Abstract
MicroRNAs (miRNAs) are small RNAs (sRNAs) that regulate gene expression in development and adaptive responses to the environment. The early days in the sRNA field was one of the most exciting and promising moments in modern biology, attracting large investments to the understanding of the underlining mechanisms and their applications, such as in gene therapy. miRNAs and other sRNAs have since been extensively studied in animals and plants, and are currently well established as an important part of most gene regulatory processes in animals and as master regulators in plants. Here, this review presents the critical discoveries and early misconceptions that shaped our current understanding of RNA silencing by miRNAs in most eukaryotes, with a focus on plant miRNAs. The presentation and language used are simple to facilitate a clear comprehension by researchers and students from various backgrounds. Hence, this is a valuable teaching tool and should also draw attention to the discovery processes themselves, such that scientists from various fields can gain insights from the successful and rapidly evolving miRNA field.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
In most eukaryotes, RNA silencing is triggered by double-stranded RNA (dsRNA) that is processed into sRNAs by a member of the dicer protein family (Meister and Tuschl 2004). These sRNAs are in turn loaded into a member of the argonaute (AGO) protein family to form the catalytic core of the RNA-induced silencing complex (RISC) (Hutvagner and Simard 2008). Endogenous or exogenous transcripts carrying complementary sequences to the AGO-loaded sRNA are targeted by RISC, which, depending on the AGO family member at its catalytic core, mediates expression inhibition at either the transcriptional or posttranscriptional level (Hutvagner and Simard 2008). Plant sRNAs also require the activity of the sRNA-specific methyltransferase, HUA ENHANCER1 (HEN1), to stabilise the sRNA post dicer processing and prior to AGO loading, via 2′-O-methylation of the 3′ terminal nucleotide (Yu et al. 2005). sRNAs can be derived from various sources of dsRNAs, defining different silencing pathways such as antisense RNA, small-interfering RNAs (siRNAs) and miRNAs, each being a substrate for a dicer protein upon formation of dsRNA through base pairing with sense RNA, formation of antisense RNA (synthesised by host enzymatic amplification) and stem loop interactions, respectively. Here, the main discoveries that have led to our current understanding of RNA silencing by miRNAs are discussed, with a focus on plant miRNAs.
Co-suppression and homology-dependent virus resistance in plants
RNA silencing was probably first reported in 1928 (reviewed by Baulcombe 2004), when tobacco plants infected with Tobacco ring spot virus (TRV) became progressively less symptomatic over time. Intriguingly, new leaves that emerged post TRV infection appeared to have ‘recovered’ from the initial infection and were shown to be resistant to the secondary infection with either the same, or to closely related, viruses (Wingard 1928). Half a century later, plant virologists started to uncover the molecular mechanisms behind virus-induced resistance. Two related theories led to the first advance towards our current understanding of RNA silencing: co-suppression and homology-dependent virus resistance.
Silencing of a selectable marker gene, introduction to the plant via Agrobacterium tumefaciens (Agrobacterium)-mediated transformation was observed upon the introduction of a second plant expression vector, carrying a different selectable marker gene, when the expression of both selectable marker genes was driven by the same strong viral promoter (Matzke et al. 1989). In addition, the constitutive expression of reintroduced copies of endogenous genes via the use of viral promoters was shown to result in silencing of both the endogenous and transgene-introduced copy (Napoli et al. 1990; van der Krol 1990). Further variations of these initial co-suppression experiments were repeatedly confirmed in subsequent studies (Smith et al. 1990; de Carvalho et al. 1992; Vaucheret et al. 1995). Similarly, homology-dependent virus resistance was observed with transformed plants harbouring viral-derived transgenes, which, upon plant genome integration, mediated resistance to viruses with homologous sequences to those present in the integrated transgene (Lindbo et al. 1993; Mueller et al. 1995). It did not take long for researchers to realise that a similar mechanism was underpinning the phenomena observed in silencing endogenous genes as well as with plant-acquired resistance to viruses (Ratcliff 1997). Both phenomena shared high target specificity leading to the hypothesis that they may be guided by a nucleic acid molecule. Several independent groups concluded that ‘silenced’ plants failed to accumulate gene products encoded by homologous genes, even though the corresponding loci remained transcriptionally active. This suggested that the observed silencing was occurring at the posttranscriptional level, and was thus termed posttranscriptional gene silencing, or PTGS (Carr and Zaitlin 1991; Baulcombe 1996; Metzlaff et al. 1997; Ratcliff 1997; Angell and Baulcombe 1997; Ruiz 1998). At this time, however, it remained a matter of debate as to whether it was a DNA- or RNA-based molecule that was directing the observed posttranscriptional regulation (reviewed by Baulcombe and English 1996).
Antisense RNAs in Animals
By the mid-1990s, the use of antisense RNA as a tool to repress complementary gene expression was commonplace in animal research, but an understanding of the molecular mechanism(s) that led to this repression remained unknown (Nellen and Lichtenstein 1993). It was well-established that natural antisense RNAs, endogenous transcripts with sequences complementary to sense transcripts were widely distributed amongst prokaryote genomes, and that they controlled numerous biological functions, including transposition, plasmid replication and regulation of gene expression (reviewed by Wagner and Simons 1994). Moreover, in eukaryotes, additional evidence of natural antisense RNAs strongly suggested that antisense RNA was part of a general, evolutionary-conserved mechanism for the control of gene expression, as opposed to solely acting as a defence mechanism against invading exogenous nucleic acids (Vanhée-Brossollet and Vaquero 1998).
dsRNA: the trigger of RNA silencing
The breakthrough that has enabled our current understanding of RNA silencing as a widespread eukaryote regulatory mechanism came in the year 1998. Fire et al. (1998) demonstrated that dsRNA was the sole trigger required to initiate RNA silencing. The authors reported highly robust and specific RNA silencing of complementary genes that was readily and reproducibly achievable following the injection of dsRNA into the nematode Caenorhabditis elegans (C. elegans). The gene silencing by dsRNA-triggered RNA silencing was, hence, termed RNA interference (RNAi). Interestingly, this discovery was inspired by the puzzling observation that sense and antisense RNA are equally effective in RNA silencing (Guo and Kemphues 1995). The paradox was resolved by showing that the preparations of sense and antisense RNA contained small amounts of dsRNA, enough to trigger RNA silencing (Fire et al. 1998). Therefore, the authors concluded that the observed RNA silencing was likely a consequence of dsRNA formation in the cell.
By the mid 1990s, an understanding of the molecular mechanisms of RNA silencing in plants had already started to form. The identification and characterisation of a tomato RNA-DEPENDENT RNA POLYMERASE (RDR) (Schiebel et al. 1993) led to the hypothesis that the role of RDRs was to transcribe complementary RNAs (cRNAs) from transgene-encoded transcripts (Baulcombe 1996). The RDR-transcribed cRNA could hybridise with a corresponding target RNA to form a hybrid substrate for dsRNA-specific RNases, leading to the arrest of translation (Baulcombe 1996; Waterhouse et al. 1998). In this context, the breakthrough demonstration in plants was provided by Waterhouse et al. (1998), showing that PTGS is also induced by a dsRNA trigger. In the same year that the Fire and Waterhouse studies were published, molecules of dsRNA were shown to be also effective triggers of RNA silencing in other organisms, such as flies (Kennerdell and Carthew 1998) and protozoa (Ngô et al. 1998). It is interesting to note that dsRNAs, as silencing triggers, were the starting pistols for a race that led to the creation of one of the most exciting and promising fields in modern times.
The discovery of small-interfering RNAs (siRNAs)
Although dsRNA was rapidly established as the trigger for eukaryote RNA silencing, the molecular mechanisms that led to the repression of gene expression remained unknown. The first definitive piece of this puzzle came from plants: the identification of small-interfering RNAs (siRNAs). Hamilton and Baulcombe (1999) showed that transgene- or virus-induced PTGS resulted in the accumulation of small RNA (sRNA) molecules of an approximately uniform length of 25 nucleotides (nt). Furthermore, the authors showed that the level of 25 nt sRNA accumulation tightly correlated with the degree of RNA silencing. However, it remained uncertain whether these 25 nt sRNAs were responsible for directing the observed silencing itself, or whether they were just byproducts resulting from the RNA silencing process. Long molecules of dsRNA were later shown to be processed into a population of 21–23 nt sRNAs in vitro, and targeted mRNA was only cleaved in regions complementary to the triggering dsRNA (Zamore et al. 2000). Moreover, the mRNA was cleaved at approximately 21–23 nt intervals, the same size as the detected sRNAs. These findings suggested that sRNAs, or siRNAs, derived from processing of the triggering dsRNA, were able to direct cleavage of complementary mRNAs. Several subsequent studies revealed siRNA-directed repression of gene expression across eukaryotes (Wianny and Zernicka-Goetz 2000; Parrish et al. 2000; Elbashir et al. 2001a). Thus, a siRNA was derived from a dsRNA molecule and, further, it could silence transcripts with complementary sequence. Consequently, there should be at least three enzymatic steps to be uncovered: the production of dsRNAs, their cleavage to produce siRNAs and siRNA-guided cleavage of target transcripts.
RNA-dependent RNA polymerases unites the kingdoms
The first cellular component required for siRNA-directed RNA silencing was identified in a screen for mutants defective in transgene-induced RNA silencing in the filamentous fungus Neurospora crassa, with the identified mutants termed q uelling- de fective (qde) (Cogoni and Macino 1997; Cogoni and Macino 1999). The gene product encoded by QDE1 (the mutated locus in the qde1 mutant background) was found to be similar to the previously characterised RNA-dependent RNA polymerase (RDR) in tomato (Cogoni and Macino 1999). Furthermore, the silencing-impaired C. elegans and Arabidopsis thaliana (Arabidopsis) mutants ego-1 and sgs2/sde1, respectively, were also determined to harbour mutations in genes encoding orthologs of the tomato RDR (Smardon et al. 2000; Dalmay et al. 2000; Mourrain et al. 2000). The identification of RDR orthologs, as conserved components in the RNA silencing pathways, provided experimental evidence for the previously proposed model based on RDR-catalysed cRNA production (Baulcombe 1996; Waterhouse et al. 1998). More importantly, RDR gene identification across eukaryotes established that PTGS and RNAi phenomena were mechanistically related (Cogoni and Macino 2000). The RDR-based model did, however, raise three major questions: (i) Are RDRs necessary to produce large molecules of dsRNA from aberrant single-stranded RNA (ssRNA) templates (ii) How are long dsRNA molecules processed into siRNAs (siRNA biogenesis) and (iii) How are siRNAs effective in repressing gene expression (siRNA activity).
In plants, the demonstration that the RDR, SDE1/SGS2, produces dsRNA using the targeted RNA as a template also revealed spreading of the siRNA silencing signal from the original target site of the triggering dsRNA into adjacent 5ˈ and 3ˈ regions (Vaistij et al. 2002). This work also further identified RDRs as central components of siRNA-directed RNA silencing mechanisms across diverse species. It is worth noting that these findings were critical to validate the potent silencing achieved using minute amounts of dsRNA Fire et al. (1998), as RDRs amplify and spread silencing signals.
siRNA biogenesis: dicer proteins
Bernstein et al. (2001) showed in an elegant experiment that siRNA production and siRNA action are separate processes, and that a RNase III-like endonuclease is required for siRNA production from the dsRNA trigger. The authors applied differential centrifugation to show that the activity of the previously identified RISC (Hammond et al. 2000) and the siRNA generating enzyme of the same RNA silencing pathway did not co-fractionate. Therefore, a nuclease specific for processing of the triggering dsRNA, such as an RNase III endonuclease, was suggested to be a central requirement for dsRNA processing and siRNA production. The Drosophila melanogaster (Drosophila) RNase III CG4792 was demonstrated to produce siRNA guide sequences of approximately 22 nt in length from much longer, almost perfectly dsRNA triggers. Due to the ability of CG4792 to ‘dice up’ the dsRNA trigger into siRNAs, CG4792 was renamed Dicer (Bernstein et al. 2001). Dicer was subsequently shown to be evolutionarily conserved across several eukaryote species, including Arabidopsis (SIN1/SUS1/CAF), C. elegans (K12H4.8) and mammals (Helicase-MOI) (Bernstein et al. 2001). In the tale of the blind men and an elephant, a group of blind men touch an elephant to learn what it is like, but each one feels a different and unique part in such a way that they come to different conclusions as to the nature of an elephant. This tale vividly describes the identification of a Dicer homologue in Arabidopsis (Schauer et al. 2002). The SHORT INTEGUMENTS1 (SIN1), SUSPENSOR1 (SUS1) and CARPEL FACTORY (CAF) alleles had been previously extensively studied in embryo, ovule and flower development, respectively, as individual loci thought to encode different proteins (Errampalli et al. 1991; Robinson-Beers et al. 1992; Jacobsen et al. 1999). However, it was later determined that SIN1/SUS1/CAF were all mutant alleles of a single locus encoding a RNase III-like endonuclease similar to the Drosophila Dicer protein, and that this locus was therefore renamed DICER-LIKE1 (DCL1) (Golden et al. 2002; Finnegan et al. 2003).
Dicer proteins often belong to multimember families, with each family member characterised by RNase III, PAZ, RNA helicase and dsRNA binding domains. For example, the Arabidopsis genome encodes four Dicer proteins, DCL1 to DCL4, which differ in the size of the protein and the presence and organisation of each functional domain (Schauer et al. 2002; Liu et al. 2009). The diversity in sRNA biogenesis proteins in Arabidopsis and other eukaryotes suggested that the dsRNAs processed, as well as the resulting sRNA species produced, act through multiple parallel RNA silencing pathways. The first experimental evidence that Dicer proteins have distinct roles in RNA silencing was obtained in an a study on Arabidopsis where DCL1 was shown to be not an essential protein component for PTGS or siRNA production (Finnegan et al. 2003).
sRNA activity: argonaute proteins
The core protein component of the siRNA effector complex, RNA-induced silencing complex (RISC), was revealed via a biochemical approach. A Drosophila ribonucleoprotein complex (~500 kDa) with RISC activity was purified and micro-sequenced to reveal the presence of an AGO protein (Hammond et al. 2001). However, the first AGO to be isolated was the Arabidopsis AGO1. As outlined for Arabidopsis DCL1, the Arabidopsis AGO1 gene was initially identified via a mutagenesis screening, and was named after the appearance that resembles the molluscs known as argonaut (a group of pelagic octopuses) (Bohmert et al. 1998). Although, members of the AGO protein family had been shown to affect the dsRNA response in Neurospora (QDE-1), C. elegans (RDE-1) and Arabidopsis (AGO1) (Tabara et al. 1999; Fagard et al. 2000; Macino et al. 2000), the ‘slicer’ activity of an AGO protein was not realised until after the crystal structure of the Pyrococcus furiosus AGO protein was resolved (Song et al. 2004) and an extensive mutational analysis of human Ago2 had been performed (Liu et al. 2004).
AGO family members are characterised by the presence of three conserved functional domains, namely, the PAZ (similar to Dicer), MID and PIWI domains. Arabidopsis and human genomes are known to encode ten and four AGO proteins, respectively, indicating large functional diversification of the action stage of the parallel RNA silencing pathways in eukaryotes (Song et al. 2004). Indeed, by the time AGO’s slicer activity had been experimentally validated, Dicer and AGO mutant characterisation had already revealed that sRNA-directed RNA silencing was central to a diverse array of biological processes (reviewed by Carmell et al. 2002).
miRNAs: a specific class of small RNAs
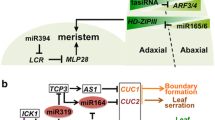
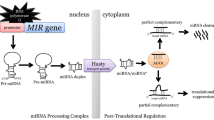
The C. elegans lin-4 RNA is recognised as the founding member of an extensive and highly specific class of small regulatory RNAs, termed microRNAs (miRNAs) (Bartel 2004). Curiously, the approximately 22 nt lin-4 (Lee et al. 1993) and let-7 (Reinhart et al. 2000; Slack et al. 2000) sRNAs had been studied as the only two examples of small temporal RNA (stRNA) products for almost a decade until the identification of over one hundred stem loop-structured RNAs that, upon Dicer cleavage, generate 21 to 24 nt non-coding small regulatory RNAs similar to stRNAs, which were later collectively called miRNAs (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001). miRNAs have been shown to be evolutionary conserved, similar to the lin-4 and let-7 stRNAs, and are typically (i) derived from independent transcriptional units; (ii) processed from stem loop precursor RNAs by Dicers, and (iii) able to regulate the expression of a large set of genes via RISC-mediated mechanisms (Fig. 1) (Bartel 2004; Budak and Akpinar 2015).
Plant miRNA guide either cleavage or translation inhibition. Primary miRNAs (pri-miRNAs) are processed by DCL1 into a miRNA duplex (not shown) with the assistance of its partnering proteins, DRB1 or DRB2. Through a yet unknown mechanism, DRB1 promotes loading of the mature miRNA onto an argonaute (AGO) protein that targets transcript for cleavage. DRB2, however, promotes translation inhibition and represses transcription of DRB1
In contrast to siRNAs, miRNAs do not trigger the amplification and spreading of secondary silencing signals via the activity of an RDR (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001). Furthermore, miRNA-directed RNA silencing was initially shown to lead exclusively to translation repression in animals, and to solely mediate mRNA cleavage in plants (Llave et al. 2002; Tang et al. 2003). In animals and flies, siRNA- and miRNA-loaded RISC, termed siRISC and miRISC, have different complementarity requirements for target transcript recognition. siRISC recognises and regulates the expression of target transcripts that harbour target sequences with high complementarity to the loaded siRNA. miRISC target regulation, on the other hand, has been shown to be based on low miRNA:mRNA complementarity requirements (Elbashir et al. 2001a; Elbashir et al. 2001b). Furthermore, it has since been demonstrated that mammalian miRNAs bearing high complementarity to their targeted transcript(s) also guide mRNA cleavage and, conversely, that exogenously supplied siRNAs can inhibit the expression of lowly complementary mRNAs without inducing any detectable transcript cleavage (Doench et al. 2003; Zeng et al. 2003). It is now well established that the AGO protein, assembled with either a miRNA or siRNA, determines which mechanism of RNA silencing the loaded sRNA will direct in animals and insects (Filipowicz et al. 2005).
Plant miRNAs were initially thought to act through a mechanism similar to siRNAs, because of the extensive miRNA:mRNA base pairing requirement and, as it was thought then, to solely mediate mRNA cleavage (Tang et al. 2003). However, the authors concluded that plant miRNAs, since they lacked RdRP-dependent amplification and spreading steps, act through a similar mechanism to that in animals and insects. Nevertheless, it was widely accepted by the plant sRNA research community at that time that AGO1, at the catalytic core of plant miRISCs, was mechanistically similar to cleavage-competent human Ago2, as opposed to directing translation inhibition (Tang et al. 2003; Filipowicz et al. 2005). Thus, although plant and animal miRNA pathways were clearly related (RdRP-independent), they were thought to be mechanistically different (Millar and Waterhouse 2005). This paradigm that plant miRNAs direct only transcript cleavage was initially challenged by Xuemei Chen’s work on the Arabidopsis miR172-AP2 silencing module. miR172 was shown to regulate the expression of its targeted gene, APETELA2 (AP2), predominantly via translation repression (Chen 2004). Later, other workers demonstrated that translation repression is a widespread silencing mechanism directed by either miRNAs or siRNAs (Brodersen et al. 2008; Reis et al. 2015a). It is currently well established that plant miRNAs can act through either transcript cleavage or translation repression, a process determined by DRB1 and DRB2, DCL1 partnering proteins. It has been shown that the dependence of DCL1 on DRB1 for miRNA biogenesis is only required for miRNA-guided transcript cleavage, whereas, DRB2 determines miRNA-guided translational inhibition and represses DRB1 expression, thereby allowing the active selection of miRNA regulatory action (Reis et al. 2015a) (Fig. 1).
It is interesting to note that, although RDRs were the first cellular component to exhibit the evolutionary conservation of RNA silencing, the demonstration that miRNAs act independently of RDR activity in plants, in which AGO1 was considered impaired in translation inhibition, was also responsible for setting apart the plant miRNA pathway from those of other organisms. It is therefore not surprising that plant miRNA-guided translation repression had been discounted for many years.
Plant miRNAs as a highly specific gene silencing tool
In plants, RNA silencing has been artificially achieved since the early 1990s via the introduction of exogenous sequences into their genome through the use of modified Agrobacterium transfer-DNA (T-DNA) expression vectors. Initially, PTGS was achieved via the use of T-DNA constructs encoding either antisense (Hamilton et al. 1990) or co-suppression RNAs (Jorgensen 1995). However, such an approach typically resulted in a low efficiency of silenced individuals within generated transformant populations. The identification of dsRNA-triggered PTGS led to the subsequent development of much more powerful tools that offered almost 100 % PTGS efficiency via the introduction of T-DNA constructs encoding hairpin RNAs (hpRNAs) (Chuang and Meyerowitz 2000; Wesley et al. 2001). Later, the expression of modified miRNA stem loop precursor transcripts, which incorporate artificial miRNA (amiRNA) sequences targeting genes of interest, has enabled the silencing of highly specific target genes (Schwab et al. 2006).
To date, the over-expression of endogenous miRNA precursor transcripts has been widely documented to be a useful tool for the characterisation of native miRNA target genes and to study the effects of miRNA misexpression (Llave et al. 2002). Conversely, studying the consequences of miRNA target gene misexpression has been largely achieved via expression of endogenous miRNA target genes harbouring silent mutations within the miRNA binding site (Baker et al. 2005; Mallory et al. 2005). More recently, via an indirect approach, the over-expression of non-cleavable miRNA target mimic sequences, to either sequester (Franco-Zorrilla et al. 2007) or completely degrade (Yan et al. 2012a) the regulating endogenous miRNA, has been used to study miRNA/mRNA target interactions in vivo. In contrast to animal miRNAs, the requirement of plant miRNAs for extensive base pairing to their target mRNA(s), has enabled such a specific transgene-based approach for the determination of their biological function. Together, these approaches have revealed that plant miRNAs play an important role throughout plant development (Rubio-Somoza and Weigel 2011), as well as to mediate tolerance or adaptation responses to biotic and abiotic stress (Ding et al. 2013; Khraiwesh and Zhu 2012; Sunkar et al. 2012). Therefore, plant miRNAs are obvious targets for molecular modification of plants to increase current crop yield and improve food security (Li et al. 2013). Indeed, several successful examples of biotechnological applications of amiRNAs have been reported (reviewed by Khraiwesh et al. 2012).
Conclusion and current challenges in the plant miRNA field
Silencing triggered by dsRNA (siRNA, RNAi and miRNA) is a process found in both animals and plants. Their discovery led to a revolution in biology and great promises for clinical treatments and crop improvement. Interestingly, the discovery race initially had both animal and plant scientists benefiting from each other’s findings until they, wrongly, realise that the miRNA pathway is fundamentally different between these two kingdoms. For almost a decade, which coincided with a peak in interest and use of miRNAs in crop improvement, scientists assumed that plant miRNAs only lead to target cleavage, whereas translation inhibition was exclusive to animals. It is now known that plant miRNAs can guide either cleave or translation inhibition, but its consequence for crop improvement and our mechanistic knowledge is poorly understood (Fig. 1) largely as a consequence of the ‛split’ between plant and animal scientists in this field.
A recent work demonstrated that these two mode of actions (cleavage or translation inhibition) play different roles, and that miRNA-guided translational repression appears biased towards response to environmental stresses (Reis et al. 2015b). In addition, a large-scale transfection of protoplast using amiRNAs and epitope-tagged targets showed that optimal amiRNAs predominantly mediated highly specific translational repression with limited mRNA decay or cleavage (Li et al. 2013). These and other works go against the still persistent paradigm that miRNA-guided translation inhibition is the secondary and minor silencing mechanism, and transcript cleavage can almost solely be used to study and modify plants—a paradigm particularly persistent among researchers working with crop plants. This can be largely explained by three main factors, i.e, (1) the widespread use of RNA-Seq, (2) very limited availability of antibodies against plant proteins and (3) challenges to obtain transgenic crop plants expressing tagged proteins. This context makes it difficult to study posttranscriptional regulations that do not alter transcript levels, as it is normally the case with translation inhibition. A thorough study of miRNAs and amiRNAs effects on translation of their targets is, thus, the main challenge that plant researchers are currently facing in this field.
References
Angell SM, Baulcombe DC (1997) Consistent gene silencing in transgenic plants expressing a replicating potato virus X RNA. EMBO J 16:3675–3684. doi:10.1093/emboj/16.12.3675
Baker CC, Sieber P, Wellmer F, Meyerowitz EM (2005) The early extra petals1 Mutant Uncovers a Role for MicroRNA miR164c in Regulating Petal Number in Arabidopsis. Curr Biol 15:303–315. doi:10.1016/j.cub.2005.02.017
Bartel D (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Baulcombe D (2004) RNA silencing in plants. Nature 431:356–363. doi:10.1038/nature02874
Baulcombe DC (1996) Mechanisms of pathogen-derived resistance to viruses in transgenic plants. Plant Cell 8:1833–1844. doi:10.1105/tpc.8.10.1833
Baulcombe DC, English JJ (1996) Ectopic pairing of homologous DNA and post-transcriptional gene silencing in transgenic plants. Curr Opin Biotechnol 7:173–180. doi:10.1016/S0958-1669(96)80009-7
Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363–366
Bohmert K, Camus I, Bellini C, et al. (1998) AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J 17:170–180. doi:10.1093/emboj/17.1.170
Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, et al. (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320:1185–1190. doi:10.1126/science.1159151
Budak H, Akpinar BA (2015) Plant miRNAs: biogenesis, organization and origins. Funct Integr Genomics. 15:523–531. doi:10.1007/s10142-015-0451-2
Carmell MA, Xuan Z, Zhang MQ, Hannon GJ (2002) The argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev 16:2733–2742. doi:10.1101/gad.1026102
Carr JP, Zaitlin M (1991) Resistance in transgenic tobacco plants expressing a nonstructural gene sequence. Mol Plant-Microbe Interact 4:579–585
Chen X (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303:2022–2025. doi:10.1126/science.1088060
Chuang CF, Meyerowitz EM (2000) Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci U S A 97:4985–90. doi: 10.1073/pnas.060034297
Cogoni C, Macino G (1997) Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc Natl Acad Sci U S A 94:10233–10238
Cogoni C, Macino G (1999) Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399:166–169. doi:10.1038/20215
Cogoni C, Macino G (2000) Post-transcriptional gene silencing across kingdoms. Curr Opin Genet Dev 10:638–643
Dalmay T, Hamilton A, Rudd S, et al. (2000) An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101:543–553
de Carvalho F, Gheysen G, Kushnir S, et al. (1992) Suppression of beta-1, 3-glucanase transgene expression in homozygous plants. EMBO J 11:2595–2602
Ding C, Chan DW, Liu W, et al (2013) Proteome-wide profiling of activated transcription factors with a concatenated tandem array of transcription factor response elements. Proc Natl Acad Sci U S A 110:6771–6776. doi:10.1073/pnas.1217657110
Doench JG, Petersen CP, Sharp PA (2003) siRNAs can function as miRNAs. Genes Dev 17:438–442. doi:10.1101/gad.1064703
Elbashir SM, Lendeckel W, Tuschl T (2001a) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 15:188–200. doi:10.1101/gad.862301
Elbashir SM, Martinez J, Patkaniowska A, et al. (2001b) Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J 20:6877–6888. doi:10.1093/emboj/20.23.6877
Errampalli D, Patton D, Castle L, et al. (1991) Embryonic Lethals and T-DNA insertional mutagenesis in Arabidopsis. Plant Cell 3:149–157. doi:10.1105/tpc.3.2.149
Fagard M, Boutet S, Morel J-B, et al. (2000) AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc Natl Acad Sci U S A 97:11650–11654
Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS (2005) Post-transcriptional gene silencing by siRNAs and miRNAs. Curr Opin Struct Biol 15:331–341. doi:10.1016/j.sbi.2005.05.006
Finnegan EJ, Margis R, Waterhouse PM (2003) Posttranscriptional gene silencing is not compromised in the Arabidopsis CARPEL FACTORY (DICER-LIKE1) mutant, a homolog of dicer-1 from drosophila. Curr Biol 13:236–240
Fire A, Xu S, Montgomery MK, et al. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811. doi:10.1038/35888
Franco-Zorrilla JM, Valli A, Todesco M, et al (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39:1033–7. doi:10.1038/ng2079
Golden TTAT, Schauer SES, Lang JJD, et al. (2002) SHORT INTEGUMENTS1/SUSPENSOR1/CARPEL FACTORY, a dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol 130:808–822. doi:10.1104/pp.003491.in
Guo S, Kemphues KJ (1995) Par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 81:611–620
Hamilton AJ, Lycett GW, Grierson D (1990) Antisense gene that inhibits synthesis of the hormone ethylene in transgenic plants. Nature 346:284–287. doi:10.1038/346284a0
Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional Gene silencing in plants. Science 286:950–952. doi:10.1126/science.286.5441.950
Hammond SM, Bernstein E, Beach D, Hannon GJ (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in drosophila cells. Nature 404:293–296. doi:10.1038/35005107
Hammond SM, Boettcher S, Caudy AA, et al. (2001) Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293:1146–1150. doi:10.1126/science.1064023
Hutvagner G, Simard MJ (2008) Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 9:22–32. doi:10.1038/nrm2321
Jacobsen SE, Running MP, Meyerowitz EM (1999) Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126:5231–5243
Jorgensen RA (1995) Cosuppression, flower color patterns, and metastable gene expression States. Science 268:686–91. doi:10.1126/science.268.5211.68
Kennerdell JR, Carthew RW (1998) Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95:1017–1026
Khraiwesh B, Zhu JJ-K (2012) Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta 1819:137–48. doi:10.1016/j.bbagrm.2011.05.001
Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294:853–858. doi:10.1126/science.1064921
Lau NC, Lim LP, Weinstein EG, Bartel DP (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294:858–862. doi:10.1126/science.1065062
Lee RC, Ambros V (2001) An extensive class of small RNAs in Caenorhabditis elegans. Science 294:862–864. doi:10.1126/science.1065329
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854
Li J-F, Chung HS, Niu Y, et al. (2013) Comprehensive protein-based artificial microRNA screens for effective gene silencing in plants. Plant Cell 25:1507–1522. doi:10.1105/tpc.113.112235
Lindbo JA, Silva-Rosales L, Proebsting WM, Dougherty WG (1993) Induction of a highly specific antiviral state in transgenic plants: implications for regulation of Gene expression and virus resistance. Plant Cell 5:1749–1759. doi:10.1105/tpc.5.12.1749
Liu J, Carmell MA, Rivas FV, et al. (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science 305:1437–1441. doi:10.1126/science.1102513
Liu Q, Feng Y, Zhu Z (2009) Dicer-like (DCL) proteins in plants. Funct Integr Genomics. 9:277–286. doi:10.1007/s10142-009-0111-5
Llave C, Xie Z, Kasschau KD, Carrington JC (2002) Cleavage of scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297:2053–2056. doi:10.1126/science.1076311
Macino G, Cogoni C, Catalanotto C, Azzalin G (2000) Transcription: gene silencing in worms and fungi. Nature 404:245
Mallory AC, Bartel DP, Bartel B (2005) MicroRNA-Directed Regulation of Arabidopsis AUXIN RESPONSE FACTOR17 Is Essential for Proper Development and Modulates Expression of Early Auxin Response Genes. Plant Cell 17:1360–1375. doi:10.1105/tpc.105.031716
Matzke MA, Primig M, Trnovsky J, Matzke AJ (1989) Reversible methylation and inactivation of marker genes in sequentially transformed tobacco plants. EMBO J 8:643–649
Meister G, Tuschl T (2004) Mechanisms of gene silencing by double-stranded RNA. Nature 431:343–349. doi:10.1038/nature02873
Metzlaff M, O’Dell M, Cluster P, Flavell R (1997) RNA-mediated RNA degradation and Chalcone synthase a silencing in petunia. Cell 88:845–854. doi:10.1016/S0092-8674(00)81930-3
Millar AA, Waterhouse PM (2005) Plant and animal microRNAs: similarities and differences. Funct Integr Genomics 5:129–135. doi:10.1007/s10142-005-0145-2
Mourrain P, Béclin C, Elmayan T, et al. (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101:533–542
Mueller E, Gilbert J, Davenport G, et al. (1995) Homology-dependent resistance: transgenic virus resistance in plants related to homology-dependent gene silencing. Plant J 7:1001–1013. doi:10.1046/j.1365-313X.1995.07061001.x
Napoli C, Lemieux C, Jorgensen R (1990) Introduction of a chimeric Chalcone synthase Gene into petunia results in reversible Co-suppression of homologous genes in trans. Plant Cell 2:279–289. doi:10.1105/tpc.2.4.279
Nellen W, Lichtenstein C (1993) What makes an mRNA anti-sense-itive? Trends Biochem Sci 18:419–423
Ngô H, Tschudi C, Gull K, Ullu E (1998) Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc Natl Acad Sci U S A 95:14687–14692
Parrish S, Fleenor J, Xu S, et al. (2000) Functional anatomy of a dsRNA trigger: differential requirement for the two trigger strands in RNA interference. Mol Cell 6:1077–1087
Ratcliff F (1997) A similarity between viral defense and gene silencing in plants. Science 276:1558–1560. doi:10.1126/science.276.5318.1558
Reinhart BJ, Slack FJ, Basson M, et al. (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403:901–906. doi:10.1038/35002607
Reis RS, Hart-Smith G, Eamens AL, et al. (2015a) Gene regulation by translational inhibition is determined by dicer partnering proteins. Nat Plants 1. doi:10.1038/nplants.2014.27
Reis RS, Hart-Smith G, Eamens AL, et al (2015b) MicroRNA Regulatory Mechanisms Play Different Roles in Arabidopsis. J Proteome Res. 151001114936009. doi: 10.1021/acs.jproteome.5b00616
Robinson-Beers K, Pruitt RE, Gasser CS (1992) Ovule development in wild-type Arabidopsis and two female-sterile mutants. Plant Cell 4:1237–1249. doi:10.1105/tpc.4.10.1237
Rubio-Somoza I, Weigel D (2011) MicroRNA networks and developmental plasticity in plants. Trends Plant Sci 16:258–64. doi:10.1016/j.tplants.2011.03.001
Ruiz MT (1998) Initiation and maintenance of virus-induced gene silencing. Plant Cell 10:937–946. doi:10.1105/tpc.10.6.937
Schauer SE, Jacobsen SE, Meinke DW, Ray A (2002) DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci 7:487–491
Schiebel W, Haas B, Marinkovic S, et al. (1993) RNA-directed RNA polymerase from tomato leaves. II. Catalytic in vitro properties. J Biol Chem 268:11858–11867
Schwab R, Ossowski S, Riester M, et al (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18:1121–1133. doi: 10.1105/tpc.105.039834.1
Slack FJ, Basson M, Liu Z, et al. (2000) The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell 5:659–669
Smardon A, Spoerke JM, Stacey SC, et al. (2000) EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr Biol 10:169–178
Smith CJS, Watson CF, Bird CR, et al. (1990) Expression of a truncated tomato polygalacturonase gene inhibits expression of the endogenous gene in transgenic plants. Mol Gen Genet. 224:477–481. doi:10.1007/BF00262443
Song J-J, Smith SK, Hannon GJ, Joshua-Tor L (2004) Crystal structure of argonaute and its implications for RISC slicer activity. Science 305:1434–1437. doi:10.1126/science.1102514
Sunkar R, Li Y-F, Jagadeeswaran G (2012) Functions of microRNAs in plant stress responses. Trends Plant Sci 17:196–203. doi:10.1016/j.tplants.2012.01.010
Tabara H, Sarkissian M, Kelly WG, et al. (1999) The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99:123–132
Tang G, Reinhart BJ, Bartel DP, Zamore PD (2003) A biochemical framework for RNA silencing in plants. Genes Dev 17:49–63. doi:10.1101/gad.1048103
Vaistij F, Jones L, Baulcombe D (2002) Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 14:857–867. doi:10.1105/tpc.010480.radation
van der Krol AR (1990) Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell 2:291–299. doi:10.1105/tpc.2.4.291
Vanhée-Brossollet C, Vaquero C (1998) Do natural antisense transcripts make sense in eukaryotes? Gene 211:1–9
Vaucheret H, Palauqui J-C, Elmayan T, Moffatt B (1995) Molecular and genetic analysis of nitrite reductase co-suppression in transgenic tobacco plants. Mol Gen Genet 248:311–317. doi:10.1007/BF02191598
Wagner E, Simons R (1994) Antisense RNA control in bacteria, phages, and plasmids. Annu Rev Microbiol 48:713–742. doi:10.1146/annurev.mi.48.100194.003433
Waterhouse PM, Graham MW, Wang MB (1998) Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci U S A 95:13959–13964
Wesley S V, Helliwell C a, Smith N a, et al (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27:581–90
Wianny F, Zernicka-Goetz M (2000) Specific interference with gene function by double-stranded RNA in early mouse development. Nat Cell Biol 2:70–75. doi:10.1038/35000016
Wingard SA (1928) Hosts and symptoms of ring spot, a virus disease of plants. J Agric Res 37:127–153
Yan J, Gu Y, Jia X, et al (2012) Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 24:415–27. doi: 10.1105/tpc.111.094144
Yu B, Yang Z, Li J, et al. (2005) Methylation as a crucial step in plant microRNA biogenesis. Science 307:932–935. doi:10.1126/science.1107130
Zamore PD, Tuschl T, Sharp PA, Bartel DP (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101:25–33. doi:10.1016/S0092-8674(00)80620-0
Zeng Y, Yi R, Cullen BR (2003) MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci U S A 100:9779–9784. doi:10.1073/pnas.1630797100
Acknowledgments
I thank Andrew L. Eamens and Thomas H. Roberts for their critical reading of the manuscript, and the Australian Postgraduate Award scheme for financial support.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article forms part of a special issue of Functional & Integrative Genomics entitled “miRNA in model and complex organisms” (Issue Editors: Hikmet Budak and Baohong Zhang)
Rights and permissions
About this article
Cite this article
Reis, R.S. The entangled history of animal and plant microRNAs. Funct Integr Genomics 17, 127–134 (2017). https://doi.org/10.1007/s10142-016-0513-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-016-0513-0