the form of heat map (Fig. 7).
The SAAPs identified in present investigation were characterized for their involvement in different metabolic processes. We observed increase in the expression of SAAPs involved in metabolic processes like glutamine metabolism, protein metabolism, starch metabolism, and carbohydrate metabolism in both the wheat cvs. under HS. Thermosusceptible cv. HD2329 showed downregulation of SAAPs associated with metabolic processes like carbohydrate and lipid metabolism under HS. Under ambient condition, we observed upregulation of SAAPs associated with ammonia metabolism and starch degradation. We also identified ~ 19 α/β amylases to be involved in starch degradation as presented in supplementary file online (Online Resource 12).
Transcript profiling for the complementation of identified SAAPs through qRT-PCR
We randomly selected defense-related genes (HSP70, HSP17, CDPK, Cu/Zn SOD), carbon assimilatory pathway-associated genes (RuBisCo, Rca, OEEP, Suc Syn), and metabolic pathway-associated genes (AGPase, SSS, SBE, and α-amylase) for the complementation in different tissues of developing wheat (root, stem, leaves, and spike) under control (C: 22 ± 3 °C) and HS-treated (T1: 30 °C, 2 h; T2: 38 °C, 2 h) conditions.
Expression analysis in spike of wheat cv. HD2985 showed maximum increase in the relative fold expression of HSP17 (8.2-fold) followed by CDPK (2.2-fold) in response to T2 (38 °C, 2 h), as compared to control (Fig. 8a). Most of the SAAPs like Cu/Zn SOD, HSP70, and CDPK showed increase in the expression with the differential HS in the spike, whereas pathway-associated genes like AGPase (1.4-fold) and SSS (1.25-fold) showed downregulation in response to T2 treatment, as compared to T1 in the spike during grain-filling stage.
Complementation study of randomly selected SAAPs identified using iTRAQ in different tissues of wheat cv. HD2985 (thermotolerant) through quantitative real-time PCR; expression analysis of SAAPs associated with defense/stress in a spike, b leaves, c stem, and (d) root under differential HS; C: 22 ± 3 °C, T1: 30 °C, 2 h, T2: 38 °C, 2 h; β-actin gene was used as endogenous control for normalizing the Ct value; relative fold expression was calculated using Pfaffl method. Three biological and three technical replicates were used for the expression analysis. Vertical bars indicate s.e (n = 3)
Similar pattern of expression of SAAPs was observed in the leaves of wheat cv. HD2985 in response to T1 and T2, as compared to control (Fig. 8b). Analysis of differentially abundant SAAPs in leaves showed maximum relative fold expression of HSP17 (22.5-fold) followed by RuBisCo (4.5-fold), Rca (4.3-fold), and OEEP (4.1-fold) in response to T2. All the randomly selected SAAPs involved in defense, carbon assimilation, and starch metabolism showed upregulation in leaves in response to differential HS. Similar pattern of expression was observed in stem of wheat cv. HD2985; relative fold expression was observed less, as compared to leaves and spike (Fig. 8c). Expression analysis of defense-related genes like HSP70 (1.55-fold), HSP17 (13.1-fold), CDPK (1.8-fold), and Cu/ Zn SOD (1.6-fold) showed increase in the relative fold expression in root under HS, as compared to control. We observed significant upregulation of defense-related genes in root under T1 and T2 treatments (Fig. 8d).
Expression analysis of randomly selected differentially abundant SAAPs in wheat cv. HD2329 (thermosusceptible) showed maximum relative fold expression of HSP17 in spike (4.2-fold), leaves (9.8-fold), stem (4.3-fold), and root (8.2-fold) under T2 treatment, as compared to control (Fig. 9). In spike, we observed increase in the expression of differentially abundant SAAPs involved in defense and downregulation of starch metabolic pathway associated genes under T2 treatments (Fig. 9a). Similarly, we observed upregulation of defense-related, carbon assimilation, and starch metabolism pathway-related genes in leaves in response to differential HS (Fig. 9b). Expression analysis in stem and root showed upregulation of differentially abundant SAAPs under HS (Fig. 9c, d). Overall, the abundance of differentially abundant SAAPs involved in different pathways was observed maximum in wheat cv. HD2985, as compared to HD2329 under differential HS.
Complementation study of randomly selected SAAPs identified using iTRAQ in different tissues of wheat cv. HD2329 (thermosusceptible) through quantitative real-time PCR; expression analysis of SAAPs associated with defense/stress in a spike, b leaves, c stem, and d root under differential HS; C: 22 ± 3 °C, T1: 30 °C, 2 h, T2: 38 °C, 2 h; β-actin gene was used as endogenous control for normalizing the Ct value. Relative fold expression was calculated using Pfaffl method. Three biological and three technical replicates were used for the expression analysis. Vertical bars indicate s.e (n = 3)
Characterization of randomly selected differentially abundant SAAPs identified from wheat under HS
We used iTRAQ to explore the galaxy of proteins imparting tolerance to the wheat under HS. SAAPs involved in different metabolic pathways were identified, and their expression patterns were analyzed under HS. We selected three potential SAAPs based on their functional and expressional properties for in silico protein characterization. The proteins selected for the analysis are sucrose synthase, ADP glucophosphorylase, and RuBisCo. Sucrose synthase is involved in synthesis of photosynthates to be transported to endospermic tissues for the accumulation. AGPase is the regulatory enzyme of starch biosynthesis pathway involved in synthesizing the starch in endospermic tissues and RuBisCo is the key regulatory enzymes involved in photosynthesis. Based on the information available on public domain, we simulated the three-dimensional protein structure of all the three proteins. 3D structure of protein was observed very complex in case of sucrose synthase, as compared with AGPase and RuBisCo (Fig. 10a–c). The sterochemical quality of the above mentioned proteins was analyzed by Ramachandran plot analysis. We observed total of 3141 residues in case of sucrose synthase, 1741 in AGPase, and 465 in RuBisCo (Fig. 10d–f). Protein-protein interaction (PPI) study of all the three identified proteins showed very complex network in case of sucrose synthase and RuBisCo, whereas AGPase showed simpler network structure (Fig. 10g–i). Sucrose synthase was observed to interact with some of the important proteins like trehalose phosphate synthase, starch synthase, isoforms of sucrose synthase, probable trehalose-phosphate phosphatase E, hydrolase, etc.
Characterization of randomly selected SAAPs identified in HS-treated wheat using iTRAQ. Three dimensional structure of a sucrose synthase, b ADP-glucophosphorylase, and c RuBisCo; Ramachandran plot showing stereo-chemical quality of three-dimensional structure of d sucrose synthase, e ADP-glucophosphorylase, and f RuBisCo; protein-protein interactions (PPIs) study of g sucrose synthase, h ADP-glucophosphorylase, and i RuBisCo
Accumulation pattern and activity assay of differentially abundant SAAPs
We randomly selected signaling molecule (CDPK) and catalytic chaperone (Rca) for analyzing the protein accumulation pattern. Similarly, metabolic pathway-associated enzymes (AGPase and SSS) and defense/stress responsive proteins (SOD and CAT) were selected for the activity assay under differential HS (T1: 30 °C, 2 h; T2: 38 °C, 2 h). Immunoblot analysis of signaling molecule showed increase in the accumulation of CDPK protein with differential HS; maximum CDPK protein accumulation was observed in response to T2 in wheat cv. HD2985, as compared to HD2329 (Fig. 11). Similarly, expression analysis of Rca showed similar pattern of increase in the accumulation of protein under differential HS. Maximum accumulation of Rca was observed in wheat cv. HD2985 in response to T2 treatment, as compared to HD2329 (Fig. 11).
Protein accumulation pattern of randomly selected SAAPs identified in wheat through immunoblot assay. a Signaling molecule (calcium-dependent protein kinase) associated with defense/stress and b Catalytic chaperone (RuBisCo activase) associated with carbon assimilation was selected for the immunoblotting; C: 22 ± 3 °C, T1: 30 °C, 2 h, T2:38 °C, 2 h; based on the size predicted using pre-stained marker, membrane of specific size was cut after blotting and used for the incubation with primary and secondary antibodies; Gel Doc Easy (Bio Rad, UK) was used to capture the image
Activity assay of AGPase involved in starch biosynthesis pathway showed decrease in the activity in both the cvs. under HS; minimum activity was observed in wheat cv. HD2329 (1.45 U/mg proteins) in response to T2, as compared to control (Fig. 12a). Similar pattern of activity of SSS was observed in both the cultivars under differential HS. Percent decrease in the activity was observed more in thermosusceptible cv. HD2329, as compared to thermotolerant cv. HD2985 under HS.
Activity assay of randomly selected SAAPs in contrasting wheat cultivars under differential heat stress. a Activities of ADP-glucophosphorylase (AGPase) and soluble starch synthase (SSS) involved in starch biosynthesis pathway. b Activities of superoxide dismutase (SOD) and catalase (CAT) involved in antioxidant defense network; wheat cvs. HD2985 and HD2329 were selected for the study; C: 22 ± 3 °C, T1: 30 °C, 2 h, T2: 38 °C, 2 h. Three biological and three technical replicates were used for the expression analysis. Vertical bars indicate s.e (n = 3)
The activity of defense/stress-associated enzymes (SOD and CAT) showed increase in the activity with the treatments (Fig. 12b). The maximum SOD activity in wheat cvs. HD2985 (14.1 U/mg proteins) and HD2329 (9.5 U/mg proteins) was observed in response to T2. Similar pattern of CAT activity was observed in both the cultivars under differential HS. The CAT activity was observed higher in HD2985 (6.2 U/mg proteins), as compared to HD2329 (5.6 U/mg proteins) under T2 treatment.
Model for characterizing the tolerance of source and sink of wheat under terminal HS
Here, we identified novel and hypothetical SAAPs involved in modulating the tolerance of photosynthesis (source) and starch metabolism (sink) under terminal HS using iTRAQ. A model was simulated using the information generated in present investigation (Fig. 13). We observed candidate SAAPs like CDPK, MAPK, HSPS17, and HSC70 to be highly upregulated in both the cultivars. Carbon assimilatory pathway showed downregulation of RuBisCo and Rca along with sucrose phosphate synthase under HS, as compared to control. Most of the signaling molecules and defense/stress-associated proteins showed upregulation in leaves of wheat under HS. Because of denaturation of catalytic chaperone like Rca, there is increase in the abundance of inactive RuBisCo inside the chloroplastic causing drastic reduction in the photosynthetic rate and accumulation of photosynthates. The transportation of sucrose is hampered because of denaturation/aggregation of sucrose phosphorylase and sucrose transporter under HS. An increase in the accumulation of stem reserve has been reported in wheat under HS, reducing the amount of photosynthates accumulating inside the endospermic tissues. Further, starch biosynthesis-associated enzymes like AGPase, GBSS, SS-isoforms, starch branching enzyme (SBE), and starch debranching enzyme (SDE) showed downregulation in the expressions and activities under terminal HS. Because of denaturation/aggregation of these enzymes, the pace of starch synthesis slows down causing formation of pleated, fragmented, distorted, and small starch granules with large empty pockets. The grain becomes shriveled and the quality is compromised under terminal HS. We observed some of the defense-related SAAPs like HSPs and antioxidant enzyme network to activate inside the endospermic tissue modulating the tolerance of pathway under HS. Even we observed different isoforms of α/β amylases in the endospermic tissues involved in hydrolysis of starch affecting the quality of the grains.
Discussions
Terminal HS is one of the major problems in wheat growth and yield. Though various attempts have been made to mitigate this problem, very limited success has been achieved in the past. The information regarding the SAAPs is very limited in case of wheat in spite of advent in technology. Here, we have used the fourth generation proteomic tools, i.e., iTRAQ to identify SAAPs in contrasting wheat cvs. under terminal HS in order to characterize the defense network associated with thermotolerance.
iTRAQ has been used in the past to identify novel proteins involved in different pathways in plant and animal systems. Here, we identified ~ 4272 SAAPs in wheat cvs. HD2985 and HD2329 under HS. Wang et al. (2014) identified 313 proteins responsive to NaCl and NaHCO3 in tomato roots using iTRAQ and reported 70 and 114 proteins to be upregulated by salt and alkali stress, respectively. Same group also identified 106 differentially abundant proteins associated with aluminum toxicity in root of rice through iTRAQ and reported that most of the SAAPs are involved in energy and stress/defense (Wang et al. 2014). Most of the identified SAAPs in present investigation were observed to be involved in carbon assimilatory pathway. The findings are in conformity with the report of Liu et al. (2014) who identified 174 SAAPs in grapevine leaves under HS and stress recovery phase using iTRAQ and reported most of the proteins to be involved in carbon assimilatory pathway. Budak et al. (2013) identified 75 differentially abundant proteins in leaves of wild and modern wheat playing important role in drought stress tolerance.
We identified some of the unique proteins like Cu-Zn SOD, RuBisCo, ATP synthase, gamma gliadin, peroxidase, HSP70 showing very high relative fold abundance in thermotolerant cv., as compared to one thermosusceptible under HS. Ge et al. (2013) reported 3425 proteins in wheat through iTRAQ and showed 157 to be SAAPs and 44 to be H2O2-responsive proteins. Similarly, Ji et al. (2016) identified 278 and 440 proteins through iTRAQ with significantly altered abundances in leaves and roots of soybean under salt stress. Characterization of identified SAAPs showed the majority of proteins to act as carboxylase enzyme followed by phosphatase and phosphorylase. Carboxylation was observed most abundant reactions inside the plant system under HS followed by phosphorylation/dephosphorylation reaction (regulatory mechanism of key enzymes in different pathways). Hu et al. (2015) characterize the phosphoprotein and ABA-regulated phophoprotein in maize leaves under osmotic stress and observed 4052 phosphopeptides, corresponding to 3017 phosphoproteins through iTRAQ-based quantitative proteomic and highlighted the role of ABA in key carbon assimilatory pathways. We observed maximum (39%) proteins to be involved in photosynthesis which is in conformity with the observation of Xie et al. (2016) who reported 5570 proteins in tobacco leaves including 466 SAAPs to be associated with photosynthesis and stress response. The role of small ubiquitin-like modifier (SUMO) in regulation of transcriptome under stress was established using iTRAQ in Arabidopsis (Miller et al. 2013).
Here, we observed very high expression of storage proteins like oleosin, Globulin 3A, 3B, and gamma gliadin in contrasting wheat cvs. under HS. Ma et al. (2014) used the iTRAQ to characterize the proteome of wheat endospermic tissue and reported 1146 non-redundant proteins with 421 showing more than a two-fold increase in the relative abundance. Most of the identified SAAPs in present investigation were involved in starch biosynthesis, storage, and defense/stress-related showing higher accumulation during late grain development stages. Similarly, Zhang et al. (2017) identified 38 SAAPs associated with high night temperature (HNT) in rice and reported that HNT affects the redox equilibrium of plant cells and triggers the CDPK and COP9 signalosome involved in modulating the tolerance of the plant under HNT.
We observed SAAPs involved in carbon assimilatory pathway to be most altered under HS in both the contrasting wheat cultivars. Guo et al. (2017) identified 58 SAAPs in leaves of Potentilla fruticosa under HS and reported severe destruction of cell membrane and thylakoids affecting the photosynthesis of the plants. Similarly, Gong et al. (2017) identified key proteins associated with cadmium toxicity and NO-induced tolerance in cucumber using iTRAQ and reported NO-induced Ca2+ signaling transduction as most important mechanism of Cd detoxification. Zhang et al. (2017) identified 256 SAAPs in wheat cv. Jing411 under HS using iTRAQ. Most of the proteins identified were associated with growth and metabolism, energy metabolism, defense-related, signal transduction, etc. Chen et al. (2018) characterize the mechanism underlying dehydration of wheat seeds during middle and late stages using iTRAQ and reported trehalose-6-phosphate synthase 7 as key enzymes associated with dehydration tolerance.
We observed very high fold increase in the expression of SAAPs involved in metabolic processes like glutamine metabolism, protein metabolism, starch metabolism, carbohydrate metabolism, etc. Ding et al. (2017) identified 120 SAAPs in response to NaHS under drought stress in seedling of wheat using iTRAQ. Most of the proteins were observed to be related to carbohydrate metabolism, photosynthesis, signal transduction, etc. Present investigation showed most of the SAAPs to be localized in chloroplast involved in carbon assimilatory process. Du et al. (2018) identified 3872 protein in terminal buds of alfalfa using iTRAQ and reported key proteins associated with the growth of dormant and non-dormant alfalfa. Similarly, iTRAQ was used to identify 167 and 172 SAAPs and phosphorylated sites in Arabidopsis under osmotic stress, and further, H(+)-ATPase 2 and cysteine-rich repeat secretory protein 38 were observed as novel regulators of shoot growth under osmotic stress (Nikonorova et al. 2018). We observed upregulation of signaling and transport proteins like protein kinase, transmembrane protein, calmodulin, etc. in contrasting wheat cvs. under HS. Similarly, Vu et al. (2018) first time reported the identification of large number of a phospoprotein in leaf and spikelet of wheat under HS using iTRAQ and showed the conserved regulatory mechanisms underlying thermotolerance. There is a need to further explore the phosphoproteome and cys-proteome (the thiol moiety of cysteine acts as switch regulating the biological structure and their reactivity by undergoing diverse modifications in response to environment) of wheat under HS in order to unravel the interaction of exposomes with SAAPs—the information generated will help us to understand the tolerance mechanism and use it for the development of ‘climate resilient’ wheat.
Conclusions
Here, we have identified ~ 4272 SAAPs in wheat using iTRAQ. Some of the differentially abundant SAAPs identified are RuBisCo, Rca, OEEP, HSP17, SOD, CAT, and CDPK. Pathway analysis showed carbon assimilatory pathway followed by starch metabolism to be most perturbed under HS. Most of the SAAPs were localized in chloroplast. We observed abundance of defense/stress-related SAAPs, whereas metabolic pathway-associated enzymes showed decrease in the activities under HS. We identified few unique SAAPs like Fe/Zn-SOD, gamma gliadin, and peroxidase showing upregulation in thermotolerant cv. and downregulation in thermosusceptible cultivar. A positive correlation was established between the expression of SAAPs at transcript and protein level in wheat under HS. An increase in the isoforms of catabolic enzyme was observed, as compared to anabolic enzymes in developing wheat under HS. The identified differentially abundant SAAPs will enrich the proteome resources of wheat available on public domain and can be used in protein targeted precision breeding program for the development of ‘climate-smart’ crop.
References
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. https://doi.org/10.1016/0003-2697(76)90527-3
Budak H, Akpinar BA, Unver T, Turktas M (2013) Proteome changes in wild and modern wheat leaves upon drought stress by two-dimensional electrophoresis and nanoLC-ESI-MS/MS. Plant Mol Biol 83(1–2):89–103. https://doi.org/10.1007/s11103-013-0024-5
Canovas FM, Dumas-Gaudot E, Recorbet G, Jorrin J, Mock HP, Rossignol M (2004) Plant proteome analysis. Proteomics 4(2):285–298
Chance B, Maehly AC (1955) Assay of catalases and peroxidases. Methods Enzymol 2:773–775
Chen L, Wang Z, Li M, Ma X, Tian E, Sun A, Yin Y (2018) Analysis of the natural dehydration mechanism during middle and late stages of wheat seeds development by some physiological traits and iTRAQ-based proteomic. J Cereal Sci 80:102–110. https://doi.org/10.1016/J.JCS.2017.12.015
Chevalier F (2010) Highlights on the capacities of “gel-based” proteomics. Proteome Sci 8:23. https://doi.org/10.1186/1477-5956-8-23
Ding H, Han Q, Ma D, Hou J, Huang X, Wang C, Xie Y, Kang G, Guo T (2017) Characterizing physiological and proteomic analysis of the action of H2S to mitigate drought stress in young seedling of wheat. Plant Mol Biol Report 36:1–13. https://doi.org/10.1007/s11105-017-1055-x
Du H, Shi Y, Li D et al (2018) Proteomics reveals key proteins participating in growth difference between fall dormant and non-dormant alfalfa in terminal buds. J Proteome 173:126–138. https://doi.org/10.1016/j.jprot.2017.11.029
Farooq M, Bramley H, Palta JA, Siddique KHM (2011) Heat stress in wheat during reproductive and grain-filling phases. CRC Crit Rev Plant Sci 30:491–507
Feller U, Vaseva II (2014) Extreme climatic events: impacts of drought and high temperature on physiological processes in agronomically important plants. Front Environ Sci 2:1–17. https://doi.org/10.3389/fenvs.2014.00039
Finnie C (2007) Plant proteomics. Wiley-Blackwell publishing ltd, pp 1–253. https://doi.org/10.1002/9780470988879
Ge P, Hao P, Cao M, Guo G, Lv D, Subburaj S, Li X, Yan X, Xiao J, Ma W, Yan Y (2013) iTRAQ-based quantitative proteomic analysis reveals new metabolic pathways of wheat seedling growth under hydrogen peroxide stress. Proteomics 13(20):3046–3058. https://doi.org/10.1002/pmic.201300042
Giannopolitis CN, Ries SK (1977) Superoxide Dismutases: I. occurrence in higher plants. Plant Physiol 59(2):309–314. https://doi.org/10.1104/pp.59.2.309
Gong B, Nie W, Yan Y, Gao Z, Shi Q (2017) Unravelling cadmium toxicity and nitric oxide induced tolerance in Cucumis sativus: insight into regulatory mechanisms using proteomics. J Hazard Mater 336:202–213. https://doi.org/10.1016/j.jhazmat.2017.04.058
Guo Y, Wang Z, Guan X, Hu Z, Zhang Z, Zheng J, Lu Y (2017) Proteomic analysis of Potentilla fruticosa L leaves by iTRAQ reveals responses to heat stress. PLoS One 12(8):e0182917. https://doi.org/10.1371/journal.pone.0182917
Hu X, Li N, Wu L, Li C, Li C, Zhang L, Liu T, Wang W (2015) Quantitative iTRAQ-based proteomic analysis of phosphoproteins and ABA-regulated phosphoproteins in maize leaves under osmotic stress. Sci Rep 5:15626. https://doi.org/10.1038/srep15626
Jeandroz S, Lamotte O (2017) Editorial: plant responses to biotic and abiotic stresses: lessons from cell signaling. Front Plant Sci 8:1772. https://doi.org/10.3389/fpls.2017.01772
Ji W, Cong R, Li S, Li R, Qin Z, Li Y, Zhou X, Chen S, Li J (2016) Comparative proteomic analysis of soybean leaves and roots by iTRAQ provides insights into response mechanisms to short-term salt stress. Front Plant Sci 7:573. https://doi.org/10.3389/fpls.2016.00573
Kleczkowski LA, Villand P, Preiss J, Olsen OA (1993) Kinetic mechanism and regulation of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves. J Biol Chem 268:228–6233
Kosová K, Vitamvas P, Prasil IT, Renaut J (2011) Plant proteome changes under abiotic stress - contribution of proteomics studies to understanding plant stress response. J Proteome 74:1301–1322
Kumar RR, Rai RD (2014) Can wheat beat the heat: understanding the mechanism of thermotolerance in wheat (Triticum aestivum L.). Cereal Res Commun 42:1–18
Kumar RR, Sharma SK, Gadpayle KA et al (2012) Mechanism of action of hydrogen peroxide in wheat thermotolerance-interaction between antioxidant isoenzymes, proline and cell membrane. Afr J Biotechnol 11:14368
Kumar RR, Sharma SK, Goswami S et al (2013) Characterization of differentially expressed stress-associated proteins in starch granule development under heat stress in wheat (Triticum aestivum L.). Indian J Biochem Biophys 50(2):126–138
Kumar RR, Goswami S, Gadpayle KA et al (2014) Ascorbic acid at pre-anthesis modulate the thermotolerance level of wheat (Triticum aestivum) pollen under heat stress. J Plant Biochem Biotechnol 23:293–306
Kumar RR, Pathak H, Sharma SK, Kala YK, Nirjal MK, Singh GP, Goswami S, Rai RD (2015) Novel and conserved heat-responsive microRNAs in wheat (Triticum aestivum L.). Funct Integr Genomics 15:323–348
Kumar RR, Goswami S, Singh K, Dubey K, Singh S, Sharma R, Verma N, Kala YK, Rai GK, Grover M, Mishra DC, Singh B, Pathak H, Chinnusamy V, Rai A, Praveen S (2016) Identification of putative RuBisCo Activase (TaRca1)—the catalytic chaperone regulating carbon assimilatory pathway in wheat (Triticum aestivum) under the heat stress. Front Plant Sci 7:986. https://doi.org/10.3389/fpls.2016.00986
Kumar RR, Goswami S, Shamim M, Mishra U, Jain M, Singh K, Singh JP, Dubey K, Singh S, Rai GK, Singh GP, Pathak H, Chinnusamy V, Praveen S (2017) Biochemical defense response: characterizing the plasticity of source and sink in spring wheat under terminal heat stress. Front Plant Sci 8:1630. https://doi.org/10.3389/fpls.2017.01603
Kumar RR, Goswami S, Singh K, Dubey K, Rai GK, Singh B, Singh S, Grover M, Mishra D, Kumar S, Bakshi S, Rai A, Pathak H, Chinnusamy V, Praveen S (2018) Characterization of novel heat-responsive transcription factor (TaHSFA6e) gene involved in regulation of heat shock proteins (HSPs) — a key member of heat stress-tolerance network of wheat. J Biotechnol 279:1–12. https://doi.org/10.1016/j.jbiotec.2018.05.008
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Large EC (1954) Growth stages in cereals illustration of the Feekes scale. Plant Pathol 3(4):128–129. https://doi.org/10.1111/j.1365-3059.1954.tb00716.x
Liu B, Liu L, Tian L, Cao W, Zhu Y, Asseng S (2014) Post-heading heat stress and yield impact in winter wheat of China. Glob Chang Biol 20:372–381
Ma C, Zhou J, Chen G, Bian Y, Lv D, Li X, Wang Z, Yan Y (2014) iTRAQ-based quantitative proteome and phosphoprotein characterization reveals the central metabolism changes involved in wheat grain development. BMC Genomics 15(1):1029. https://doi.org/10.1186/1471-2164-15-1029
Miller MJ, Scalf M, Rytz TC, Hubler SL, Smith LM, Vierstra RD (2013) Quantitative proteomics reveals factors regulating RNA biology as dynamic targets of stress-induced SUMOylation in Arabidopsis. Mol Cell Proteomics 12(2):449–463. https://doi.org/10.1074/mcp.M112.025056
Nakamura Y, Yuki K, Park SY, Ohya T (1989) Carbohydrate metabolism in the developing endosperm of rice grains. Plant Cell Physiol 40:1–61. https://doi.org/10.1093/oxfordjournals.pcp.a077813
Nanjo Y, Nakamura T, Komatsu S (2013) Identification of indicator proteins associated with flooding injury in soybean seedlings using label-free quantitative proteomics. J Proteome Res 12(11):4785–4798. https://doi.org/10.1021/pr4002349
Nikonorova N, Van den Broeck L, Zhu S, et al. (2018) Early mannitol-triggered changes in the Arabidopsis leaf (phospho)proteome. J Exp Bot 69(19):4591–4607. https://doi.org/10.1101/264259
Patterson J, Ford K, Cassin A, Natera S, Bacic A (2007) Increased abundance of proteins involved in phytosiderophore production in boron-tolerant barley. Plant Physiol 144:1612–1631
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45–e445. https://doi.org/10.1093/nar/29.9.e45
Prasad PVV, Staggenborg SA, Ristic Z (2008) Impacts of drought and/or heat stress on physiological, developmental, growth, and yield processes of crop plants. In: Ahuja LR, Reddy VR, Saseendran SA, Yu Q (eds) Response of crops to limited water: understanding and modeling water stress effects on plant growth processes, Adv. Agric. Syst. Model., vol 1. ASA, CSSA, SSSA, Madison, WI, pp 301–355. https://doi.org/10.2134/advagricsystmodel1.c11
Qin D, Wu H, Peng H, Yao Y, Ni Z, Li Z, Zhou C, Sun Q (2008) Heat stress-responsive transcriptome analysis in heat susceptible and tolerant wheat (Triticum aestivum L.) by using wheat genome Array. BMC Genomics 9:432
Rodziewicz P, Swarcewicz B, Chmielewska K, Wojakowska A, Stobiecki M (2014) Influence of abiotic stresses on plant proteome and metabolome changes. Acta Physiol Plant 36:1–19
Vu LD, Zhu T, Verstraeten I, et al (2018) Temperature-induced changes in wheat phosphoproteome reveal temperature-regulated interconversion of phosphoforms. J Exp Bot 69(19):4609–4624. https://doi.org/10.1101/261065
Wang D, Xie SZ, Yang J, Wang QF (2014) Molecular characteristics and expression patterns of Rubisco activase, novel alternative splicing variants in a heterophyllous aquatic plant, Sagittaria graminea. Photosynthetica 52:83–95. https://doi.org/10.1007/s11099-014-0013-1
Wiese S, Reidegeld KA, Meyer HE, Warscheid B (2007) Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics 7(3):340–350. https://doi.org/10.1002/pmic.200600422
Xie H, Yang D-H, Yao H, Bai G, Zhang YH, Xiao BG (2016) iTRAQ-based quantitative proteomic analysis reveals proteomic changes in leaves of cultivated tobacco (Nicotiana tabacum) in response to drought stress. Biochem Biophys Res Commun 469(3):768–775. https://doi.org/10.1016/j.bbrc.2015.11.133
Zhang B, VerBerkmoes NC, Langston MA et al (2006) Detecting differential and correlated protein expression in label-free shotgun proteomics. J Proteome Res 5(11):2909–2918. https://doi.org/10.1021/pr0600273
Zhang Y, Pan J, Huang X et al (2017) Differential effects of a post-anthesis heat stress on wheat (Triticum aestivum L.) grain proteome determined by iTRAQ. Sci Rep 7(1):3468. https://doi.org/10.1038/s41598-017-03860-0
Acknowledgments
This work was financially supported by the grant received from Indian Council of Agriculture Research (ICAR) under the National Innovation on Climate Resilient Agriculture (NICRA) project (Grant no. 12/115 TG 3079) and CABin project (21-56) TG3064.
Author information
Authors and Affiliations
Contributions
RRK SG SP: conceptualize and plan the experiment; KS SA MT: involved in sowing, HS treatment and sample collection; RRK SG GKR: involved in sample preparation and iTRAQ run; IS SK MG DM AR: involved in bioinformatics characterization of iTRAQ data; DM KS GPS: executed the complementation study of identified DEPs; RRK SG VC: involved in manuscript drafting; VC HP AR SP: involved in editing of manuscript for language.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Electronic supplementary material
ESM 1
(XLSX 860 kb)
Rights and permissions
About this article
Cite this article
Kumar, R.R., Singh, K., Ahuja, S. et al. Quantitative proteomic analysis reveals novel stress-associated active proteins (SAAPs) and pathways involved in modulating tolerance of wheat under terminal heat. Funct Integr Genomics 19, 329–348 (2019). https://doi.org/10.1007/s10142-018-0648-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-018-0648-2