Abstract
The eastern oyster Crassostrea virginica is a major aquaculture species for the USA. The sustainable development of eastern oyster aquaculture depends upon the continued improvement of cultured stocks through advanced breeding technologies. The Eastern Oyster Breeding Consortium (EOBC) was formed to advance the genetics and breeding of the eastern oyster. To facilitate efficient genotyping needed for genomic studies and selection, the consortium developed two single-nucleotide polymorphism (SNP) arrays for the eastern oyster: one screening array with 566K SNPs and one breeders’ array with 66K SNPs. The 566K screening array was developed based on whole-genome resequencing data from 292 oysters from Atlantic and Gulf of Mexico populations; it contains 566,262 SNPs including 47K from protein-coding genes with a marker conversion rate of 48.34%. The 66K array was developed using best-performing SNPs from the screening array, which contained 65,893 oyster SNPs including 22,984 genic markers with a calling rate of 99.34%, a concordance rate of 99.81%, and a much-improved marker conversion rate of 92.04%. Null alleles attributable to large indels were found in 13.1% of the SNPs, suggesting that copy number variation is pervasive. Both arrays provided easy identification and separation of selected stocks from wild progenitor populations. The arrays contain 31 mitochondrial SNPs that allowed unambiguous identification of Gulf mitochondrial genotypes in some Atlantic populations. The arrays also contain 756 probes from 13 oyster and human pathogens for possible detection. Our results show that marker conversion rate is low in high polymorphism species and that the two-step process of array development can greatly improve array performance. The two arrays will advance genomic research and accelerate genetic improvement of the eastern oyster by delineating genetic architecture of production traits and enabling genomic selection. The arrays also may be used to monitor pedigree and inbreeding, identify selected stocks and their introgression into wild populations, and assess the success of oyster restoration.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The eastern oyster Crassostrea virginica is native to the northwestern Atlantic coast, ranging from the Gulf of St. Lawrence in Canada to the Gulf of Mexico and West Indies in the south (Galtsoff 1964). As a reef-builder and filter-feeder, the eastern oyster provides critical ecological services to coastal and estuarine ecosystems (Loren et al. 2007; Beck et al. 2011; Grabowski et al. 2012). The eastern oyster also is economically important. Historically, the eastern oyster was highly abundant in estuaries and supported important fishing industries in the USA. Since the early 1900s, however, eastern oyster populations along much of the mid-Atlantic coast have declined as a consequence of overfishing, habitat destruction, and two major diseases: MSX caused by the parasite Haplosporidium nelsoni and Dermo caused by the parasite Perkinsus marinus (Ford and Tripp 1996). The decline of oyster resources has caused economic hardship in coastal fishing communities. Major efforts have been made to restore oyster populations, with limited success (Mann and Powell 2007). Oyster aquaculture has emerged as an important industry and the main source of oysters for consumption.
Eastern oyster aquaculture in the USA has grown rapidly during the past decade, with the value of farmed eastern oysters increasing from US $45 million in 2009 to $162 million in 2019 (FAO 2022). Although some of the growth in oyster aquaculture is attributed to the development and introduction of disease-resistant and triploid oysters, disease and environmental stress continue to cause mortalities and pose challenges to eastern oyster farming. Continued genetic improvement of cultured stocks is essential for the sustainable development of the eastern oyster aquaculture industry. The Eastern Oyster Breeding Consortium (EOBC) was formed to advance the genetics and breeding of the eastern oyster in support of the aquaculture industry. Members of EOBC have developed oyster strains with improved disease resistance through selective breeding (Frank-Lawale et al. 2014; Proestou et al. 2016; Guo 2021; Allen et al. 2021) and sequenced the genome of the eastern oyster (Gómez-Chiarri et al. 2015; Modak et al. 2021; Puritz et al. 2022a), paving the way for advanced genetic improvement through genomic selection (GS).
Genomic selection utilizes genome-wide marker information and can therefore provide more precise breeding values for quantitative traits that often are controlled by large numbers of small-effect loci. Genomic selection is particularly efficient when the breeding cycle is long, and phenotypes, such as disease resistance, are costly to measure. Genomic selection has been applied to the improvement of all major agricultural crops and livestock (Meuwissen et al. 2016; Voss-Fels et al. 2019) and is expected to also transform genetic improvement of aquacultural species.
A prerequisite for genomic selection is a high-throughput genotyping platform that can efficiently genotype a large number of markers that cover the entire genome. Although genotyping by targeted or whole-genome resequencing is possible, high-density, single-nucleotide polymorphism (SNP) arrays remain the most efficient and widely used platform for genomic selection. SNP arrays are simple to implement and can provide accurate and highly reproducible genotypes at high calling rates (> 99%) without laborious bioinformatic treatment, which is important for practical breeding. High-density SNP arrays have revolutionized genetic analyses of complex traits in humans and agriculture species through genome-wide association studies (GWAS). They are widely used in crop and livestock improvement through marker-assisted selection (MAS) and GS (Korte and Farlow 2013; Unterseer et al. 2014). High-density SNP arrays have been developed for several major aquaculture species, including catfish (Liu et al. 2014), carp (Xu et al. 2014), Atlantic salmon (Houston et al. 2014), rainbow trout (Palti et al. 2015), and the Pacific and European flat oysters (Gutierrez et al. 2017; Qi et al. 2017). In the eastern oyster, multiplex microsatellite assay and low-throughput SNP panels have been developed for pedigree assignment and population genetics studies (Wang et al. 2010; Thongda et al. 2018; Turley et al. 2019), but no high-density SNP arrays have been available.
A core objective of the EOBC has been to develop and evaluate high-density SNP arrays that can be used for GS as well as GWAS of complex traits important to oyster aquaculture and biology. We herein report on the development of two SNP arrays by means of a thorough characterization of sequence variation in the eastern oyster genome. The genome of the eastern oyster is highly polymorphic, with one SNP approximately every 20–54 bp (Zhang and Guo 2010; Eierman and Hare 2014). This high degree of polymorphism provides abundant SNPs for genotyping, but it also poses challenges for array design wherein unknown polymorphism within the probe sequence creates problems for probe hybridization on the array. To reduce problems from high polymorphism, we took a two-step strategy to develop a 600 K screening array first and then a 60 K breeders’ array based upon marker performance on the screening array. Our results show that the two-step process greatly increased the marker conversion rate of the breeders’ array, making it a highly efficient genotyping platform for GS and other applications for the eastern oyster.
Materials and Methods
Ethics Approval Statement
This study did not involve vertebrate animals. Eastern oysters (Bivalvia, Mollusca) used in this study were collected from farms or wild populations with necessary permits and handled following institutional guidelines.
Resequencing and SNP Discovery
For SNP discovery, whole-genome resequencing data (~ 15–30X coverage each) from 292 oysters covering diverse Atlantic and Gulf of Mexico populations were obtained and analyzed. The oysters included 90 oysters sequenced in a previous study (Puritz et al. 2022b) and 202 oysters sequenced in this study (Supplementary Table 1): 179 oysters sequenced by EOBC and 23 Gulf oysters sequenced by the Gulf of Mexico Oyster Genetics and Breeding Research Consortium. For the 179 oysters sequenced by EOBC, DNA was extracted with a CTAB protocol or the E.Z.N.A.® Mollusc DNA kit (Omega Bio-tek, GA, USA), quantified with NanoDrop, and submitted to Genewiz (NJ, USA) for 150 bp paired sequencing with Illumina HiSeq 4000. For the 23 Gulf oysters, DNA was extracted with Omega Bio-tek Mag-Bind Blood & Tissue DNA HDQ 96 kit, quantified with NanoDrop, and submitted to Genewiz for Illumina 150 bp paired sequencing. Raw sequence reads were submitted to National Center for Biotechnology Information (NCBI) Sequence Read Archive under BioProject PRJNA890664.
Raw sequencing reads were processed with a modified version of the dDocent pipeline (Puritz et al. 2014) with 23 Gulf of Mexico samples and 269 EOBC samples processed separately. Briefly, reads were trimmed for low quality bases and adapter sequences using fastp (Chen et al. 2018). Trimmed reads were then mapped to the haplotig-masked version of the eastern oyster genome (Puritz et al. 2022a) using bwa (Li and Durbin 2010) with modified mismatch and gap-opening parameters (-B 3 -O 5). Duplicates were marked using Picard (Institute 2016), and subsequent BAM files were filtered with samtools (Li et al. 2009) to remove low quality mappings, secondary alignments, and PCR duplicates. Samples were then analyzed for small nucleotide variants (SNPs, InDels, small complex events) using freebayes (Garrison and Marth 2012). Raw variants were filtered in parallel using a combination of bcftools (Danecek et al. 2021) and vcftools (Danecek et al. 2011). Variants were first filtered based on allelic balance at heterozygous loci (between 0.1 and 0.9) and quality to depth ratio of greater than 0.1. Variants were further filtered based on mean-depth, excluding all loci above the 95th percentile. Variants were then decomposed into SNPs, and InDels using vcflib (Garrison 2016). SNPs were further filtered to allow SNPs with less than 10% missing data, only biallelic, and with a minor allele frequency (MAF) of ≥ 0.05. Details on initial bioinformatics, including reproducible code and scripts, can be found in Puritz et al. (2022a, b).
After initial bioinformatic filtering, SNPs were further filtered to help ensure array compatibility. Decomposed InDel calls from Freebayes (Garrison and Marth 2012) were filtered to InDel variants with greater than 5% MAF and less than 10% missing data for the EOBC samples and indels from the 23 Gulf samples were more stringently filtered to less than 5% missing data and an MAF of > 0.1. The filtered InDel calls were converted to “BED” format and bedtools (Quinlan and Hall 2010) was used to “pad” or add 20 bp on both the 5′ and 3′ end of each InDEL using the “slop” function. For EOBC samples, filtered SNP calls were thinned to 1 locus per 20 bp and all loci falling within padded InDel bed file were removed using bcftools (Danecek et al. 2021). For Gulf samples, SNPs were further filtered to a MAF of > 0.1, thinned to 1 locus per 20 bp, and all loci falling within padded InDel bed file were removed using bcftools (Danecek et al. 2021). Both filtered SNP sets were converted to bed files and overlapping Gulf and EOBC SNPs were removed from the Gulf dataset using bedtools (Quinlan and Hall 2010). Outlier loci from preliminary analyses from Puritz et al. 2022b) were also added into the target list and the combined target list was filtered once again for any overlap with InDel intervals.
SNP Selection and Array Design
The Affymetrix Axiom platform was chosen for its accuracy, reproducibility, and ease to use and customize. We chose SNPs for the 600 K screening array from resequencing data after quality control, first excluding SNPs with MAF < 0.05, and then excluding SNPs with another polymorphism within 20 bp. In addition to SNPs discovered from resequencing, we included genic SNPs that were identified in previous studies as associated with growth, disease, and stress resistance (Yu et al. 2011; He et al. 2012; Zhang and Guo 2010; Thongda et al. 2018; Turley et al. 2019; Zeng and Guo 2022; Li et al. 2022). Also included are 33 previously identified SNPs from the mitochondrial genome (18 from Guo Lab, 14 from Hare Lab, and one from Turley et al. 2019). We prioritized genic SNPs on the screening array and filled in the remaining array with random SNPs for even distribution on chromosomes. All probes selected had a p-convert score (probability of conversion) of > 0.6. SNPs for the 60 K breeders’ array were selected based upon polymorphism and high performance on the screening array (see results).
Inclusion of Pathogen Probes
To test if the SNP arrays can genotype oysters and simultaneously detect pathogen presence in the oyster sample, we included in both arrays non-polymorphic sequences from 13 oyster and human pathogens as probes. The 13 pathogens included important oyster pathogens such as Perkinsus marinus, Aliiroseovarius crassostreae, Ostreid Herpesvirus 1 (OsHV-1), as well as Haplosporidium, Bonamia, and Vibrio species, including Vibrio spp. affecting larval bivalves (e.g., V. coralliilyticus) and Vibrio spp. of human health concern, such as Vibrio vulnificus and Vibrio parahaemolyticus (Supplementary Table 2).
Samples for Array Evaluation
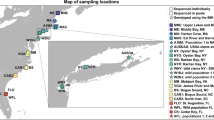
To evaluate the screening array and identify SNPs for the breeders’ array for Atlantic populations that are of interest to EOBC, we assembled 960 oysters from wild and hatchery populations along the Atlantic coast and genotyped them on the screening array (Table 1). The wild populations included two sites each in Maine, Long Island Sound, Delaware Bay, Chesapeake Bay and North Carolina, with 32 oysters from each site or 64 from each population. The hatchery populations included selected lines or families from breeding programs at the University of Maine, Rutgers University, Virginia Institute of Marine Science (VIMS) and the University of North Carolina Wilmington, as well as two private hatcheries in Maine and New York. The samples also included two parents and 94 progeny from a family derived from a wild Delaware Bay population, as well as 32 tissue samples positive for or spiked with various pathogens as positive controls.
The positive pathogen controls were included to test if the probes can detect the target pathogens. For each Bonamia species, DNA from one eastern or European flat oyster infected with each pathogen (courtesy of Ryan Carnegie, VIMS) was used as a positive control (Supplementary Table 2). For OsHV-1, gill tissue from three infected Pacific oysters from a challenge experiment (He et al. 2015) was used. For Haplosporidium costale and Haplosporidium nelsoni, tissue samples from three infected oysters were used as positive controls. For P. marinus, A. crassostreae, and Vibrio species, oyster tissue was spiked with pathogen cells before DNA extraction. No negative controls were included, as many of the genotyped oysters were expected to be negative.
To evaluate the breeders’ array and test its efficacy with different sample types, we assembled and genotyped 384 oysters (Table 1), including oysters or spat from wild New Jersey populations (Cape Shore and Hope Creek in Delaware Bay, and Rose Cove in Barnegat Bay, 48 each), Rutgers selected NEH® (n = 48), parents and progeny from a full-sib family (n = 98), biopsied hemolymph from NEH® (n = 48), small spat with and without shells (n = 36), archived samples that were fixed with ethanol for 0.25–18 years (n = 40), DNA samples (n = 8, extracted with the Kurabo system, Japan), and 10 duplicated samples to test genotyping concordance. The population of origin for the 384 oysters was not a major concern because the polymorphism of all markers was characterized with 960 oysters on the 566K array. We used oysters from one selected and three wild populations from New Jersey to show how informative the 66K array was for a given site or breeding program. Biopsied hemolymph (~ 1 ml) was taken from the adductor muscle through a notch at the edge of the shell and fixed in absolute ethanol (1 ml). The fixative was changed once within 2 h. After fixing, salt crystals formed in most samples. To assess any effects of these crystals upon DNA extraction and genotyping, half of the samples were dissolved in distilled water and re-pelleted to remove the salt crystals, and half of the samples were processed with the crystals. To validate mitochondrial SNPs (mtSNPs) and characterize differences between Atlantic and Gulf of Mexico populations, mtSNP genotypes were extracted from 263 oysters from a Florida wild population (Goose Point, Gulf of Mexico, collected under the Florida Fish and Wildlife Conservation Commission special activity license number SAL-22–2391-SR) and 50 from a selected line from Auburn University that is derived from Florida, Alabama, Louisiana, and Texas oysters (Scott Rikard, personal communication). The Gulf oysters were genotyped on the breeders’ array for another project, and only mtSNP data are included here as Gulf reference.
For the screening array, genomic DNA was extracted at Rutgers from ethanol-fixed tissue with the Qiagen DNeasy 96 Blood and Tissue Kit following protocols provided by the manufacturer. DNA was quantified with PicoGreen to ensure DNA concentration above 10 ng/μl. For the breeders’ array, genomic DNA was extracted at Center for Aquaculture Technology (CA, USA) using the Mag-Bind Blood and Tissue DNA Kit (Omega BioTek, Norcross, GA) according to manufacturer’s guidelines. Automated processing and liquid handling steps associated with the extraction protocol were performed using a PurePrep unit (Molgen, San Diego, CA) according to instrument-specific recommended guidelines. Resulting DNAs were assessed for yields and quality by NanoDrop and 2% agarose gels; with targeted requirements of 35 μl at > 15–20 ng/μl of largely intact DNA (minimum 5 Kb) for Axiom Array SNP genotyping. Genotyping of the screening SNP array was conducted at Thermofisher, and genotyping of the breeders’ array was conducted at Neogen (NE, USA), on Affymetrix GeneTitan.
SNP Array Data Analyses
SNP array data were analyzed with Axiom Analysis Suite 5.0 software (Thermofisher, CA) following the Best Practices Workflow with recommended threshold settings (DishQC ≥ 0.82, QC call rate ≥ 97%, average call rate for passing samples ≥ 98.5%). A marker-conversion rate was calculated as the percentage of SNPs on the SNP arrays that were polymorphic, passed QC thresholds, and were classified as BestAndRecommended by the Best Practice Workflow. Genotype data were exported in text or the PLINK format, which was converted to other formats for downstream applications.
Concordance rate was calculated to estimate genotyping accuracy. Parent and progeny genotypes from the full-sib family were used to detect genotypes unexpected from Mendelian inheritance. Principal component analysis (PCA) was conducted to assess genetic differentiation between selected populations and their wild, progenitor populations with genotype data for markers on the screening array and the breeders’ array to compare the two platforms. All genotype data for PCA were from the screening array with one analysis using all markers on the screening array and another using the markers on the breeders’ array. As non-random association between alleles at different loci (i.e., linkage-disequilibrium LD) can bias estimates of population structure (Lotterhos 2019), we performed LD-pruning (default settings, r < 0.2) to reduce statistical non-independence between SNPs using SNPRelate (Zheng et al. 2012). In addition, genetic relatedness among individuals was estimated using KING software (Manichaikul et al. 2010). The PC-AiR algorithm implemented in the GENESIS package was used to partition the samples into unrelated subsets to reduce confounding signals from family structure, with a default kinship threshold of 0.022 (Conomos et al. 2015). The ancestry divergence calculated from KING was used to select which individuals from a set of relatives that carried the most unique ancestry information and should be given priority for inclusion in the unrelated subset (Conomos et al. 2015). The PC-AiR function from GENESIS was then used to perform PCA analysis using LD-pruned SNPs and unrelated individuals (202 of 219).
For pathogen probes, we conducted a fixed-region, pseudo-copy number analysis where each pathogen was considered a genomic region, and signal strength from all probes was used to infer the copy number of the pathogen. A median Log2 signal ratio was calculated for all samples, which was expected to follow a normal distribution around a population mean of background signals. The positive pathogen controls and putatively infected samples were expected to have signals above this normal distribution.
Results
SNP Discovery and Design of the Screening Array
From the 269 resequenced oysters from Atlantic populations, we obtained 10,628,193 biallelic SNPs that were called in > 90% of individuals (minimum of 5 reads to call) and had a MAF of 0.05 or higher. Removing SNPs that had another SNP or indel variant within 20 bp brought the number down to 2.74 million SNPs. From the Gulf of Mexico dataset, 913,412 SNPs were obtained at a 95% call rate, MAF > 0.10, and no other variant within 20 bp. A final set of 3,661,861 SNPs was submitted to Thermofisher for evaluation and probe design, including ~ 32 K SNPs that were identified as Fst outliers based upon population genetics analysis and ~ 11 K genic SNPs that were identified in previous studies as associated with growth, disease, and stress resistance. Also submitted for array design were 33 previously identified SNPs from the mitochondrial genome, and 399 non-polymorphic sequences from 13 oyster pathogens collected from GenBank. For each SNP, a 71-mer sequence with 35 bp on either side of the SNP was submitted for probe design.
All designed probes were assessed for genomic duplication, interaction with other probes, distance from known polymorphism, and the probability of probe conversion. The evaluation produced 2,691,166 recommended SNPs that had a probability of conversion > 0.60, and no genomic duplication or other interfering polymorphism. Except for some SNPs of special interest from previous studies, SNPs of the A/T and C/G type were not used because they require two probe positions on the array. All recommended genic SNPs (two per gene) were tiled on the screening array and then the array was filled with intergenic SNPs with a MAF > 0.10. Intergenic SNPs with MAF 0.05–0.09 were used only to fill large gaps for even distribution across the genome. The final design of the screening array contained 568,396 probes covering 567,018 markers, including 566,262 SNPs from the eastern oyster (566,229 nuclear and 33 mitochondrial) and 756 probes from 13 pathogens (Table 2). The number of probes per pathogen placed on the array can be found in Supplementary Table 2.
Of the nuclear SNPs, 47,426 were genic SNPs located in the body of 23,936 protein-coding genes and representing ~ 69.2% of all genes in the genome. The nuclear SNPs were distributed evenly across chromosomes (Fig. 1), with an average interval of 1207 bp ranging from 1185 to 1216 bp among chromosomes (Supplementary Table 3). The maximum interval ranged from 576,288 to 1,200,005 bp on the 10 chromosomes. Because the screening array contains 566,262 SNPs from the eastern oyster, it is hereafter referred to as the 566K SNP array. The official name of the eastern oyster 566K SNP array is Applied Biosystems Axiom CvSNP600 (96-plate format).
Performance of the Screening Array
To evaluate the performance of the 566K screening array and identify best SNPs for the breeders’ array, 960 wild and selected oysters were genotyped. Of the samples tested, 842 (87.9%) passed the genotyping QC (DQC > 0.82 and QC call rate > 97%). Most of the failed samples came from three sample sets that were apparently not fixed well (some discoloration) or archived pathogen control samples that had low or no eastern oyster DNA. The other groups had high sample passing rates ranging from 93.8 to 100% (Table 3). For all passing samples, the average cluster call rate of the screening array was 98.89%, and the sample reproducibility was 99.14%.
After genotype calling with the Best Practices Workflow, the 566K SNPs on the screening array were classified into six categories: (1) PolyHighResolution, polymorphic SNPs with well-separated genotype clusters; (2) NoMinorHom, polymorphic SNPs with one of the homozygous genotype missing; (3) MonoHighResolution, monomorphic SNPs with one well-defined genotype cluster; (4) CallRateBelowThreshold, SNPs with a call rate below the threshold < 0.97; (5) OffTargetVariant (OTV), SNPs with a possible OTV cluster; and (6) other, SNPs with more than one problematic issue. We identified 223,433, 50,277 and 26,189 SNPs belonging to the PolyHighResolution, NoMinorHom, and MonoHighResolution clusters, respectively, producing a total number of 299,899 BestandRecommended markers (52.96% of the total; Table 4). The number of polymorphic and recommended SNPs was 273,710, corresponding to a marker conversion rate of 48.34%. The other markers were either below the call rate threshold (58,087 or 10.26%), OTVs (32,269 or 5.7%), or others (176,007 or 31.08%) that had one or more clusters below threshold. Examples of genotyping clustering are given in Supplementary Fig. 1. Of all SNPs genotyped in the 842 oysters passing genotyping QC, 142,652 SNPs (25.2%) had a MAF < 0.05, and 241,665 SNPs (42.7%) had a MAF < 0.10 (Fig. 2).
SNPs for the Breeders’ Array
For the breeders’ array, candidate SNPs were selected based upon their MAF and performance on the 566K array. Only recommended markers from the 566K array that are polymorphic in the panel of 960 oysters (excluding 118 failed genotyping QC) were considered. A set of 42,823 priority SNPs was selected based upon quality/performance, polymorphism, and perceived usefulness for the consortium. The priority SNPs consisted of 17,275 SNPs from the VIMS population, 6,019 SNPs showing large Fst from a population study, and 22,984 genic SNPs from protein-coding genes (with 3,455 overlapping). The genic SNPs included 922 in genes associated with shell growth, disease, and stress resistance identified in previous studies. Most of the selected SNPs had a call rate ≥ 98% and FLD (Fisher’s linear discriminant measuring cluster quality) ≥ 5 (63.8%), and a MAF ≥ 0.05 (92.5%). In addition to these priority SNPs, 49,806 best-performing SNPs were selected to fill the breeders’ array. All best-performing SNPs had a call rate ≥ 99%, FLD ≥ 5, no call and OTV calls ≤ 2%, and MAF ≥ 0.10. The two sets of SNPs were submitted to Thermofisher for array design. The final design consisted of 66,649 markers, including 65,893 SNPs from the eastern oyster (65,862 nuclear and 31 mitochondrial) and 756 probes from 13 pathogens (Table 2). The nuclear markers covered 22,984 protein-coding genes, with one SNP per gene. The markers were distributed evenly across chromosomes (Fig. 1), with an average interval per chromosome of 10,465 bp, ranging from 9048 to 12,097 bp (Supplementary Table 3). The maximum interval was 1,538,165 bp for chromosome 1 and ranged from 720,746 to 1,031,408 bp for chromosomes 2–9. Because the breeders’ array contains 65,893 SNPs from the eastern oyster, it is hereafter referred to as the 66K SNP array. The official name of the eastern oyster 66K SNP array is Applied Biosystems Axiom OysterCv array (384-plate format). Of all SNPs on the 66K array, 3189 SNPs (4.8%) had a MAF < 0.05, and 15,308 SNPs (22.8%) had a MAF < 0.10 as assessed in the 842 oysters from Atlantic populations that passed genotyping QC (Fig. 2).
Performance of the Breeders’ Array
Of the 384 samples tested with the 66K array (Axiom OysterCv), one sample failed because of insufficient amount of DNA, eight had DQC values below the threshold of 0.82, and 15 had call rates below the recommended rate of 97%, leading to an overall sample passing rate of 93.8% (Table 5). Adductor muscle samples fixed for up to 18 years had a passing rate of 83–100%. The passing rate was not determined by the age of the samples, but by whether or not the samples were fixed well or had enough cells. Nine of the 10 samples that were fixed for 18 years passed genotyping QC, along with all 6 samples that were fixed but dried following evaporation of ethanol. Mantle or gill tissues fixed for about 1 year had a 100% passing rate (Table 5). Small spat tissue, with or without shells, had lower sample pass rates probably because of small size, insufficient fixing, and/or the presence of shells. Biopsied hemolymph had an overall lower passing rate, likely attributable to insufficient DNA, and samples that contained salt crystals had lower sample passing rate than those without salt crystals (Table 5). Seven of the eight DNA samples extracted by the Kurabo system passed genotyping QC.
In the 360 samples passing QC, the 66K array yielded 57,136, 3,512 and 25 SNPs belonging to the PolyHighResolution, NoMinorHom, and MonoHighResolution clusters, respectively, producing a total number of 60,673 BestandRecommended markers (92.08% of the total; Table 4). The number of polymorphic and recommended SNPs was 60,648, corresponding to a marker conversion rate of 92.04%. The other markers were either below the call rate threshold (1468 or 2.23%), OTVs (938 or 1.42%), or others (2814 or 4.27%) that had one or more clusters below threshold (Table 4). Of the 10 samples that were replicated on the 66K array, the average concordance rate was 99.81%, which means only 0.19% of the SNPs yielded different genotypes if they were genotyped twice. Most of the 10 samples had high concordance rates of 99.94–100.00%; whereas, two had relatively lower concordance rates of 99.02% and 99.34 (Table 6), probably because of variations in DNA quality.
Non-Mendelian Segregation
The high concordance rates suggest that array genotyping is highly reproducible with error rates mostly below 0.2%. Analysis of trio segregation patterns in the full-sib family, however, revealed significant proportions of loci showing unexpected progeny genotypes that were inconsistent with parental genotypes and Mendelian segregation. For example, some loci with parental genotypes of AB × AA produced the BB genotype in approximately 25% of the progeny, which can be explained only by a null allele and the parental genotypes of AB × A-. Such segregation patterns often were observed at two–three neighboring loci on the chromosome flanked by loci showing perfect Mendelian segregation (Table 7), suggesting that they were indeed caused by large deletions on the chromosome, not by genotyping error in parents, as the same error occurring randomly at two adjacent loci should be extremely rare. In two of the examples in Table 7, two and three consecutive SNPs on chromosomes 1 and 5 had ~ 25% BB (B-) genotypes that are possible only with a null allele or deletion in the father, and the deletion spanned at least 132,686 bp and 139,520 bp, respectively. The presence of null alleles or deletions was further supported by the excess of AA genotypes which included A- genotypes. For the 566K array, null alleles were observed in 13.1% of the loci. By selecting high-quality SNPs, the proportion of loci showing unexpected genotypes was reduced to 6.7% in the same family for markers on the 66K array.
Stock Identification
A large number of samples from diverse populations was used to evaluate the 566K array, primarily for characterizing SNP variation and performance for the design of the breeders’ array. Analyses of all populations genotyped are complex and beyond the scope of this paper. To demonstrate the utility of the SNP arrays in stock identification, we conducted PCA of two pairs of wild and selected populations: wild samples from Long Island Sound population (LIW1 and LIW2), selected lines derived from LIW (NEH1 and NEH2), wild samples from Delaware Bay population (DBW1 and DBW2), and a selected line derived from DBW (DBX2). Data from both the 566K and the 66K SNPs produced similar patterns, indicating that the 66K SNPs are as informative as the 566K SNPs in differentiating selected from wild populations (Fig. 3). The two wild populations showed some genetic differentiation and formed two overlapping clusters. Data from the 566K SNPs produced slightly better separation of the two wild populations than data from the 66K SNPs (Fig. 3). Within each wild population, the two locations sampled were indistinguishable. On the other hand, the selected populations were clearly separated from corresponding wild progenitor populations and from each other with marker sets for both arrays (Fig. 3).
Mitochondrial SNPs
Of the 31 mitochondrial SNPs (mtSNPs) on both arrays, three were monomorphic in all samples screened in this study, including 263 from a Florida wild population (Goose Point, Gulf of Mexico) and 50 from a selected line derived from Gulf populations. Oysters from Gulf populations were included to confirm that some of the alleles were indeed Gulf-specific. No heterozygotes were observed in any individuals across all mtSNPs, confirming the haploid nature of mitochondrial markers and an absence of heteroplasmy. Most polymorphic mtSNPs included on the arrays showed distinctive allele frequencies between Atlantic and the Gulf populations, with 11 loci fixed for different alleles, providing useful diagnostic markers (Supplementary Table 4).
Analysis with these mtSNPs revealed the presence of Gulf-specific mitochondrial alleles in some Atlantic populations. No Gulf-specific alleles were observed in Atlantic wild populations, except in the Barnegat Bay wild population from New Jersey, wherein three of the 48 spat (6.25%) had a Gulf mitochondrial genome (Supplementary Table 4). The classification is unambiguous because the three spat had Gulf alleles at all 11 diagnostic mtSNPs. In some Atlantic wild populations, dominant Gulf alleles were observed at some non-diagnostic loci in a few individuals (< 3%), which does not mean that the oysters were derived from the Gulf, as all other loci were distinctively Atlantic. The rare occurrence of Gulf alleles at some loci in Atlantic individuals is likely attributable to independent substitution events leading to the same nucleotide.
Oysters with Gulf-specific alleles at all 11 diagnostic mtSNPs were detected in three selected populations with known history of crossbreeding with oysters from the Gulf of Mexico. The VIMS population, which has been crossed with oysters from the Gulf, had Gulf mtSNP alleles in 23% of the 139 oysters genotyped (Supplementary Table 4). The NEG line from Rutgers was derived from Gulf female × NEH male crosses that subsequently were backcrossed to mostly NEH males for four generations. Expectedly, most of the NEG oysters (13 of 18, or 72.2%) had Gulf mitochondria. In one NEH subline, 3.1% of the oysters contained diagnostic Gulf mtSNP alleles, probably arising from rare hatchery cross-contamination from NEG.
Pathogen Probes
To assess the efficacy of the pathogen probes, we analyzed two plates of samples (n = 192) from the 566K array in which the positive pathogen controls were located. Probes for all 13 pathogens were effective in detecting the presence of pathogens in positive controls, although some of the probes lacked species specificity (Fig. 4).
Probes for the three Bonamia species detected 2–16 fold higher signals in Bonamia ostreae and Bonamia perspora positive controls than the median background signal. Although there was cross-reaction between probes for the two species, B. ostreae probes produced more than two-fold stronger signals in B. ostreae positive controls than in the B. perspora control (Fig. 4). Conversely, B. perspora probes produced two-fold stronger signals in B. perspora than in B. ostreae positive controls, indicating some species discrimination. The positive control for Bonamia exitiosa showed weak signals under probes for all three species, probably because the lone positive control sample did not contain enough parasites. Other than the positive controls, oysters screened in this study did not show a higher signal than the narrow and balanced distribution of MedianLog2Ratio, suggesting that the screened oysters either did not have these parasites or their parasite load was significantly lower than the positive controls.
Probes for Haplosporidium costale showed higher signals in H. costale positive controls, and Haplosporidium nelsoni showed higher signals in H. nelsoni positive controls (Fig. 4). Signals from H. nelsoni controls were overall weak, probably because the positive controls were not fixed well or contained parasite DNA below the detection limit. Signal distribution for H. costale and H. littoralis was narrow and balanced, suggesting no parasite infection in the samples screened or infection below detection limits. For H. nelsoni, the signal distribution of test samples is heavily skewed toward positives, passing beyond the positive controls, indicating that some of the samples might be infected with H. nelsoni. Samples showing signals above that of positive controls were mostly from North Carolina (n = 13), Long Island Sound (n = 10), Chesapeake Bay (n = 3), and Delaware Bay (n = 2), accounting for 20%, 24%, 15%, and 6% of samples screened in the respective populations. None of the 16 samples from Maine appeared positive.
Positive controls for Perkinsus marinus, Aliiroseovarius crassostreae, and Vibrio species were oyster samples spiked with isolated/purified pathogens and thus produced high signals relative to other samples (Fig. 4). Signal MedianLog2Ratio distribution indicated that some of the oysters screened might be positive for these pathogens. For P. marinus, 23 oysters had signals > 2 fold higher than the median. These samples were from Delaware Bay (n = 13), Chesapeake Bay (n = 6), and North Carolina (n = 4), accounting for 48%, 30%, and 6% of the samples in the respective populations. None of the samples from Maine or Long Island Sound showed levels above the threshold for P. marinus. For A. crassostreae, most of the samples with signals above the normal distribution were from Delaware Bay and North Carolina.
Probes for Vibrio coralliilyticus did not show differences in signal strength between V. coralliilyticus and V. parahaemolyticus positive samples, but V. parahaemolyticus probes produced stronger signals in V. parahaemolyticus than in V. coralliilyticus samples (Fig. 4). The Vibrio tubiashii probes also showed stronger signals in V. parahaemolyticus than in V. coralliilyticus samples (Fig. 4), as did V. vulnificus probes (data not shown here). These patterns may be explained by some species preference of the probes or higher bacterium concentrations in the V. parahaemolyticus positive controls. Because of the cross-reaction among the Vibrio probes and the possible presence of other Vibrio species, we did not analyze the Vibrio results further.
OsHV-1 probes recognized OsHV-1 positive controls and seemed to suggest that some of the samples were positive (Fig. 4). Fourteen of the 15 positives showing 2-fold increase over the median signal were all from the panel of 32 pathogen controls that were assembled and processed for DNA extraction in one batch at the same time, which opens the possibility of cross-contamination in the lab. One of the positive samples was from North Carolina in a different batch.
Discussion
Array Performance
Genomes of oysters and other marine bivalves are highly polymorphic, and the high polymorphism poses a challenge for SNP array design because of difficulties identifying probes free of unwanted polymorphism. We used a two-step process, first developing a high-density screening array and then a breeders’ array using best-performing probes from the screening array. Screening with a large number of oysters (n = 960 minus 118 failed QC) from diverse Atlantic populations allowed us to identify polymorphic and high-performance SNPs for the breeders’ array. Consequently, the marker conversion rate increased greatly from 48.33% for the screening array to 92.04% for the 66K breeders’ array, and the latter is higher than that of the two SNP arrays for the Pacific oyster that did not go through the two-step process (70.4% and 68.2%, respectively) (Gutierrez et al. 2017; Qi et al. 2017). The two-step process also improved genotyping accuracy enabled by the selection of high-quality SNPs and resulted in a concordance rate of 99.81%, compared with the 96.6% reported for one of the Pacific oyster arrays (Qi et al. 2017). The low conversion rate of the screening array (48.33%) is probably caused by high polymorphism, less stringent filtering and the inclusion of SNPs (~ 25%) from the genetically distinctive Gulf population. Our results together with results from previous studies show that marker conversion rate is low in species with high polymorphism, and the two-step process can greatly improve marker conversion rate and the accuracy of the SNP array. Improved marker conversion rate and accuracy increase the efficiency of the SNP array and reduce per-locus genotyping cost, which is important for the breeders’ array. The cost of the arrays and genotyping services may vary over time and depending on sample volume and the service provider. Currently, the array cost including all necessary reagents is US $70/sample for the CvSNP600 screening array (96-plate format) and US $18/sample for the 66K OysterCv breeders’ array (384-plate format). Genotyping service from tissue to genotype costs another US $12–$15/sample depending on the array (with the screening array being more expensive because of longer machine time) and service provider.
Molluscs are known to be rich in mucopolysaccharides, polyphenolic proteins, and pigments that may hinder DNA extraction and subsequent sequencing or genotyping applications (Adema 2021). Our results show that the two SNP arrays worked well with oyster DNA extracted from various tissue types, including those fixed with ethanol (95 or 100%) for many years. Although aged samples were more likely to fail on the array, failed samples often were associated with poorly-fixed (discolored) tissue rather than years fixed. A common mistake is fixing too much tissue in a small volume of ethanol. In our experience, it is important to keep a 1:5 (v:v) ratio of tissue to ethanol (absolute) or change fixative within 24 h. If fixed well, samples fixed for 12–18 years (stored at room temperature) can be genotyped successfully on the SNP arrays, including samples that were dry following ethanol evaporation. Sampling live oysters by biopsy is important for GS. Biopsied hemolymph samples had a higher failure rate than other samples, probably because there were not enough hemocytes in some of the samples (~ 1 ml). Biopsied hemolymph should be centrifuged and pelleted before adding ethanol to prevent the formation of salt crystals.
It should be noted that the 66K array is designed for use with Atlantic populations that are of primary interest to EOBC. SNPs on the 66K array were selected based upon performance in 842 oysters from Atlantic populations only. As the Gulf of Mexico and Atlantic populations are genetically distinct (Reeb and Avise 1990; Karl and Avise 1992; Hoover and Gaffney 2005), some of these markers may not be polymorphic or informative in Gulf populations, leading to lower marker efficiency or ascertainment bias. In another study, the 66K array yielded 54,754 BestandRecommended markers (83.1%) and 51,723 recommended and polymorphic markers, corresponding to a marker conversion rate of 78.5% in 677 wild Gulf oysters from Florida (Supplementary Table 5), which is lower than that for the Atlantic populations but still higher than the marker conversion rate for the Pacific oyster arrays. The 566K screening array contains SNPs from both Atlantic and Gulf populations, which can be used to identify best-performing markers for a Gulf array if it is needed. Also, the 66K array contains ~ 17 K SNPs selected from the VIMS population to accommodate the family-based selection program (Allen et al. 2021). Some of these SNPs may be less polymorphic in some other Atlantic populations, which may create bias when comparing genetic diversity of the VIMS population with that of other populations, but not among non-VIMS populations. Selecting SNPs based on MAF in particular populations, while useful for breeding, may create ascertainment bias in genetic analysis of natural populations, which should be assessed in future studies.
Null Alleles
The finding of a large proportion of loci (13.1%) showing unexpected progeny genotypes was surprising. We show that the unexpected progeny genotypes were not caused by genotyping error (which was only 0.19%), but by null alleles. Null alleles cause the false appearance of non-Mendelian segregation in families and heterozygote deficiency in populations. Null alleles have been observed in both allozyme and microsatellite data. For microsatellites, null alleles have been attributed to polymorphism at the PCR priming site (Hedgecock et al. 2004; Wang and Guo 2007). Our results suggest that deletions, rather than polymorphism at priming sites, are the main cause of null alleles with DNA markers. Polymorphism was stringently filtered out during probe design, and the chance of polymorphism occurring in two–three adjacent probes should be extremely small. Our null allele observations may be related to the high genetic load that has been documented in C. gigas (Launey and Hedgecock 2001; Li and Guo 2004; Plough and Hedgecock 2011; Plough et al. 2016) and high fecundity invertebrates in general (Plough 2016). In the eastern oyster, segregation distortion attributable to genetic load is variable and lower in selected strains, possibly because of purging by selection (Yu and Guo 2003; Reece et al. 2004; Wang and Guo 2007; Wang et al. 2009; Zhang and Guo 2010). As shown in the two examples in Table 6, not all null alleles cause segregation distortion, but deletion of one allele could affect fitness by altering gene expression and/or exposing recessive lethal mutations.
Heterozygote deficiency is a common phenomenon in marine bivalves, which has been explained by nonrandom mating, selection, population structure (Wahlund effect), and null alleles (Zouros and Foltz 1984; Foltz 1986; Gaffney et al. 1990). The finding of a high proportion of loci with null alleles in this study argues for a major role of null alleles or genomic deletions contributing to heterozygote deficiency. If so, the observed heterozygote deficiency may be an artifact caused by the pervasive deletions in the genome. Genomic deletions can also explain overdominance in growth and viability observed in marine bivalves (Zouros et al. 1988).
Genomic analysis in the eastern oyster has revealed extensive duplications that account for 16.5% of the genome (Modak et al., 2021). These duplications are copy number variations (CNVs) that are 161–1,489,627 bp in length and present in varying copy numbers in different individuals. Individuals with fewer copies can be considered having deletions at the loci. Thus, our finding of massive null alleles (13.1%) caused by deletions is consistent with Modak et al.’s (2021) finding of extensive duplications, suggesting a large portion of the eastern oyster genome has CNVs. The 13.1% observed in this study is likely an underestimate of CNVs, as CNVs with more than two copies cannot be detected by null alleles or presence-absence variations observed in this study.
In randomly mated populations, single-locus deviations from Hardy–Weinberg equilibrium (HWE) are often interpreted as an indication of selection. SNPs with null alleles may falsely appear to deviate from HWE. The presence of null alleles should be taken into consideration when inferring selection from HWE deviations. Here, analyses of trio segregation data provided insights that may generally help minimize problems with null alleles when novel samples are analyzed. Null alleles were often associated with loci showing lower quality scores, such as lower FLD, lower HetSO (heterozygous strength offset), lower call rates, or higher numbers of OTVs, which probably is attributable to signal reduction from a null allele. The proportion of SNPs with null alleles was greatly reduced in the 66K array following the selection of higher-quality SNPs. Similarly, for any sample on any array, calling following the Best Practices Workflow and subsequent filtering of genotypes with stringent quality criteria may help filter out loci with null alleles. Even with stringent calling and filtering, some loci with null alleles will persist, and caution is needed when inferring selection based upon HWE deviations alone.
Identification of Selected Stocks
The PCA analysis easily separated selected and wild populations without ambiguity, similar to results reported for C. gigas using SNPs (Hedgecock and Pan 2021), demonstrating the power of genome-wide SNPs. Data from the two marker sets (566K and 66K) produced nearly identical patterns (Fig. 3), indicating that the 66K array can be as effective as the 566K array at a reduced cost. Understanding recruitment contributions from aquaculture stocks is important for fishery management. The mitochondrial genome is powerful in tracking genetic introgression because of its lack of recombination and LD decay. The finding that three oysters from Barnegat Bay (6.25%) have the Gulf mitochondrial genome is surprising. These individuals clearly have some Gulf ancestry, as their Gulf mitochondrial genome was implicated by multiple diagnostic markers, and no Gulf mitochondrial genome was detected in any other wild Atlantic populations, albeit with the limited sampling conducted so far. These oysters certainly had ancestors that carried Gulf mtDNA and were introduced by human activities. We are not aware of any historical introduction of Gulf oysters to Barnegat Bay; however, there are oyster farms and oyster restoration projects in Barnegat Bay that have used selected, disease-resistant stocks (Rutgers NEH, NEG and VIMS DEBY) that contain some Gulf mitochondrial genome (Thompson et al. 2022). Because mtDNA does not recombine when backcrossed into the local wild oysters, Gulf genomes observed today could have been introduced at any time and maintained in the population. Thus, unlike the estimate of recruitment from Gulf oysters by Milbury et al. (2004) based on a single large planting, it is difficult to estimate contributions to recruitment from selected strain introductions in Barnegat Bay that may have happened over time. Nevertheless, Barnegat Bay oysters with Gulf mitochondria are undoubtedly derived from human introductions. Assuming the mitochondrial genomes were from selected, disease-resistant stocks, the observed 6.25% Gulf type may represent a serious underestimate of the overall contribution to oyster recruitment in Barnegat Bay from these stocks, because only a fraction of the disease-resistant stocks carried the Gulf mitochondrial genome (ranging from 3.1 to 72.2%, Supplementary Table 4). The successful detection of significant human-mediated recruitment in Barnegat Bay, a small and enclosed system where natural recruitment of oysters is limited, is not entirely unexpected. This study provides a clear case of aquaculture, restoration or other human activities contributing to oyster recruitment, but additional study will be needed to quantify that contribution.
Pathogen Detection
Diseases are a major concern for the oyster aquaculture industry, and disease resistance is a major target trait for eastern oyster breeding (Ford and Tripp 1996; Guo and Ford 2016; Guo 2021). Ideally, breeding programs should determine the infection status of genotyped oysters when breeding for disease resistance. Most of the pathogen probes were able to detect high signals in positive control samples. Test samples showed a range of values, from non-detectable to levels comparable to the positive controls. These results suggest that the arrays have the potential to provide data on pathogen abundance in genotyped oysters. For B. exitiosa and H. nelsoni, the lower than expected signals may be a consequence of low parasite counts in these positive control samples below detection limits. Probes for congeneric species showed some species specificity, but cross-reaction is evident, especially for Vibrio species. For H. nelsoni, P. marinus, A. crassostreae, and some Vibrio species, the distribution of the signal MedianLog2Ratio is clearly skewed toward high signals (Fig. 4). Signals several folds above the normal range and close to that of positive controls strongly indicate the presence of parasites or infections of increasing intensity. The distribution of samples that likely contained H. nelsoni, P. marinus, A. crassostreae, and Vibrio species DNA is consistent with known occurrences of these species in our region. Although preliminary, the results of this study suggest that these pathogen probes can detect most positive control samples and are likely useful in detecting pathogens beyond a certain threshold. We should note that, other than the few positive controls, we did not independently verify the infection status of the oysters on the array. Samples with exceptionally high signals are likely positive, but the presence of low amounts of DNA does not equate directly to infection (Piesz et al. 2022). Oysters with low pathogen signals may have pathogens at levels below the detection limits of the arrays. Further studies are needed to improve species specificity, determine detection limits, validate infection status, and establish the relationship between parasite load and signal strength for possible quantification of parasite infections. This preliminary assessment indicates that detection of positive controls is possible, but extensive testing and validation are needed for each pathogen before the arrays can be used for diagnosis.
Conclusions
This study developed two high-density SNPs arrays for the eastern oyster, an important aquaculture species in North America. The 566K SNP array contains SNPs from both Atlantic and Gulf of Mexico populations, which can be used for marker screening and high-density genotyping. The 66K SNP array is highly reproducible and efficient, providing a useful tool for genomic research and selection. Our study shows that marker conversion rate is initially low in this highly-polymorphic species, and the two-step process greatly improved the performance and efficiency of the 66K SNP array. Null alleles were observed in a significant proportion of SNPs because of pervasive deletions or copy number variations in the genome; this was reduced on the 66K array but may still affect some downstream applications. The 66K array can discriminate selected lines from wild populations, with mtSNPs providing unambiguous detection of introgression of Gulf genotypes into Atlantic populations. The two arrays will advance genomic research and accelerate genetic improvement of eastern oyster strains by delineating genetic architecture of production traits and enabling genomic selection. The arrays also will be useful for monitoring pedigree and the level of inbreeding in hatchery stocks, identifying selected stocks and their introgression into wild populations, and assessing the successful retention of genetic variation in hatchery-supported oyster restoration.
Data Availability
All DNA sequence reads generated from this study were submitted to National Center for Biotechnology Information (NCBI) Sequence Read Archive under BioProject PRJNA890664.
References
Adema CM (2021) Sticky problems: extraction of nucleic acids from molluscs. Phil Trans R Soc B Biol Sci 376:20200162
Allen SK Jr, Small JM, Kube PK (2021) Genetic parameters for Crassostrea virginica and their application to family-based breeding in the mid-Atlantic, USA. Aquaculture 538:736578
Beck MW, Brumbaugh RD, Airoldi L, Carranza A, Coen LD, Crawford C, Defeo O, Edgar GJ, Hancock B, Kay MC, Lenihan HS, Luckenbach MW, Toropova CL, Zhang G, Guo X (2011) Oyster reefs at risk and recommendations for conservation, restoration, and management. Bioscience 61:107–116
Chen S, Zhou Y, Chen Y, Gu J (2018) fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890
Conomos MP, Miller MB, Thornton TA (2015) Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet Epidemiol 39:276–293
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker GL, Marth GT, Sherry ST, McVean G, Durbin R (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158
Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, Li H (2021) Twelve years of SAMtools and BCFtools. GigaScience 10:giab008
Eierman LE, Hare MP (2014) Transcriptomic analysis of candidate osmoregulatory genes in the eastern oyster Crassostrea virginica. BMC Genomics 15:503
Fao (2022) Global aquaculture production. Fisheries and aquaculture division [online]. Rome
Foltz DW (1986) Null alleles as a possible cause of heterozygote deficiencies in the oyster Crassostrea virginica and other bivalves. Evolution 40:869–870
Ford SE, Tripp MR (1996) Diseases and defense mechanisms. In: Newell RIE, Kennedy VS, Eble AF (eds) The Eastern Oyster Crassostrea virginica. Maryland Sea Grant College
Frank-Lawale A, Allen SK, Dégremont L (2014) Breeding and domestication of eastern oyster (Crassostrea virginica) lines for culture in the Mid-Atlantic, USA: line development and mass selection for disease resistance. J Shellfish Res 33:153–166
Gaffney PM, Scott TM, Koehn RK, Diehl WJ (1990) Interrelationships of heterozygosity, growth rate, and heterozygote deficiencies in coot clam, Mulinia lateralis. Genetics 124:687–699
Galtsoff PS (1964) The American Oyster, Crassostrea virginica Gmelin. Fish Bull 64:1–488
Garrison E, Marth G (2012) Haplotype-based variant detection from short-read sequencing. arXiv Preprint arXiv:1207.3907
Garrison E (2016) Vcflib, a simple C++ library for parsing and manipulating VCF files
Gómez-Chiarri M, Warren WC, Guo X, Proestou D (2015) Developing tools for the study of molluscan immunity: the sequencing of the genome of the eastern oyster, Crassostrea virginica. Fish Shellfish Immunol 46:2–4
Grabowski JH, Brumbaugh RD, Conrad RF, Keeler AG, Opaluch JJ, Peterson CH, Piehler MF, Powers SP, Smyth AR (2012) Economic valuation of ecosystem services provided by oyster reefs. Bioscience 62:900–909
Guo X (2021) Genetics in shellfish culture. In: Shumway SE (ed) Molluscan Shellfish Aquaculture: A Practical Guide. 5m Books Ltd, Essex
Guo X, Ford SE (2016) Infectious diseases of marine molluscs and host responses as revealed by genomic tools. Phil Trans R Soc B 371:20150206
Gutierrez AP, Turner F, Gharbi K, Talbot R, Lowe NR, Peñaloza C, Mccullough M, Prodöhl PA, Bean TP, Houston RD (2017) Development of a medium density combined-species SNP array for Pacific and European oysters (Crassostrea gigas and Ostrea edulis). G3 Genes|Genomes|Genetics 7:2209–2218
He Y, Yu H, Bao Z, Zhang Q, Guo X (2012) Mutation in promoter region of a serine protease inhibitor confers Perkinsus marinus resistance in the eastern oyster (Crassostrea virginica). Fish Shellfish Immunol 33:411–417
He Y, Jouaux A, Ford SE, Lelong C, Sourdaine P, Mathieu M, Guo X (2015) Transcriptome analysis reveals strong and complex antiviral response in a mollusc. Fish Shellfish Immunol 46:131–144
Hedgecock D, Pan FTC (2021) Genetic divergence of selected and wild populations of Pacific oysters (Crassostrea gigas) on the West Coast of North America. Aquaculture 530:735737
Hedgecock D, Li G, Hubert S, Bucklin K, Ribes V (2004) Wild spread null alleles and poor cross-species amplification of microsatellite DNA loci cloned from the Pacific oyster, Crassostrea gigas. J Shellfish Res 23:379–385
Hoover CA, Gaffney PM (2005) Geographic variation in nuclear genes of the eastern oyster, Crassostrea virginica Gmelin. J Shellfish Res 24:103–112
Houston RD, Taggart JB, Cézard T, Bekaert M, Lowe NR, Downing A, Talbot R, Bishop SC, Archibald AL, Bron JE, Penman DJ, Davassi A, Brew F, Tinch AE, Gharbi K, Hamilton A (2014) Development and validation of a high density SNP genotyping array for Atlantic salmon (Salmo salar). BMC Genomics 15:90
Institute B (2016) Picard tools. Broad Institute, GitHub repository. https://broadinstitute.github.io/picard/
Karl SA, Avise JC (1992) Balancing selection at allozyme loci in oysters: implications from nuclear RFLPs. Science 256:100–102
Korte A, Farlow A (2013) The advantages and limitations of trait analysis with GWAS: a review. Plant Methods 9:29
Launey S, Hedgecock D (2001) High genetic load in the pacific oyster Crassostrea gigas. Genetics 159:255–265
Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595
Li L, Guo X (2004) AFLP-based genetic linkage maps of the Pacific oyster Crassostrea gigas Thunberg. Mar Biotechnol 6:26–36
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Li C, Wang H, Guo X (2022) Regulation of the cell cycle, apoptosis, and proline accumulation plays an important role in the stress response of the eastern oyster Crassostrea virginica. Front Mar Sci 9:921877
Liu S, Sun L, Li Y, Sun F, Jiang Y, Zhang Y, Zhang J, Feng J, Kaltenboeck L, Kucuktas H (2014) Development of the catfish 250 K SNP array for genome-wide association studies. BMC Res Notes 7. https://doi.org/10.1186/1756-0500-7-135
Loren DC, Robert DB, David B, Ray G, Mark WL, Martin HP, Sean PP, Tolley SG (2007) Ecosystem services related to oyster restoration. Mar Ecol-Prog Ser 341:303–307
Lotterhos KE (2019) The effect of neutral recombination variation on genome scans for selection. G3 Genes|Genomes|Genetics 9:1851–1867
Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen W-M (2010) Robust relationship inference in genome-wide association studies. Bioinformatics 26:2867–2873
Mann R, Powell EN (2007) Why oyster restoration goals in the Chesapeake Bay are not and probably cannot be achieved. J Shellfish Res 26:905–917
Meuwissen T, Hayes B, Goddard M (2016) Genomic selection: a paradigm shift in animal breeding. Anim Front 6:6–14
Milbury CA, Gaffney PM, Meritt DW, Newell RIE (2004) Mitochondrial DNA markers allow monitoring of oyster stock enhancement in the Chesapeake Bay. Mar Biol 145:351–359
Modak TH, Literman R, Puritz JB, Johnson KM, Roberts EM, Proestou D, Guo X, Gomez-Chiarri M, Schwartz RS (2021) Extensive genome-wide duplications in the eastern oyster (Crassostrea virginica). Phil Trans R Soc B Biol Sci 376:20200164
Palti Y, Gao G, Liu S, Kent M, Lien S, Miller M, Rexroad C, Moen T (2015) The development and characterization of a 57 K single nucleotide polymorphism array for rainbow trout. Mol Ecol Resour 15. https://doi.org/10.1111/1755-0998.12337
Piesz J, Scro AK, Corbett R, Lundgren KM, Smolowitz R, Gomez-Chiarri M (2022) Development of a multiplex qPCR for the quantification of three protozoan parasites of the eastern oyster Crassostrea virginica. Dis Aquat Org. https://doi.org/10.3354/dao03694. in Press
Plough LV (2016) Genetic load in marine animals: a review. Curr Zool 62:567–579
Plough LV, Hedgecock D (2011) Quantitative trait locus analysis of stage-specific inbreeding depression in the Pacific oyster Crassostrea gigas. Genetics 189:1473–1486
Plough LV, Shin G, Hedgecock D (2016) Genetic inviability is a major driver of type III survivorship in experimental families of a highly fecund marine bivalve. Mol Ecol 25:895–910
Proestou DA, Vinyard BT, Corbett RJ, Piesz J, Allen SK, Small JM, Li C, Liu M, Debrosse G, Guo X (2016) Performance of selectively-bred lines of eastern oyster, Crassostrea virginica, across eastern US estuaries. Aquaculture 464:17–27
Puritz JB, Hollenbeck CM, Gold JR (2014) dDocent: a RADseq, variant-calling pipeline designed for population genomics of non-model organisms. PeerJ 2:e431
Puritz JB, Guo X, Hare MP, He Y, Hillier L, Lotterhos K, Liu M, Mix P, Proestou D, Rice E, Tomlinson C, Warren W, Zhao H, Gomez-Chiarri M (2022a) A second unveiling: haplotig masking of the eastern oyster genome improves population-level inference. BioArXiv
Puritz JB, Zhao H, Guo X, Hare MP, He Y, Peyre JL, Lotterhos KE, Lundgren KM, Proestou D, Rawson P, Robledo JaF, Wedop B, Witkop E, Gomez-Chiarri M (2022b) Nucleotide and structural polymorphisms of the eastern oyster genome paint a mosaic of divergence, selection, and human impacts. BioArXiv
Qi H, Song K, Li C, Wang W, Li B, Li L, Zhang G (2017) Construction and evaluation of a high-density SNP array for the Pacific oyster (Crassostrea gigas). PLoS ONE 12:e0174007
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842
Reeb CA, Avise JC (1990) A genetic discontinuity in a continuously distributed species - mitochondrial-DNA in the American oyster, Crassostrea virginica. Genetics 124:397–406
Reece KS, Ribeiro WL, Gaffney PM, Carnegie RB, Allen SK (2004) Microsatellite marker development and analysis in the eastern oyster (Crassostrea virginica): confirmation of null alleles and non-Mendelian segregation ratios. J Hered 95:346–352
Thompson CM, Evert S, Baez JA (2022) Comparing oyster population structure and disease prevalence between a disease-resistant and transplanted wild population in Barnegat Bay, NJ (U.S.A.). Restor Ecol 30:e13628
Thongda W, Zhao H, Zhang D, Jescovitch LN, Liu M, Guo X, Schrandt M, Powers SP, Peatman E (2018) Development of SNP panels as a new tool to assess the genetic diversity, population structure, and parentage analysis of the Eastern oyster (Crassostrea virginica). Mar Biotechnol 20:385–395
Turley B, Reece K, Shen J, Lee J-H, Guo X, Mcdowell J (2019) Multiple drivers of interannual oyster settlement and recruitment in the lower Chesapeake Bay. Conserv Genet 20:1057–1071
Unterseer S, Bauer E, Haberer G, Seidel M, Knaak C, Ouzunova M, Meitinger T, Strom TM, Fries R, Pausch H, Bertani C, Davassi A, Mayer KF, Schön C-C (2014) A powerful tool for genome analysis in maize: development and evaluation of the high density 600 k SNP genotyping array. BMC Genomics 15:823
Voss-Fels KP, Cooper M, Hayes BJ (2019) Accelerating crop genetic gains with genomic selection. Theor Appl Genet 132:669–686
Wang Y, Guo X (2007) Development and characterization of EST-SSR markers in the eastern oyster Crassostrea virginica. Mar Biotechnol 9:500–511
Wang Y, Shi Y, Guo X (2009) Identification and characterization of 66 EST-SSR markers in the Eastern oyster Crassostrea virginica (Gmelin). J Shellfish Res 28:227–234
Wang Y, Wang X, Wang A, Guo X (2010) A 16-microsatellite multiplex assay for parentage assignment in the eastern oyster (Crassostrea virginica Gmelin). Aquaculture 308:S28–S33
Xu J, Zhao Z, Zhang X, Zheng X, Li J, Jiang Y, Kuang Y, Zhang Y, Feng J, Li C (2014) Development and evaluation of the first high-throughput SNP array for common carp (Cyprinus carpio). BMC Genomics 15:307
Yu Z, Guo X (2003) Genetic linkage map of the eastern oyster Crassostrea virginica Gmelin. Biol Bull 204:327–338
Yu H, He H, Wang X, Zhang Q, Bao Z, Guo X (2011) Polymorphism in a serine protease inhibitor gene and its association with disease resistance in the eastern oyster (Crassostrea virginica Gmelin). Fish Shellfish Immunol 30:757–762
Zeng D, Guo X (2022) Mantle transcriptome provides insights into biomineralization and growth regulation in the eastern oyster (Crassostrea virginica). Mar Biotechnol 24:82–96
Zhang L, Guo X (2010) Development and validation of single nucleotide polymorphism markers in the eastern oyster Crassostrea virginica Gmelin by mining ESTs and resequencing. Aquaculture 302:124–129
Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS (2012) A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28:3326–3328
Zouros E, Foltz D (1984) Possible explanations of heterozygote deficiency in bivalve molluscs. Malacologia 25:583–591
Zouros E, Romero-Dorey M, Mallet AL (1988) Heterozygosity and growth in marine bivalves: further data and possible explanations. Evolution 42:1332–1341
Acknowledgements
We thank Adam Weygandt and Christofer Bertani for assistance with SNP array design and Fig. 1; Michael Whiteside, Samuel Ratcliff, Jillian Jamieson, Sydney Tyburski, Liam Abrams, Vienna Luu, Iris Burt, Jennifer Gius, Leslie Sturmer, and Eric Haffey for assistance with sample collection and processing; Ryan Carnegie, Emily McGurk, Daphne Munroe, Jason Morson, Carter Newell, and Jose Fernandez-Robledo for providing pathogen or oyster samples.
Funding
The study is conducted by the Eastern Oyster Breeding Consortium under support from the National Oceanic and Atmospheric Administration (NOAA), United States Department of Commerce through the Atlantic States Marine Fisheries Commission (award number NA18NMF4720321). The sequencing of 23 Gulf oysters is sponsored by NOAA through the Gulf States Marine Fisheries Commission (award number NA15NMF4720399 subaward to USM number ACQ-210–039-2019-USM). Genotype data for the Gulf oysters used for array validation are from work supported by the Defense Advanced Research Projects Agency (DARPA) under Contract No. HR0011122C0136. Guo and Bushek are also supported by USDA NIFA Animal Health projects 1021665/NJ30401 and 1009201/NJ32114, respectively. The statements, opinions, findings, conclusions and recommendations are those of the authors and don't necessarily reflect the views of NOAA, Department of Commerce, DARPA or USDA.
Author information
Authors and Affiliations
Contributions
X.G., S.K.A., J.P., D.P., B.A., M.G., M.H., M.L., K.E.L., L.P., P.R., J.S., G.W., and A.W. conceived the study as PIs of the EOBC project. X.G., Z.W., D.P., J.S., J.H., K.L., B.A., D.B., M.H., C.H., J.L.P., M.L., L.P., P.R., S.R., E.S., R.V., and A.W. collected and processed samples or contributed sequence/genotype data. X.G., J.P., Z.W., K.V., H.Z., J.H., N.C., D.Z., C.H., and K.E.L. conducted data and bioinformatic analyses. X.G., J.P., and H.Z. wrote the manuscript with input from all authors especially M.H., G.W., M.G., S.K.A., P.R., and K.E.L. All authors reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Guo, X., Puritz, J.B., Wang, Z. et al. Development and Evaluation of High-Density SNP Arrays for the Eastern Oyster Crassostrea virginica. Mar Biotechnol 25, 174–191 (2023). https://doi.org/10.1007/s10126-022-10191-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-022-10191-3