Abstract
Our understanding of the pathogenesis of immune thrombocytopenia (ITP) remains limited due to the complexity and heterogeneity of the disease. Recently, we observed that bone marrow mesenchymal stem cells (MSCs) derived from ITP patients exhibited growth defects and functional abnormalities that might be involved in the breakdown of self-tolerance. However, the underlying mechanism remains unclear. In this study, we profiled the expression of both mRNAs and miRNAs by utilizing the microarray technique and deciphered the mechanism underlying the impairment of MSCs derived from ITP patients (MSC-ITP). In total, we identified 740 genes and 32 miRNAs that were differentially expressed between ITP patients and controls. A compromised unfolded protein response (UPR) and decreased DNA transcription were shown to be significantly related to MSC-ITP. The interaction of miRNA with mRNA suggested that the cellular stress response, the UPR, and DNA transcription may be involved in the defects observed in MSC-ITP. Key differentially expressed genes were further validated by RT-PCR. Our results highlight that defects in the cellular stress response, as shown by a compromised UPR and differential DNA transcription, play key roles in causing the abnormalities observed in MSC-ITP. These data might contribute to a better understanding of the abnormal bone marrow niche and provide new insights into the pathogenesis of ITP.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Immune thrombocytopenia (ITP) is an autoimmune disorder characterized by accelerated platelet destruction and decreased platelet production due to the breakdown of self-tolerance (Cines et al. 2014). Platelet and megakaryocyte dysfunction orchestrated by abnormalities of the adaptive immune system plays a crucial role in ITP pathogenesis (Semple et al. 2010).
Immune-mediated acceleration of platelet destruction and decrease in platelet production can be ameliorated by immunosuppressants or agents that prevent reticulo-endothelial system phagocytosis, such as IVIgG, corticosteroids, or calmodulin-dependent protein kinase antagonists. Splenectomy was thought to permanently resolve the issue (Provan et al. 2010). However, these standard therapeutics targeting the mainstream pathogenesis are not effective for every patient, and a proportion of patients suffer refractory or relapse course (Gernsheimer et al. 1989; Louwes et al. 2001). The differing outcomes indicate that ITP is a heterogeneous disease and that, in addition to peripheral antibody- or T cell-mediated platelet and megakaryocyte abnormalities, additional pathogeneses might be involved (Riviere et al. 2015).
Mesenchymal stem cells (MSCs) are multipotent cells that play an important role in immune tolerance (Trento and Dazzi 2010). Despite these cells’ direct immunosuppressive effects on adaptive immune system (Li and Hua 2017), they are capable of modulating T cell responses by promoting the tolerogenic properties of dendritic cells (DCs) (Ma et al. 2013).
Nevertheless, MSCs have been observed to function abnormally in patients with ITP. Several studies have shown that proliferation is impaired and the immunosuppressive capacity is compromised in MSCs derived from ITP patients (Perez-Simon et al. 2009; Zhang et al. 2014). In addition, the ability to induce tolerogencity in mature DCs was impaired in MSCs from ITP patients (Ma et al. 2013). In our previous study, we observed that MSCs from ITP patients displayed enhanced senescence and apoptosis along with an impaired ability to inhibit T cell proliferation, induce Tregs, and suppress anti-GPIIb-IIIa antibody production (Zhang et al. 2016). However, treatment with growth factors, which improve proliferation and ameliorate excess senescence and apoptosis, restored the immunosuppressive capacity of MSCs from ITP patients (Zhang et al. 2016). Taken together, these data suggested that impairment of MSCs could result in defective immunoregulative abilities, which might be involved in ITP pathogenesis by aggravating the breakdown of self-tolerance in ITP patients. Although the manifestation of senescence and apoptosis was observed in MSCs from ITP patients, the underlying molecular mechanism is not fully understood.
In this study, we used microarray analysis to report the mRNA and miRNA expression profiles of MSCs derived from newly diagnosed ITP patients and to identify the biological processes associated with the differentially expressed genes. These analyses might contribute to elucidating the underlying molecular mechanisms associated with the impairment of MSCs in ITP patients and provide new insights into the pathogenesis of ITP.
Results
Characterization of MSCs
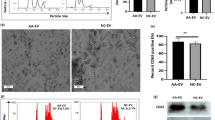
MSCs were successfully derived from ITP patients (MSC-ITP) or healthy individuals (MSC-control) as previously described. Prior to their application in the microarray, the cultured MSCs underwent characterization of surface markers and differentiation capacity according to the criteria of the International Society for Cellular Therapy. MSC-control expanded and acquired the typical spindle morphology during culture. In contrast, MSC-ITP expanded slower and appeared larger and flattened. Surface marker analysis showed that both MSC-control and MSC-ITP expressed CD105, CD73, and CD90 but not CD14, CD19, CD34, CD45, or HLA-DR (Fig. 1a). After induction, the MSCs could differentiate in vitro into osteoblasts, adipocytes, and chondroblasts, as shown in Fig. 1b.
Characterization of MSCs. a Surface antigens characterized by flow cytometry showed that MSC-control and MSC-ITP express CD73, CD90, and CD105 but not CD14, CD19, CD34, CD45, or HLA-DR. b MSC-control and MSC-ITP presented similar differentiation capacities toward osteoblast, adipocyte, and chondroblast lineages. Scale bar represents 100 μm
Overview of the gene expression profile and the function and disease analysis of MSC-ITP
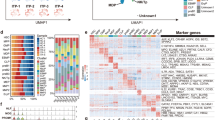
To uncover the molecular mechanisms involved in the defects observed in MSC-ITP, we first compared the overall genomic profiles of MSC-ITP and MSC-control. Bioinformatic analysis revealed that the expression of 740 genes was significantly changed between MSC-ITP and MSC-control (Fig. 2a). Two-dimensional hierarchical clustering of the 740 genes showed the degree of separation between the two groups (Fig. 2b). Next, we performed function and disease analysis. Differentially expressed genes were associated with 57 classes of biological processes. The top 20 enriched pathways are shown in Fig. 2c. Interestingly, the enrichment analysis revealed that the differentially expressed genes are most significantly involved in the processes of cell death and survival, cellular development, cell growth and proliferation and cell cycle (Fig. 2c). This finding is in agreement with findings from our previous study, which demonstrated excessive apoptosis and senescence in MSCs from ITP patients.
Compromised unfolded protein response (UPR) in MSC-ITP
To evaluate the pathways underlying cell signaling defects in MSC-ITP, we performed a canonical pathway analysis. Canonical pathway annotation enabled us to classify differentially expressed genes into 57 pathways. The top 8 enriched pathways included the UPR, mitotic roles of Polo-Like kinase, ERK5 signaling, cell cycle, G2/M DNA damage checkpoint regulation, cyclins and cell cycle regulation, endoplasmic reticulum (ER) stress pathway, PI3K/AKT signaling, and ATM signaling. These enriched pathways have been shown to be involved in cell proliferation, cell cycle, and cell apoptosis, which might underlie the differences between MSC-ITP and MSC-control. The top 20 enriched pathways are shown in Fig. 3a. Notably, the UPR, which is a defense response to protect cells from ER stress-induced damage (Carreras-Sureda et al. 2017; Yang et al. 2016), was identified as the most significantly changed pathway (−log(p value) = 8.32). Next, rigorous gene set enrichment analysis (GSEA) was performed to detect the correlation. GSEA identified subsets of 103 UPR-specific genes that were negatively correlated with MSC-ITP (Fig. 3b; p = 0.033, FDR q = 0.038). Taken together, these data suggested that a deficient UPR might play a key role in MSC-ITP.
Decreased DNA transcription and upstream regulators in MSC-ITP
Upstream analysis of the identified genes revealed significantly decreased transcription of DNA (Fig. 4a), which might account for the attenuated proliferation and cell cycle arrest of MSCs derived from ITP patients shown in our previous study. Further enrichment analysis identified a stress response gene, nuclear protein transcriptional regulator 1 (Nupr1). Nupr1 is a chromatin-binding protein that converts stress signals into a gene expression program and plays a central role in the pro-survival process of the ER stress pathway (Emma et al. 2016; Galichon et al. 2017). As shown in Fig. 4b, most Nupr1 target genes were negatively regulated, indicating that Nupr1 signaling was strongly inhibited in MSC-ITP. In addition, TGFβ-1 signaling was shown to be down-regulated (Fig. 4c). Previous studies have shown that both Nupr1 and TGFβ-1 signaling plays roles in ER stress and transcription, which corroborates our findings (Ozkaya et al. 2017; Tang et al. 2015). Therefore, our results indicate that decreased overall DNA transcription and a deficient UPR may play vital roles in the regulation of MSC-ITP and that these combined effects probably result in the observed defects in MSC-ITP.
Potential cross talk between the UPR and the p53 pathway
We utilized gene network analysis to target the key “cross-road” regulator in MSC-ITP (Fig. 5). Network analysis illustrated that JUN and CDKN1A are two key cross regulators with multiple roles in the biological process of MSC-ITP (score = 36). JUN is a critical downstream regulator that is activated upon initiation of the UPR and subsequently mediates a series of biological processes (Yang et al. 2016). CDKN1A is involved in the p53-mediated inhibition of cellular proliferation in response to DNA damage (Galluzzi et al. 2016). This network suggests potential cross talk between the UPR and the p53 pathway, with the latter having been verified in MSC-ITP in our previous study (Zhang et al. 2016).
miRNA-mRNA interactions are involved in the cellular stress response
miRNAs are small noncoding RNA molecules that regulate gene expression. To investigate the role of miRNAs in MSC-ITP, we detected 62 miRNAs that differed significantly between ITP patients and controls. The TargetScan and Miranda algorithms were used and Kolmogorov-Smirnov statistics were applied to evaluate if these were associated with changes in the mRNA expression of the target genes. This analysis identified 34 miRNAs that were significantly associated with mRNA expression in the database. To better understand the role of the regulated miRNAs, bioinformatics analysis was performed with predicted target genes. Reactome enrichment analysis indicated that the cellular response to stress, TGF-β receptor complex signaling, and genetic transcriptional pathways were significantly dysregulated in MSC-ITP (Fig. 6), which was in agreement with the implications of the mRNA microarray results.
Then, the predicted target genes of the regulated miRNAs were cross-referenced against mRNAs that were significantly different between MSC-ITP and MSC-control, resulting in 163 overlapped genes. Mapping all the overlapped genes to the GO term database enabled annotation of the 163 genes into cellular component, molecular function, and biological process categories (Fig. 7a). The UPR was again identified by reactome enrichment analysis as the process most affected by the differentially expressed genes between MSC-ITP and MSC-control (Fig. 7b), which further demonstrated the association between UPR and the defects observed in MSC-ITP.
Gene Ontology (GO) analysis (a) and reactome enrichment (b) analysis of the overlapped genes
Validation of differentially expressed mRNAs
To establish the validity of gene expression levels determined by microarray analysis, we performed RT-PCR to measure the expression levels of differentially expressed genes. This analysis included UPR-associated genes (GRP78, PERK, ATF6, and DDIT3), the upstream regulators Nupr1 and TGF-β1, and the cross regulators identified by network analysis (JUN, CDKN1A, DKK1, and CHEK1) (Fig. 8). Three key regulators of UPR, GRP78, PERK, and ATF6 were significantly down-regulated, which was consistent with the trend revealed by microarray. Similarly, the mRNA expression levels of DDIT3 and JUN, two crucial downstream molecules in the UPR, were significantly decreased. The mRNA expression levels of TGFβ-1 and Nupr1 were down-regulated according to RT-PCR. The network cross regulators JUN, CDKN1A, and DKK1 were also down-regulated while CHEK1 was up-regulated in MSC-ITP compared with MSC-control; this further validated the accuracy and the implications of the microarray results.
Discussion
In the present study, we performed genome-wide expression analyses of mRNA and miRNA in MSCs from ITP patients and healthy individuals. We identified cellular stress response defects in MSCs from ITP patients, as indicated by deficiencies in the UPR and DNA transcription in MSC-ITP. This finding revealed potential molecular mechanisms underlying the impairments observed in MSC-ITP and might explain the abnormalities in the bone marrow niche and the breakdown of self-tolerance in ITP patients.
ITP is an autoimmune disorder characterized by platelet destruction and impaired platelet production due to the breakdown of self-tolerance (Cines et al. 2014). Abnormal polarization between T helper (Th)1 cells and Th2 cells in the peripheral blood has been thought to play an important role in the pathogenesis of ITP (Semple et al. 2010). In addition, decreased levels and functional defects of Tregs are involved in the breakdown of self-tolerance in ITP (Liu et al. 2007).
Although several pathological mechanisms underlying the development of ITP have been well established and targeted treatments have been developed (Provan et al. 2010), numerous patients suffer relapse or refractory courses of ITP (Neunert et al. 2011), which lead to the question of whether there are alternative pathogeneses. However, due to the complexity of the immune system and heterogeneity of the disease, our understanding of ITP pathogenesis remains limited.
In our previous study, bone marrow MSCs, which are potent immunoregulators and are responsible for the maintenance of self-tolerance in the bone marrow niche, were found to be functionally abnormal in ITP patients (Zhang et al. 2016). MSCs have long been documented as important immunoregulators due to their robust immunosuppressive properties and their roles in regulating both adaptive and innate immune responses (Li and Hua 2017). MSCs from healthy individuals may exert immunosuppressive effects by either directly inhibiting the conversion of Th2 to Th1 and inducing the generation of Tregs (Li and Hua 2017) or indirectly modulating the T cell response by promoting the tolerogenic properties of DCs (Li et al. 2008; Spaggiari et al. 2009), thus contributing to the maintenance of self-tolerance.
Consistent with the manifestations observed in many other autoimmune diseases, MSCs from ITP patients exhibited loss of their conventional proliferative capacity and were defective in immunoregulation (Zhang et al. 2016). In contrast, in either animal models or ITP patients, transplantation of MSCs derived from healthy individuals partly restored megakaryocytic function and platelet production by reversing the shift in the Th1/Th2 cytokine balance (Fang et al. 2012; Ma et al. 2012; Xiao et al. 2012). These data suggested that MSC impairment might contribute to the breakdown of self-tolerance and be involved in the pathogenesis of ITP.
Although we demonstrated excessive apoptosis and senescence along with compromised immunosuppressive properties of MSCs derived from ITP patients (Zhang et al. 2016), the underlying molecular mechanism remains unclear.
Genome-wide measurement of disease-specific alterations is widely used to characterize the underlying molecular mechanisms, and this technique may help obtain a more comprehensive understanding of the disease. In the present study, we used microarrays to identify both the mRNA and miRNA expression profile changes in MSC-ITP and to unveil the intracellular mechanisms leading to defects. Differentially expressed genes were detected and analyzed in the subsequent bioinformatic analysis.
First, function and disease analysis was carried out to identify the global biological processes that differed between the two groups. The most significantly enriched biological processes were related to cell proliferation, death, and survival, which are in line with our previous experimental data demonstrating increased apoptosis, cell cycle arrest, and attenuated proliferation in MSC-ITP (Zhang et al. 2016). Additionally, microarray studies of MSCs derived from SLE patients showed a similar trend, with genes involved in cell cycle control presenting significant differences in their gene expression profiles (Tang et al. 2012). This finding suggested that the defect in cell cycle progression and proliferation of bone marrow MSCs might be a common manifestation in autoimmune disease.
Next, pathway and upstream analysis was utilized to further elucidate the intracellular mechanism underlying the defects. In these analyses, inhibition of the UPR and DNA transcription was shown to be significantly associated with MSC-ITP.
The UPR is a highly conserved defense system that protects cells from ER stress resulting from genetic or environmental insults (Grootjans et al. 2016). The UPR is critical for maintaining cell homeostasis under physiological and pathological conditions (Yang et al. 2016). To escape adverse cellular stress, the ER activates the stress sensor pathway, namely, the UPR, through a complex signaling network of PERK-eIF2a, IRE1-XBP1, and ATF6-CREBH transducers (Yang et al. 2016). This signaling network initiates changes in the expression of hundreds of genes to alleviate stress-induced deregulation and damage and to restore cellular homeostasis (Carreras-Sureda et al. 2017). Inability to respond to cellular stress would result in the accumulation of cell damage and in irreversible consequences (Galluzzi et al. 2016). Given these indications, our data highlighted that a deficient UPR might lead to an inability to initiate defenses against ER stress and thus contribute to cell impairment.
Additional evidence of deficient cellular stress responses in MSC-ITP was the decreased level of overall DNA transcription identified by upstream analysis. Cells are exposed to diverse stresses. Cells react to potential perturbations of the intracellular or extracellular microenvironment by activating rapid mechanisms or delayed and robust adaptive systems to cope with stress and to attempt to restore homeostasis; alternatively, cells may actively engage in cellular suicide (Galluzzi et al. 2016). Among the defense systems, transcription factors involved in the reprogramming of gene expression are highlighted as an essential mechanism for cellular stress responses (Ljungman 2007). Numerous lines of evidence show that transcription factors allow cells to coordinate unified responses to transmitted signals (Staby et al. 2017). These responses modulate pathway-specific gene expression and organize transcriptomic responses to stresses to maintain homeostasis (Alasiri et al. 2017; Sykiotis and Bohmann 2010). MSC-ITP exhibited attenuated global transcription levels, and the expression levels of many stress response-related transcriptional regulators, such as Nupr1 and TGF-β1, were down-regulated. This finding also indicated that MSC-ITP exhibited an inability to respond to cellular stress and restore cellular homeostasis, which may contribute to cellular impairment or death. Overall, the compromised UPR and decreased DNA transcription identified by mRNA profile changes in MSC-ITP implied a defective defense response to cellular stress.
For further validation of our mRNA findings, we simultaneously detected the expression profiles of miRNAs. miRNAs are small noncoding RNA molecules that regulate gene expression. The effect of miRNA on mRNA is mediated through the binding of the miRNA to the ribonucleoprotein complex RNA-induced silencing complex that also binds to the 3’ untranslated region of complementary mRNAs (Schwarz et al. 2003). The double-stranded complex between the miRNA and mRNA is later degraded, which leads to decreased protein translation (Meister and Tuschl 2004). We then identified the potential target genes of the differentially expressed miRNAs by reactome enrichment analysis. Consistent with the results of the mRNA expression analysis, the cellular response to stress and transcriptional pathways were among the most differentially regulated pathways, thus confirming the implications of the mRNA profiles. Analysis with the cross-referenced genes further validated the association between a dysregulated UPR and MSC-ITP. Suppression of the UPR was also detected by RT-PCR, which confirmed the accuracy of the microarray results.
Taken together, by integrating the mRNA and miRNA profiles, we demonstrated a defective cellular stress response in MSC-ITP. These observations may help elucidate the mechanism underlying the defects observed in MSC-ITP and might provide new insights in the pathogenesis of ITP.
Materials and methods
Patients
BM samples were obtained from four patients with newly diagnosed ITP who met the previously reported criteria (Provan et al. 2010), and four age- and sex-matched healthy donors were included as normal controls. The body mass index distribution was similar between the ITP patients and the healthy donors (control). All the patients and controls provided consent to participate in the study, which was approved by the Ethics Committee of the Peking University People’s Hospital and conducted in accordance with the Declaration of Helsinki.
Isolation, expansion, and characterization of MSCs
Bone marrow mononuclear cells from the patients and normal controls were isolated by Ficoll gradient and cultured in Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific Life Sciences, Waltham, MA, http://www.thermofisher.com), with 10% defined fetal bovine serum (Thermo Fisher Scientific Life Sciences) and 100 U/ml penicillin/streptomycin. The cultures were maintained at 37 °C in a 5% CO2 incubator, and the medium containing nonadherent cells was replaced every 3–4 days of the culture period. When the cultures reached 80% confluence, the cells were detached using 0.25% trypsin-EDTA. The cells were seeded in flasks at 1 × 106 cells per 25 cm2 and cultured for another 4–5 days to obtain the next passage of MSCs.
To confirm the human MSCs phenotype, plastic adherent cells were analyzed for the expression of surface-specific antigens using flow cytometry. The cells were stained with the following fluorescein isothiocyanate (FITC)-conjugated, allophycocyanin (APC)-conjugated, peridinin chlorophyll protein (PerCP)-conjugated, or phycoerythrin (PE)-conjugated monoclonal antibodies: CD14, CD19, CD34, CD45, CD105, CD90, CD73, and human leukocyte antigen (HLA)-DR. The FITC-, PE-, APC-, and PerCP-conjugated isotypes were used as negative controls. The capacity of the MSCs to differentiate along osteogenic, chondrogenic, and adipogenic lineages was assessed, as described previously (Dominici et al. 2006), using commercially available kits (osteogenesis differentiation kit, chondrogenesis differentiation kit, and adipogenesis differentiation kit (Thermo Fisher Scientific Life Sciences)), according to the manufacturer’s instructions.
RNA extraction and microarray
MSCs were placed in Trizol (Invitrogen, USA) and processed for RNA extraction using the RNeasy kit according to the manufacturer’s instructions (Qiagen, Valencia, CA). The integrity of the RNA was assessed using denaturing RNA agarose gel electrophoresis. The quantity of the RNA samples was assessed by absorbance spectrometry using a NanoDrop 2000 (Thermo, Waltham, MA). For the miRNA microarray experiment, total RNA was purified using the mirVana™ miRNA Isolation Kit (CapitalBio Corp, Beijing, China) and labeled with polyA tailing and biotin. The labeled RNA was hybridized with Affymetrix miRNA microarray (CapitalBio Corp, Beijing, China). For mRNA microarray experiments, total RNA was purified and subjected to first-strand cDNA and second-strand cDNA synthesis. cDNA was generated and labeled with biotin and then fragmented to a suitable size. Hybridization was performed with Affymetrix mRNA microarray (CapitalBio Corp, Beijing, China) according to the manufacturer’s instructions. After washing and staining, arrays were scanned and the imaging data were extracted with Affymetrix GeneChip Command Console Software (CapitalBio Corp, Beijing, China). The significantly changed genes were selected based on p value < 0.05 and > 2-fold as criteria.
Ingenuity pathway analysis (IPA)
To assess biological relationships among genes, we used the Ingenuity Pathway Analysis software (IPA, Ingenuity System, Redwood City, CA, USA; http://www.ingenuity. com). The pathway, upstream, and network analysis was performed using IPA. The canonical pathways generated by IPA are the most significant for the uploaded data set. Fischer’s exact test with FDR option was used to calculate the significance of the canonical pathway. IPA computes a score for each network according to the fit of the set of supplied focus genes. These scores indicate the likelihood of focus genes to belong to a network versus those obtained by chance. A score > 2 indicates a ≤ 99% confidence that a focus gene network was not generated by chance alone.
Gene set enrichment analysis (GSEA)
Normalized RNA-seq expression data were preranked based on the fold change between the two groups. The Hallmark curated Gene sets in MSigDB database 5.0 were used for GSEA analysis. GSEA3.0 was used to perform the analysis. Gene sets were tested for enrichment in rank ordered lists via GSEA using a classic statistics and compared to enrichment results from 1000 random permutations of the gene set to obtain p values. A corrected p value was obtained from the analysis using the FDR q value correction. On the basis of this correction, the cutoff for significance was established at a p value < 0.05.
MiRNA analysis
The mirBase (http://www.mirbase.org) was used to identify miRNA functions and miRNA target mRNA using TargetScan and Miranda algorithms. Target genes from the miRNA were then cross-referenced against the list of significantly regulated mRNA between ITP patients and controls.
Quantitative RT-PCR analysis
Total RNA isolated from MSCs derived five ITP patients and five healthy individuals were reverse transcribed to cDNA. Gene expressions were examined in triplicate by real-time RT-PCR performed by ABI 7500 real-time PCR detection system (Applied Biosystems, USA) using SYBR Green detection mix (TaKaRa, Japan). The expressions of GRP78, PERK, ATF6, DDIT3, NUPR1, TGF-β1, JUN, CDKN1A, DKK1, and CHEK1 were analyzed. The 2-ΔΔCt method was used to analyze the relative quantification data. GAPDH was used as the internal control gene. Differences between two groups were estimated by Student’s t test and p < 0.05 was considered significant.
Abbreviations
- ITP:
-
immune thrombocytopenia
- MSCs:
-
mesenchymal stem cells
- UPR:
-
unfolded protein response
- ER:
-
endoplasmic reticulum
- GSEA:
-
gene set enrichment analysis
- GO:
- DCs:
-
dendritic cells
References
Alasiri G, Fan LY, Zona S, Goldsbrough IG, Ke HL, Auner HW, Lam EW (2017) ER stress and cancer: the FOXO forkhead transcription factor link. Mol Cell Endocrinol. https://doi.org/10.1016/j.mce.2017.05.027
Carreras-Sureda A, Pihan P, Hetz C (2017) The unfolded protein response: at the intersection between endoplasmic reticulum function and mitochondrial bioenergetics. Front Oncol 7:55
Cines DB, Cuker A, Semple JW (2014) Pathogenesis of immune thrombocytopenia. Presse Med 43(4):e49–e59. https://doi.org/10.1016/j.lpm.2014.01.010
Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop D, Horwitz E (2006) Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 8(4):315–317. https://doi.org/10.1080/14653240600855905
Emma MR, Iovanna JL, Bachvarov D, Puleio R, Loria GR, Augello G, Candido S, Libra M, Gulino A, Cancila V, McCubrey JA, Montalto G, Cervello M (2016) NUPR1, a new target in liver cancer: implication in controlling cell growth, migration, invasion and sorafenib resistance. Cell Death Dis 7(6):e2269. https://doi.org/10.1038/cddis.2016.175
Fang B, Mai L, Li N, Song Y (2012) Favorable response of chronic refractory immune thrombocytopenic purpura to mesenchymal stem cells. Stem Cells Dev 21(3):497–502. https://doi.org/10.1089/scd.2011.0231
Galichon P, Bataille A, Vandermeersch S, Wetzstein M, Xu-Dubois YC, Legouis D, Hertig A, Buob D, Placier S, Bige N, Lefevre G, Jouanneau C, Martin C, Iovanna JL, Rondeau E (2017) Stress response gene Nupr1 alleviates cyclosporin A nephrotoxicity in vivo. J Am Soc Nephrol 28(2):545–556. https://doi.org/10.1681/ASN.2015080936
Galluzzi L, Bravo-San Pedro JM, Kepp O, Kroemer G (2016) Regulated cell death and adaptive stress responses. Cell Mol Life Sci 73(11-12):2405–2410. https://doi.org/10.1007/s00018-016-2209-y
Gernsheimer T, Stratton J, Ballem PJ, Slichter SJ (1989) Mechanisms of response to treatment in autoimmune thrombocytopenic purpura. N Engl J Med 320(15):974–980. https://doi.org/10.1056/NEJM198904133201505
Grootjans J, Kaser A, Kaufman RJ, Blumberg RS (2016) The unfolded protein response in immunity and inflammation. Nat Rev Immunol 16(8):469–484. https://doi.org/10.1038/nri.2016.62
Li N, Hua J (2017) Interactions between mesenchymal stem cells and the immune system. Cell Mol Life Sci 74(13):2345–2360. https://doi.org/10.1007/s00018-017-2473-5
Li YP, Paczesny S, Lauret E, Poirault S, Bordigoni P, Mekhloufi F, Hequet O, Bertrand Y, Ou-Yang JP, Stoltz JF, Miossec P, Eljaafari A (2008) Human mesenchymal stem cells license adult CD34+ hemopoietic progenitor cells to differentiate into regulatory dendritic cells through activation of the Notch pathway. J Immunol 180(3):1598–1608. https://doi.org/10.4049/jimmunol.180.3.1598
Liu B, Zhao H, Poon MC, Han Z, Gu D, Xu M, Jia H, Yang R, Han ZC (2007) Abnormality of CD4(+)CD25(+) regulatory T cells in idiopathic thrombocytopenic purpura. Eur J Haematol 78(2):139–143. https://doi.org/10.1111/j.1600-0609.2006.00780.x
Ljungman M (2007) The transcription stress response. Cell Cycle 6(18):2252–2257. https://doi.org/10.4161/cc.6.18.4751
Louwes H, Vellenga E, Houwerzijl EJ, de Wolf JT (2001) Effects of prednisone and splenectomy in patients with idiopathic thrombocytopenic purpura: only splenectomy induces a complete remission. Ann Hematol 80(12):728–732. https://doi.org/10.1007/s002770100375
Ma L, Zhou Z, Zhang D, Yang S, Wang J, Xue F, Yang Y, Yang R (2012) Immunosuppressive function of mesenchymal stem cells from human umbilical cord matrix in immune thrombocytopenia patients. Thromb Haemost 107(5):937–950. https://doi.org/10.1160/TH11-08-0596
Ma J, Ning YN, Xu M, Hou Y, Wang N, Hou XY, Yu YY, Li H, He WD, Shao LL, Zhou H, Min YN, Liu XG, Shi Y, Qin P, Guo CS, Hou M, Peng J (2013) Thalidomide corrects impaired mesenchymal stem cell function in inducing tolerogenic DCs in patients with immune thrombocytopenia. Blood 122(12):2074–2082. https://doi.org/10.1182/blood-2013-03-491555
Meister G, Tuschl T (2004) Mechanisms of gene silencing by double-stranded RNA. Nature 431(7006):343–349. https://doi.org/10.1038/nature02873
Neunert, C., Lim, W., Crowther, M., Cohen, A., Solberg, L., Jr. & Crowther, M.A. (2011) The American Society of Hematology 2011 evidence-based practice guideline for immune thrombocytopenia. Blood, 117, 4190–4207, 16, DOI: https://doi.org/10.1182/blood-2010-08-302984
Ozkaya AB, Ak H, Aydin HH (2017) High concentration calcitriol induces endoplasmic reticulum stress related gene profile in breast cancer cells. Biochem Cell Biol 95(2):289–294. https://doi.org/10.1139/bcb-2016-0037
Perez-Simon JA, Tabera S, Sarasquete ME, Diez-Campelo M, Canchado J, Sanchez-Abarca LI, Blanco B, Alberca I, Herrero-Sanchez C, Canizo C, San Miguel JF (2009) Mesenchymal stem cells are functionally abnormal in patients with immune thrombocytopenic purpura. Cytotherapy 11(6):698–705. https://doi.org/10.3109/14653240903051558
Provan D, Stasi R, Newland AC, Blanchette VS, Bolton-Maggs P, Bussel JB, Chong BH, Cines DB, Gernsheimer TB, Godeau B, Grainger J, Greer I, Hunt BJ, Imbach PA, Lyons G, McMillan R, Rodeghiero F, Sanz MA, Tarantino M, Watson S, Young J, Kuter DJ (2010) International consensus report on the investigation and management of primary immune thrombocytopenia. Blood 115(2):168–186. https://doi.org/10.1182/blood-2009-06-225565
Riviere E, Viallard JF, Guy A, Kilani B, Vieira-Dias J, Pons AC, Couffinhal T, Pellegrin JL, James C (2015) Intrinsically impaired platelet production in some patients with persistent or chronic immune thrombocytopenia. Br J Haematol 170(3):408–415. https://doi.org/10.1111/bjh.13444
Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD (2003) Asymmetry in the assembly of the RNAi enzyme complex. Cell 115(2):199–208. https://doi.org/10.1016/S0092-8674(03)00759-1
Semple JW, Provan D, Garvey MB, Freedman J (2010) Recent progress in understanding the pathogenesis of immune thrombocytopenia. Curr Opin Hematol 17(6):590–595. https://doi.org/10.1097/MOH.0b013e32833eaef3
Spaggiari GM, Abdelrazik H, Becchetti F, Moretta L (2009) MSCs inhibit monocyte-derived DC maturation and function by selectively interfering with the generation of immature DCs: central role of MSC-derived prostaglandin E2. Blood 113(26):6576–6583. https://doi.org/10.1182/blood-2009-02-203943
Staby L, O'Shea C, Willemoes M, Theisen F, Kragelund BB, Skriver K (2017) Eukaryotic transcription factors: paradigms of protein intrinsic disorder. Biochem J 474(15):2509–2532. https://doi.org/10.1042/BCJ20160631
Sykiotis GP, Bohmann D (2010) Stress-activated cap'n'collar transcription factors in aging and human disease. Sci Signal 3:re3
Tang, Y., Ma, X., Zhang, H., Gu, Z., Hou, Y., Gilkeson, G.S., Lu, L., Zeng, X. & Sun, L. (2012) Gene expression profile reveals abnormalities of multiple signaling pathways in mesenchymal stem cell derived from patients with systemic lupus erythematosus. Clin Dev Immunol, 2012, 826182, 1, 12, DOI: https://doi.org/10.1155/2012/826182
Tang B, Li Q, Zhao XH, Wang HG, Li N, Fang Y, Wang K, Jia YP, Zhu P, Gu J, Li JX, Jiao YJ, Tong WD, Wang M, Zou QM, Zhu FC, Mao XH (2015) Shiga toxins induce autophagic cell death in intestinal epithelial cells via the endoplasmic reticulum stress pathway. Autophagy 11(2):344–354. https://doi.org/10.1080/15548627.2015.1023682
Trento C, Dazzi F (2010) Mesenchymal stem cells and innate tolerance: biology and clinical applications. Swiss Med Wkly 140:w13121
Xiao J, Zhang C, Zhang Y, Zhang X, Zhao J, Liang J, Zhong X, Chen Y (2012) Transplantation of adipose-derived mesenchymal stem cells into a murine model of passive chronic immune thrombocytopenia. Transfusion 52(12):2551–2558. https://doi.org/10.1111/j.1537-2995.2012.03642.x
Yang Y, Cheung HH, Tu J, Miu KK, Chan WY (2016) New insights into the unfolded protein response in stem cells. Oncotarget 7(33):54010–54027. https://doi.org/10.18632/oncotarget.9833
Zhang D, Li H, Ma L, Zhang X, Xue F, Zhou Z, Chi Y, Liu X, Huang Y, Yang Y, Yang R (2014) The defective bone marrow-derived mesenchymal stem cells in patients with chronic immune thrombocytopenia. Autoimmunity 47(8):519–529. https://doi.org/10.3109/08916934.2014.938320
Zhang JM, Feng FE, Wang QM, Zhu XL, Fu HX, Xu LP, Liu KY, Huang XJ, Zhang XH (2016) Platelet-derived growth factor-BB protects mesenchymal stem cells (MSCs) derived from immune thrombocytopenia patients against apoptosis and senescence and maintains MSC-mediated immunosuppression. Stem Cells Transl Med 5(12):1631–1643. https://doi.org/10.5966/sctm.2015-0360
Funding
This work was supported by National Natural Science Foundation of China (No. 81470343 and No. 81670116), Beijing Natural Science Foundation (No. 7171013), Beijing Municipal Science and Technology Commission (No. Z171100001017084), and The National Key Research and Development Progran of China (No. 2017YFA0105503).
Author information
Authors and Affiliations
Contributions
Jia-Min Zhang and Xiao-Hui Zhang conceived and designed the study; Xiao-Lu Zhu and Jing Xue performed the experiments; Ying-Jun Chang analyzed the data; Jia-Min Zhang wrote the paper; X. Long Zheng, Kai-Yan Liu, and Xiao-Jun Huang oversaw the work and critically revised the manuscript; Xiao-Hui Zhang attracted funding.
Corresponding author
Ethics declarations
All the patients and controls provided consent to participate in the study, which was approved by the Ethics Committee of the Peking University People’s Hospital and conducted in accordance with the Declaration of Helsinki.
Conflicts of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Zhang, JM., Zhu, XL., Xue, J. et al. Integrated mRNA and miRNA profiling revealed deregulation of cellular stress response in bone marrow mesenchymal stem cells derived from patients with immune thrombocytopenia. Funct Integr Genomics 18, 287–299 (2018). https://doi.org/10.1007/s10142-018-0591-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-018-0591-2