Abstract
Effects of CRISPR/Cas9 knockout of the melanocortin-4 receptor (mc4r) gene in channel catfish, Ictalurus punctatus, were investigated. Three sgRNAs targeting the channel catfish mc4r gene in conjunction with Cas9 protein were microinjected in embryos and mutation rate, inheritance, and growth were studied. Efficient mutagenesis was achieved as demonstrated by PCR, Surveyor® assay, and DNA sequencing. An overall mutation rate of 33% and 33% homozygosity/bi-allelism was achieved in 2017. Approximately 71% of progeny inherited the mutation. Growth was generally higher in MC4R mutants than controls (CNTRL) at all life stages and in both pond and tank environments. There was a positive relationship between zygosity and growth, with F1 homozygous/bi-allelic mutants reaching market size 30% faster than F1 heterozygotes in earthen ponds (p = 0.022). At the stocker stage (~ 50 g), MC4R × MC4R mutants generated in 2019 were 40% larger than the mean of combined CNTRL × CNTRL families (p = 0.005) and 54% larger than F1 MC4R × CNTRL mutants (p = 0.001) indicating mutation may be recessive. With a high mutation rate and inheritance of the mutation as well as improved growth, the use of gene-edited MC4R channel catfish appears to be beneficial for application on commercial farms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The melanocortin-4 receptor (mc4r) is a G protein-coupled receptor within the five-member melanocortin receptor family (MC1R-MC5R) (Cortés et al. 2014). This family of receptors regulates many functions, with mc4r primarily controlling energy homeostasis (Cone 2006). mc4r is expressed in the hypothalamus and is activated by α-melanocyte stimulating hormone (α-MSH), a neuropeptide derived from proopiomelanocortin (POMC) and antagonized by agouti-related peptide (AGRP) (Alrubaian et al. 2003). Mutations in mc4r result in increased appetite and decreased metabolism and energy expenditure (Liu et al. 2019). It is also located upstream of kisspeptin and downstream of leptin and ghrelin, making it an essential metabolic component (Manfredi-Lozano et al. 2016). Channel catfish mc4r gene is a single exon gene with a transcript of 4315 bp located on chromosome 20 (GenBank Accession No. LBML01001141.1).
In mammals, mc4r is predominantly expressed in the central nervous system (Liu et al. 2019). mc4r plays a key role in feeding inhibition with reduced expression leading to increased appetite (Fan et al. 1997). Homozygous MC4R-deficient mice, Mus musculus, showed hyperphagic obesity and hyperinsulinemia, while heterozygous mice exhibited an intermediate phenotype (Huszar et al. 1997). In humans, mc4r mutations are the leading genetic cause of obesity with some studies reporting up to 4% of early-onset obesity cases being caused by a missense or nonsense mutation in the gene (Yeo, et al. 1998; Vaisse et al. 1998). mc4r is a potentially valuable gene for improving growth and feed conversion traits in livestock as it can have large effects on body weight and energy homeostasis.
In fish, mc4r plays a key role in many physiological processes (Cerda-Reverter et al. 2003; Schjolden et al. 2009). For instance, in zebrafish, Danio rerio, mc4r is largely controlled by MRAP2, which is found in two forms. Larval zebrafish produce MRAP2a, which downregulates mc4r, thereby increasing appetite, and adults produce MRAP2b, which upregulates mc4r (Agulleiro et al. 2010). Similarly, clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) mc4r knockout zebrafish showed no phenotypic differences from wild-type individuals before 2.5 months post-fertilization (mpf), while adults displayed increased food consumption, increased growth, and higher body fat percentage compared to wild-type individuals (Fei et al. 2017). This appears to alter the natural growth rate of most fish, where larval fish exhibiting fast growth rate, which gradually slows down throughout its life and typically asymptotes after sexually maturity. Goldfish, Carassius auratus, and rainbow trout, Oncorhynchus mykiss, that were administered the mc4r agonist MTII showed feed inhibition, while those injected with mc4r antagonist, HSO24, had increased feed intake and subsequent growth increases (Cerda-Reverter et al. 2003; Schjolden et al. 2009). Transgenic zebrafish overexpressing the mc4r antagonist agouti-related protein (AgRP) exhibited obesity, increased linear growth, and adipocyte hypertrophy (Song and Cone 2007).
Previous studies show high mutation rates in mc4r gene-edited fish. In a study by Xie et al. (2016), the mloxP gene was inserted into the zebrafish genome at the mc4r locus. Using a novel method in which oocytes are injected in advance and incubated in storage medium before fertilization, they were able to achieve a mutation rate of 94.4% in the mc4r locus in P1 individuals. Additionally, the germline transmission for mc4r mutation was 96.7%. Kawahara et al. (2015) achieved a 95% mutation rate in the mc4r locus of medaka, Oryzias latipes, using transcription activator-like effector nucleases (TALENs).
mc4r is a useful candidate not only for its role in body weight but also in fat production. In both brown and brindle cattle, Bos taurus, the C1069G SNP of the mc4r gene was associated with increased marbling (Lee et al. 2013). However, there was no association between the same SNP and marbling in a third breed, or any effect on carcass weight in any of the three breeds, indicating that certain mutations may only have commercial value in specific populations and species (Lee et al. 2013). If mc4r mutations cause a similar increase in fat production in fish, particularly in healthy omega-3 fatty acids, it could improve its health and flavor qualities.
The objectives of this study were to knock out the mc4r gene in channel catfish, evaluate growth, test inheritance of the mutation in the F1 generation, and evaluate how zygosity affects growth rate. To our knowledge, this is the first study using CRISPR/Cas9 to induce a mutation in the mc4r gene of a commercial food-fish species.
Materials and Methods
Ethics Statement
All experiments were conducted at the Fish Genetics Research Unit, E.W. Shell Fisheries Research Center, Auburn University, AL. All experimental protocols used in this experiment were approved by the Auburn University Institutional Animal Care and Use Committee (IACUC) before the experiment was initiated and followed the Association for Assessment and Accreditation of Laboratory Animal Care protocols and guidelines.
Broodstock Selection, Husbandry, and Spawning
The Kansas strain of channel catfish was chosen as broodstock due to their superior growth and fry output when induced by injection of LHRHa. Broodstock were cultured in 0.04 ha earthen ponds averaging 1 m in depth at a density of 3,250 kgs/ha. They were fed a 32% protein catfish pellet at 1–2% of their body weight 5 days per week. Dissolved oxygen was maintained > 3 mg/L using a ½ horsepower surface aerator (Air-O-Lator). Broodstock spawning followed the procedures described by Qin et al. (2016).
Design and Preparation of sgRNA and CRISPR/Cas9 System
Three customized single guide RNAs (sgRNAs) were designed and generated using the Maxiscript T7 PCR-based method. First, three gene-specific oligonucleotides (MC4R-A, MC4R-B, MC4R-C; Table 1) containing the PAM were designed using the CRISPRscan online tool to target the channel catfish mc4r gene (GenBank Accession No. LBML01001141.1). The Universal Primer (Table 1), containing the sgRNA scaffold, was ordered through Thermo Fisher Scientific. Each oligonucleotide was reconstituted using DNase/RNase free water to 10 mM. The sgRNA templates were generated by synthesizing double stranded DNA through T7 run-off as described by Varshney et al. (2015) with modifications from Khalil et al. (2017). The two oligos were annealed using EconoTaq® Plus 2 × Master Mix (Lucigen, Middleton, WI). The sgRNA was synthesized using the Maxiscript T7 Kit (Thermo Fisher Scientific), following manufacture guidelines. The obtained sgRNAs were purified using the Zymo RNA Clean and Concentrator kit (Zymo Research). The sgRNAs were stored in a − 80 °C freezer. The Cas9 protein was acquired from PNA Bio (3541 Old Conejo Rd, Newbury Park, CA 91,320) and reconstituted in dH20 to a concentration of 1 mg/mL. Shortly (20 min) prior to fertilizing the eggs, three sets of injection solutions were prepared by mixing equal parts of Cas9 protein with each of the sgRNAs individually. The mixture was incubated on ice for 10 min before adding phenol red to a total ratio of 1:1:1 of Cas9, sgRNA, and phenol red, respectively. The final concentrations of Cas9 protein and sgRNA were 300–350 ng/µL and 150–200 ng/µL, respectively.
Gene-Edited Fish Production
Approximately 200–300-g eggs were transferred to a greased pan for fertilization. Thereafter, ~ 3 mL of sperm solution (10 mL/g of testes) was added to the eggs and mixed gently. Fresh water was added to barely cover the eggs to activate the gametes and water was swirled to form a single layer and prevent sticking. After 2 min, the eggs should be fertilized. More water was added to completely fill the pan and the eggs were left to harden for 15 min. While the embryos were hardening, 5–10 µL of the Cas9/sgRNA/phenol red mixture was loaded into 1.0 mm OD borosilicate glass capillary microinjection needles using a microloader. After 15 min, 100–200 embryos were transferred in a single layer to a greased 100-mm petri dish and covered with Holtfreter’s solution for microinjection. The microinjection procedure followed that described by Khalil et al. (2017). Each embryo was injected with 5 nL of solution at the 1 cell stage. Control embryos were injected with 5 nL of 12% phenol red solution.
After microinjection, embryos were placed in 4-L tubs of Holtfreter’s solution (Bart and Dunham 1996) with 10 mg/L doxycycline kept at 27 °C with continuous aeration. The solution was changed, and dead embryos were removed daily. After 5 days, or when the embryos were moving rapidly within the egg membrane and close to hatch, doxycycline treatment was discontinued. Fry were kept at 20 fry/L and fed Purina® AquaMax® powdered starter feed (50% crude protein, 17% crude fat, 3% crude fiber, and 12% ash) (Purina Animal Nutrition LLC, Shoreview, MN) four times a day for 2 months. At 20 dph, fry were moved to 60 L aquaria in recirculating aquaculture systems (RAS).
Culture and Growth
Fry from each genetic type were stocked into 3 replicate 50 L aquaria in RAS for growth experiments. Fish were kept at a density of 2 fish/L. Feed size was adjusted as the fish grew. Fry were fed Aquaxcel WW Fish Starter 4512 (45% crude protein, 12% crude fat, 3% crude fiber, and 1% phosphorus) (Cargill Animal Nutrition, Minneapolis, MN) twice a day for 4 months. Juvenile fish were fed with WW 4010 Transition (40% crude protein, 10% crude fat, 4% crude fiber, and 1% phosphorus) (Cargill Animal Nutrition, Minneapolis, MN) once a day. All fish were fed every day to satiation.
Mutation Analysis
At 6 months post-hatch (mph), pelvic fin-clip samples (10–20 mg) were collected in sterile 1.5-mL Eppendorf tubes and kept at − 80 °C until DNA extraction. Genomic DNA was extracted using proteinase K digestion followed by protein precipitation and iso-propanol precipitation of DNA as described by Kurita et al. (2004). DNA concentration and purity was measured using a NanoDrop 2000 spectrophotometer (Thermo Scientific) and concentration was adjusted to 500 ng/µL.
The primer set MC4R1-F and MC4R1-R (Table 1) was designed using Primer3plus to encapsulate all possible mutation sites in the mc4r gene. The Expand High FidelityPLUS PCR System (Roche) was used with 500 ng of genomic DNA. A Bio-Rad T100 thermal cycler was used to run the PCR with an initial denaturing at 95 °C for 3 min, 34 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C for 40 s with a ramp speed of − 0.2 °C/s, extension at 72 °C for 40 s, and final extension at 72 °C for 10 min. The PCR product was confirmed on a 1% TAE (Tris base, acetic acid and EDTA) agarose gel. The Surveyor® mutation detection kit (Integrated DNA Technologies, Coralville, IA) was used to detect mutations. The PCR product from the treatment fish was mixed with PCR product from a wild-type control of the same family at a 1:1 ratio. The combined product was then hybridized in a BioRad thermocycler using the following procedure: initial denaturing at 95 °C for 3 min, then 95 to 25 °C at − 0.2 °C/s. Hybridized PCR products were mixed with Nuclease S, Enhancer S, MgCl2, and Reaction Buffer (2) according to kit instructions and incubated at 42 °C for 1 h. The digested products were separated on a 1.5% TBE (Tris base, borate, and EDTA) agarose gel and compared with that of control samples.
To confirm and identify the mutations, positive samples were sequenced, and the DNA cloned using the TA cloning method. First, genomic DNA from three mutants per treatment was amplified with PCR using Expand High FidelityPLUS PCR System (Roche) using the above protocol. The PCR product was verified using a 1% TAE agarose gel and cloned into the TOPO® TA Cloning® Kit (Invitrogen) with 20 clones per sample and sent to MCLabs (320 Harbor Way, South San Francisco, CA 94,080) for sequencing. The resulting sequences were interpreted using the MAFFT sequence alignment tool.
Generation ofF1Progeny
In 2018, both male and female P1 MC4R mutants were injected with 40 µg/kg LHRHa and 1600 IU human chorionic gonadotropin (HCG) intraperitoneally, resulting in successful spawning of all pairs that were mated. A total of 8 MC4R pairings were generated. Five pairings were made between wild-type females × P1 MC4R mutant males, 2 pairings between P1 MC4R mutant females × wild-type males, and 1 pairing between a P1 MC4R mutant female × P1 MC4R mutant male. One pair of wild-type Kansas strain channel catfish from the same family was paired in identical conditions as controls but using our standard protocol of 100 µg/kg LHRHa. The system received flow through water from a source pond between 26 and 28 °C and dissolved oxygen was maintained above 5 mg/L using an air stone diffuser. Starting at 24 h after injection, the bottom of each aquarium was checked every 2 h for courtship behavior and egg masses. Egg masses were weighed and transferred to 4 L bins of Holtzfreter’s solution and maintained according to the protocol previously mentioned in the “Gene-Edited Fish Production” subsection.
In 2019, two more families of MC4R × MC4R and MC4R × CNTRL were generated using the same method as 2018. Two- and 2-year-old MC4R gene-edited channel catfish with good secondary sexual characteristics were chosen as broodstock. Both male and female P1 MC4R mutants were injected with 40 µg/kg LHRHa and 1600 IU HCG intraperitoneally and kept in 70 L glass aquaria. Two pairs of wild-type Kansas strain channel catfish from the same family were paired in identical conditions as controls. The spawning protocol was identical to that stated above.
Grow Out and Growth Sampling
Fry (n = 100 per genetic type) were stocked into 3 replicate 50 L aquaria in RAS for growth experiments. Fish in each aquarium were fed ad libitum with Aquamax powder and pelleted fish diets and catfish diets.
Pellet feed size was adjusted to a maximum of ¼ the size of the mouth as the fish grew. Fry were fed Purina® AquaMax® powdered starter feed (50% crude protein, 17% crude fat, 3% crude fiber, and 12% ash) (Purina Animal Nutrition LLC, Shoreview, MN) four times a day for 2 months until they were large enough to eat Purina® AquaMax® 100. Fingerlings were fed Aquaxcel WW Fish Starter 4512 (45% crude protein, 12% crude fat, 3% crude fiber, and 1% phosphorus) (Cargill Animal Nutrition, Minneapolis, MN) twice a day for 4 months. Juvenile fish were fed with WW 4010 Transition feed (40% crude protein, 10% crude fat, 4% crude fiber, and 1% phosphorus) (Cargill Animal Nutrition, Minneapolis, MN) once a day. All fish were fed every day to satiation. Mutants and controls were kept separately in 50 L aquaria until 12 mph, when they were pit-tagged and transferred to 3 replicate 0.04 ha confined earthen ponds and kept communally with unrelated channel catfish, blue catfish, I. furcatus, and hybrids to bring the density to a commercial level of 10,000 fish/ha and fed daily to satiation. A random sample of fish (n = 15 per genetic type) remained in the aquaria in the RAS. The fish were sampled at multiple time points in both aquaria and ponds and body weight determined. Having fish in multiple environments is necessary to determine any genotype-environment interactions to ensure increased performance is observed in a more commercial-like culture unit.
Statistical Analyses
All data were analyzed using the R programming language (R Core Team, Vienna, Austria). A paired t-test was used to calculate differences in body weight between MC4R mutants and controls. To calculate differences in body weight between F1 MC4R families and CNTRL families, a one-way ANOVA followed by Tukey’s multiple comparisons test was performed. When different treatments were kept in separate aquaria at varying densities, a regression based on density was calculated, and weights were adjusted accordingly before running the statistical analysis. Differences in mutation rate were calculated with logistic regression.
Results
Mutation Analysis
A total of 18 fish survived microinjection of CRISPR/Cas9 and sgRNA targeting the mc4r gene in 2017. The mutation rate of the survivors was 33.3% (6/18) (Table 2). Of the 6 P1 mutants, 33.3% were homozygous/bi-allelic and 66.7% were heterozygous. In 2018, a total of 398 F1 MC4R offspring within 8 families were generated by pairing P1 mutants. There was a significant difference in mutation rate between families generated in 2018 (p < 0.05). One family was generated by pairing a MC4R mutant female with a MC4R mutant male (MC4R × MC4R), 5 families were generated by pairing a wild-type female with a MC4R mutant male (CNTRL × MC4R-1,2,3,4,5), and 2 families were generated by pairing a MC4R mutant female with a wild-type male (MC4R × CNTRL-1,2). The average mutation rate for all F1 MC4R channel catfish was 42% (170/398). Of the 21 MC4R × MC4R F1 mutants, 76% (16/21) were homozygotes/bi-allelic.
In 2019, a total of 130 F1 MC4R offspring were generated by pairing a MC4R mutant female with a MC4R mutant male (MC4R × MC4R). The mutation rate was 86% (112/130) and 72% of mutants were homozygotes/bi-allelic (81/112) (Table 2). The same year, a total of 180 F1 MC4R offspring were generated by pairing a MC4R mutant female with a wild-type male (MC4R × CNTRL). The mutation rate was 61% (109/180). The average mutation rate for all F1 MC4R channel catfish was 71% (221/310). There was a significant difference in mutation rate between families generated in 2019 (p < 0.05).
Gel electrophoresis confirmed mutations in both P1 and F1 generations of MC4R mutants (Fig. 1). Multiple bands in the image corresponded to expected cut sites. In all cases, 1 band indicated a wild-type sequence, while multiple bands were associated with mutations. Each positive result was confirmed with a second gel.
Identification of edited melanocortin-4 receptor (mc4r) gene sequences in channel catfish, Ictalurus punctatus, using the Surveyor® mutation detection assay. All samples were hybridized with an equal volume of non-injected control (“–”) to detect both homozygotes/bi-allelic and heterozygotes. Wild-type sequences are indicated with a single 932 bp band, while mutations are signified by three bands. Samples 1 and 2 are MC4R × MC4R F1 progeny. Sample 3 is CNTRL × MC4R-1 F1 progeny. Samples 4 and 5 are CNTRL × MC4R-2 F1 progeny. Samples 6 and 7 are CNTRL × MC4R-3 F1 progeny. Samples 8 and 9 are CNTRL × MC4R-4 F1 progeny. Sample “ + ” is a previously identified MC4R × MC4R F1 progeny mutant. Sample “–” came from wild-type control. “M” indicates 1 kb marker
Sequence results confirmed mutations indicated by gel electrophoresis. Two F1 mutants were tested with each possessing mutations in the mc4r gene (Fig. 2). Both samples had large deletions occurring outside of the target sites. There were no insertions generated. The MC4R × MC4R sample was a homozygous/bi-allelic mutant and contained a deletion (− 641 bp) spanning nearly the entire amplicon. The CNTRL × MC4R sample was a heterozygous mutant with four large deletions (− 427 bp, − 211 bp, − 67 bp, − 127 bp) and 26 substitutions.
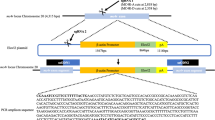
CRISPR/Cas9 induced mutations in the melanocortin-4 receptor (mc4r) gene coding sequence of channel catfish, Ictalurus punctatus, in two F1 mutants generated in 2018. The exon and introns are indicated by upper- and lowercase and the underlined bold uppercase is the start codon. The primers used in PCR are indicated in red. The guide RNA target sites are indicated in green followed by PAM (Protospacer adjacent motif, NGG) in blue. Deletion mutations are represented by a dashed line with each dash corresponding to a nucleotide that has been deleted. Double slash indicates wild-type continuation of the sequence for simplicity. Single slash indicates that there is a large deletion. Each sequence starting with 5′ and ending with 3′ came from a single reaction representing a single allele. Brackets indicate deletion/insertion/substitution value. The wild-type sequence was acquired from Genbank (Accession No. LBML01001141.1). Corresponding predicted amino acid sequence indicated by “AA.” Red letters indicate substitutions. Corresponding predicted amino acid sequence is indicated by “AA.” Red letters indicate substitutions. Predicted amino acid sequence acquired from Expasy and confirmed with NCBI ORFfinder
Growth
A total of 6 MC4R mutant P1 channel catfish and 29 control channel catfish generated in 2017 were PIT tagged and transferred into a 0.04 ha earthen pond at 12 mph (mean body weight for MC4R and CNTRL were 55.75 g and 46.90 g, respectively). MC4R mutants had an observed mean body weight between 13 and 48% larger than controls from 12 to 36 mph; however, the increase was not statistically significant (p = 0.061; Table 3; Fig. 3A).
Box and whisker plots of body weights (g). A Control (CNTRL) and melanocortin-4 receptor (MC4R) P1 knockout channel catfish, Ictalurus punctatus, generated in 2017 at 27 months post-hatch (mph). B CNTRL, MC4R F1 heterozygous knockout, and MC4R F1 homozygous/bi-allelic knockout channel catfish generated in 2018, at 21 mph. C CNTRL, MC4R F1 heterozygous knockout, and MC4R F1 homozygous/bi-allelic knockout channel catfish generated in 2019 at 18 mph. The plot shows median of the data (dark line), lower and upper quartiles (25% and 75%; top and bottom of the box, respectively), area 1.5 × interquartile range (whiskers), and outliers (circles)
A total of 21 MC4R female × MC4R male homozygous/bi-allelic MC4R mutant F1 channel catfish (MC4R × MC4R), 113 CNTRL female × MC4R male heterozygous MC4R mutant channel catfish in 5 families (CNTRL × MC4R-1, CNTRL × MC4R-2, CNTRL × MC4R-3, CNTRL × MC4R-4, CNTRL × MC4R-5), 36 MC4R female × CNTRL male heterozygous MC4R mutant channel catfish in 2 families (MC4R × CNTRL-1 and MC4R × CNTRL-2), and 20 control (CNTRL × CNTRL) channel catfish were generated in 2018. Significant differences in body weight existed among treatments at 6 mph, 12 mph, and 15 mph (p < 0.0001; Table 4). At most time points, homozygous MC4R mutants were larger than controls, while most heterozygous MC4R mutant families were smaller than controls. There were no significant differences in body weight among treatments at 21 mph when treatments were at market size (p = 0.109; Fig. 3B). Several families were not measured at 21 and 24 mph due to partial sampling of the pond. No significant differences in body weight existed among families at 28 mph (p = 0.196). However, homozygous/bi-allelic mutants were 30% larger than pooled F1 heterozygotes (p = 0.022).
A total of 112 MC4R female × MC4R male homozygous/bi-allelic MC4R mutant F1 channel catfish (MC4R × MC4R), 109 MC4R female × CNTRL male heterozygous MC4R mutant channel catfish (MC4R × CNTRL), and 108 control channel catfish in two families (CNTRL × CNTRL-1 and CNTRL × CNTRL-2) were generated in 2019. Significant differences in body weight existed among treatments at 12 and 18 mph (p < 0.0001; Table 5). At 12 mph, MC4R × MC4R mutants were 30% and 8.77 g larger than the mean of both CNTRL × CNTRL families (p < 0.0001; Table 5). MC4R × CNTRL mutants were 9% and 2.34 g smaller than the mean of both CNTRL × CNTRL families (p = 0.493). At 18 mph, MC4R × MC4R mutants were 40% and 15.75 g larger than the mean of both CNTRL × CNTRL families (p = 0.005; Fig. 3C). MC4R × CNTRL mutants were 10% and 3.73 g smaller than the mean of both CNTRL × CNTRL families (p = 0.721).
Discussion
The US farmed-raised catfish industry has suffered decline since its peak in 2003, largely due to foreign competition, particularly in Asia. Longer and warmer growing seasons, lower labor costs, and air breathing species such as Pangasianodon spp. and Clarias batrachus that can be grown in higher stocking densities make it difficult for US catfish farmers to compete. Improving the genetics of channel catfish, particularly for faster growth and feed efficiency, would help American catfish farmers increase profits and improve production. mc4r has been shown to be a valuable gene to target for improving growth in a variety of model organisms; however, its usefulness has not yet been demonstrated in commercially important aquaculture species.
This study investigated the effects of microinjection of gRNAs targeting the channel catfish mc4r gene in conjunction with Cas9 protein. Efficient mutagenesis was achieved as demonstrated by PCR, Surveyor® assay, and DNA sequencing. A total of 18 fish survived microinjection of CRISPR/Cas9 and sgRNA targeting the mc4r gene in 2017 with a mutation rate of 33% (6/18). Of the 6 P1 mutants 33% were homozygous/bi-allelic. This is substantially lower than the mutation rate of 87% achieved in both zebrafish and rats (Li et al. 2013; Xie et al. 2016). This can likely be attributed to variability in the effectiveness of different sgRNAs, variability of egg quality, period during the spawning season, timing after ovulation, and/or other unknown factors. Future experiments should exclusively use best performing sgRNAs and test family and strain effects on mutation rate. Multiple sgRNAs were used to increase likelihood of mutation. Growth was higher in P1 MC4R mutants when compared to controls at all life stages and in pond and tank environments, and the same result was observed in F1 homozygous mutants.
Successful spawning of MC4R mutants to produce F1 progeny was achieved in both 2018 and 2019, and offspring inherited the mutation at a high rate. In 2018, a total of 398 F1 MC4R offspring within 8 families were generated by pairing P1 mutants. The average mutation rate for all F1 MC4R channel catfish was 42% (170/398). Of the 21 MC4R × MC4R F1 mutants, 76% (16/21) were homozygotes/bi-allelic. In 2019, a total of 310 F1 MC4R offspring within 2 families were generated by pairing P1 mutants. The average mutation rate for all F1 MC4R channel catfish was 71% (221/310). The inheritance rates were much higher than those achieved by Hruscha et al. (2013) and Varshney et al. (2015) with zebrafish, where 11% and 28% of mutations were inherited by progeny. This indicates much less mosaicism being generated with our protocols for catfish. However, these studies looked at different species and different genes. The variability in inheritance between different years is likely due to a combination of environmental and genetic effects and different sets of parents.
Growth was generally higher in homozygous F1 MC4R mutants when compared to controls at all life stages and in both pond and tank environments. Homozygous F1 MC4R mutants grew 26.7% faster than controls in earthen ponds to 1 kg, but heterozygous F1 mutants were not different than controls for body weight. There was a positive relationship between zygosity and growth, with F1 homozygous/bi-allelic mutants growing faster than both MC4R × CNTRL and CNTRL × MC4R F1 heterozygotes. The wild-type allele appears to be dominant as body weight in heterozygous MC4R mutants was not different than that of controls, while homozygous mutants were generally larger than controls. This contradicts previous studies that found mutations in mc4r in humans are associated with a dominant form of obesity (Vaisse et al. 1998; Yeo et al. 1998; Hinney et al. 1999). Similar to our results with channel catfish, chickens with bi-allelic SNPs in the G54C locus of the mc4r gene were larger than heterozygous mutants and wild-type chickens, while heterozygotes were smaller than wild-type chickens (Li and Li, 2006). Ortega-Azorin et al. (2012) found a third genetic mechanism in play for MC4R mutants, as there was an additive effect of human mc4r polymorphisms on appetite, while Kim et al. (2004, 2006) showed that additive action of mc4r may influence growth and fat deposition in pigs. In channel catfish, it appears that only one functional copy of the mc4r wild-type allele is necessary for normal growth. Further research should evaluate whether zygosity impacts other important traits such as fertility, survival, and fat deposition in channel catfish.
Beyond the stocker stage, MC4R mutants typically grew faster than controls. This concurs with results found by Fei et al. (2017), in which MC4R knockout zebrafish only showed increased growth after 2.5 mpf as well as those by Liu et al. (2019), where zebrafish showed downregulation of mc4r in larval fish and upregulation in adults. These results compliment the natural growth rate in fish and may counteract the normal decrease in growth that occurs when fish reach sexual maturity. The current market size for catfish is between 0.5 and 1.0 kg, typically reached before sexual maturity (Green and Engle 2004). However, there is a demand for larger market-sized fish, which could be satisfied by genetically improved MC4R knockout fish (Green and Engle 2004).
This increase in growth in MC4R mutants supports previous studies on fish, chickens, pigs, and mice (Lu et al. 1994; Kim et al. 2000; Li et al. 2006; Wan et al. 2012; Yang et al. 2018; Yang et al. 2020). Homozygous F1 MC4R mutants grew 26.7% faster than controls to market size. Similarly, Holland’s carp containing a SNP in the mc4r gene grew 25% faster at 1 year of age than wild-type carp (Yang 2018). MC4R knockout zebrafish were 15.7% larger at 3 months of age than wild-type zebrafish (Fei et al. 2017). Chicken, Awassi sheep, and pigs each containing a SNP in the mc4r gene were between 1.1 and 6.7%, 17.1%, and 6.0% larger respectively than wild-type individuals (Li and Li 2006; Meidtner et al. 2006; Kubota et al. 2019; Al-Thuwaini et al. 2021). While few studies have investigated mutations of mc4r in fish, current data indicates that mutations in fish result in larger phenotypic gains than terrestrial animals. The larger gains in teleost fish are likely explained by the larger proportion of muscle to body weight in fish compared to terrestrial animals or the indeterminate growth of fish (Tlusty et al. 2018). This faster growth to harvest makes mc4r a valuable gene commercially.
The growth of individual MC4R mutants and different MC4R families varied. This can be attributed to differences in knockout patterns and subsequent expression levels of mc4r, differences in genetic background coupled with epistatic interaction with other loci. The variability can be advantageous as combining gene editing with selection could result in maximum genetic enhancement.
The improved growth indicates that the use of gene-edited MC4R channel catfish could be beneficial for commercial farms. Gene editing presents a valuable tool to increase profitability, sustainability, and industry growth. There are, however, ethical, logistical, and regulatory hurdles for the MC4R mutant channel catfish to become applied commercially in the USA, as FDA currently regulates gene-edited animals. The improvement of gene editing technologies, greater understanding of its effects, and the commercial success of genetically improved organisms, including Aquabounty’s AquaAdvantage salmon in the USA and Canada, Intrexon’s gene-edited tilapia in Argentina, and Regional Fish Institute’s gene-edited sea bream and tiger puffer fish in Japan, make this technology a viable option in the near future (Waltz et al. 2017; “Japan embraces CRISPR-edited fish” 2022). By combining mc4r gene editing with other genetic techniques, such as selection, crossbreeding, and hybridization, it is likely possible to achieve even greater growth results, shorten the grow-out period, and select for multiple traits. With an increasing human population and declining natural resources, all solutions should be evaluated to determine the most efficient and sustainable methods of food production.
References
Agulleiro MJ, Roy S, Sánchez E, Puchol S, Gallo-Payet N, Cerdá-Reverter JM (2010) Role of melanocortin receptor accessory proteins in the function of zebrafish melanocortin receptor type 2. Mol Cell Endocrinol 320:145–152
Alrubaian J, Sollars C, Danielson PB, Dores RM (2003) Evaluating the radiation of the POMC gene in teleosts: characterization of American eel POMC. Gen Comp Endocrinol 132:384–390
Al-Thuwaini TM, Al-Shuhaib MBS, Lepretre F, Dawud HH (2021) Two co-inherited novel SNPs in the MC4R gene related to live body weight and hormonal assays in Awassi and Arabi sheep breeds of Iraq. Veterinary Medicine and Science 7:897–907
Bart AN, Dunham RA (1996) Effects of sperm concentration and egg number on fertilization efficiency with channel catfish (Ictalurus punctatus) eggs and blue catfish (I. furcatus) spermatozoa. Theriogenology 45:673–682
Cerda-Reverter JM, Ringholm A, Schioth HB, Peter RE (2003) Molecular cloning, pharmacological characterization, and brain mapping of the melanocortin 4 receptor in the goldfish: involvement in the control of food intake. Endocrinology 144:36–49
Cone RD (2006) Studies on the physiological functions of the melanocortin system. Endocr Rev 27:736–749
Cortés R, Navarro S, Agulleiro MJ, Guillot R, García-Herranz V, Sánchez E, Cerdá-Reverter JM (2014) Evolution of the melanocortin system. Gen Comp Endocrinol 209:3–10
Fan W, Boston BA, Kesterson RA, Hruby VJ, Cone RD (1997) Role of melanocortinergic neurons in feeding and the agouti obesity syndrome. Nature 385:165–168
Fei F, Sun SY, Yao YX, Wang X (2017) Generation and phenotype analysis of zebrafish mutations of obesity-related genes lepr and MC4R. Acta Physiologica Sinica 69:61–69
Green BW, Engle CR (2004) Growth of stocker channel catfish to large market size in single-batch culture. J World Aquaculture Soc 35:25–32
Hinney A, Schmidt A, Nottebom K, Heibult O, Becker I, Ziegler A, Gerber G, Sina M, Görg T, Mayer H, Siegfried W, Fichter M, Remschmidt H, Hebebrand J (1999) Several mutations in the melanocortin-4 receptor gene including a nonsense and a frameshift mutation associated with dominantly inherited obesity in humans. J Clin Endocrinol Metab 84:1483–1486
Hruscha A, Krawitz P, Rechenberg A, Heinrich V, Hecht J, Haass C, Schmid B (2013) Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development 140:4982–4987
Huszar D, Lynch CA, Fairchild-Huntress V, Dunmore JH, Fang Q, Berkemeier LR, Gu W, Kesterson RA, Boston BA, Cone RD, Smth FJ, Campfield LA, Burn P, Lee F (1997) Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell 88:131–141
Japan embraces CRISPR-edited fish (2022) Nat Biotechnol 40:10. https://doi.org/10.1038/s41587-021-01197-8
Kawahara A, Yabe T, Ansai S, Takada S, Kinoshita M (2015) Genome editing in zebrafish and medaka. Targeted Genome Editing Using Site-Specific Nucleases 119–131. Springer, Tokyo
Khalil K, Elayat M, Khalifa E, Daghash S, Elaswad A, Miller M, Abdelrahman H, Ye Z, Odin R, Drescher D, Vo K, Gosh K, Bugg W, Ribinson D, Dunham R (2017) Generation of myostatin gene edited channel catfish (Ictalurus punctatus) via zygote injection of CRISPR/Cas9 system. Sci Rep 7:1–12
Kim KS, Larsen N, Short T, Plastow G, Rothschild MF (2000) A missense variant of the porcine melanocortin-4 receptor (MC4R) gene is associated with fatness, growth, and feed intake traits. Mamm Genome 11:131–135
Kim KS, Thomsen H, Bastiaansen J, Nguyen NT, Dekkers JC, Plastow GS, Rothschild MF (2004) Investigation of obesity candidate genes on porcine fat deposition quantitative trait loci regions. Obes Res 12:1981–1994
Kim KS, Lee JJ, Shin HY, Choi BH, Lee CK, Kim JJ, Cho BW, Kim TH (2006) Association of melanocortin 4 receptor (MC4R) and high mobility group AT-hook 1 (HMGA1) polymorphisms with pig growth and fat deposition traits. Anim Genet 37:419–421
Kurita K, Burgess SM, Sakai N (2004) Transgenic zebrafish produced by retroviral infection of in vitro-cultured sperm. Proc Natl Acad Sci 101:1263–1267
Kubota S, Vandee A, Keawnakient P, Molee W, Yongsawatdikul J, Molee A (2019) Effects of the MC4R, CAPN1, and ADSL genes on body weight and purine content in slow-growing chickens. Poult Sci 98:4327–4337
Lee Y, Park S, Kim H, Lee SK, Kim JW, Lee HK, Joeng DK, Lee SJ (2013) A C1069G SNP of the MC4R gene and its association with economic traits in Korean native cattle (brown, brindle, and black). Electron J Biotechnol 16:14
Li CY, Li H (2006) Association of MC4R gene polymorphisms with growth and body composition traits in chicken. Asian Australas J Anim Sci 19:763–768
Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, Li Y, Gao N, Wang L, Lu X, Zhao Y, Liu M (2013) Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol 31:681–683
Liu R, Kinoshita M, Adolfi MC, Schartl M (2019) Analysis of the role of the MC4R system in development, growth, and puberty of medaka. Front Endocrinol 10:213
Lu D, Willard D, Patel IR, Kadwell S, Overton L, Kost T, Luther M, Chen W, Woychick RP, Wilkison WO, Cone RD (1994) Agouti protein is an antagonist of the melanocyte-stimulating-hormone receptor. Nature 371:799–802
Manfredi-Lozano M, Roa J, Ruiz-Pino F, Piet R, Garcia-Galiano D, Pineda R, Zamora A, Leon S, Sanchez-Garrido MA, Romero-Ruiz A, Dieguez C, Jesus Vasquez M, Herbison AE, Pinilla L, Tena-Sempere M (2016) Defining a novel leptin–melanocortin–kisspeptin pathway involved in the metabolic control of puberty. Molecular Metabolism 5:844–857
Meidtner K, Wermter AK, Hinney A, Remschmidt H, Hebebrand J, Fries R (2006) Association of the melanocortin 4 receptor with feed intake and daily gain in F2 Mangalitsa × Piétrain pigs. Anim Genet 37:245–247
Ortega-Azorín C, Sorlí JV, Asensio EM, Coltell O, Martínez-González MÁ, Salas-Salvadó J, Covas MI, Arós F, Lapetra J, Serra-Majem L, Gómez-Gracia E, Fiol M, Sáez-Tormo G, Pintó X, Muñoz MA, Ros E, Ordovás JM, Estruch M, Corella D (2012) Associations of the FTO rs9939609 and the MC4R rs17782313 polymorphisms with type 2 diabetes are modulated by diet, being higher when adherence to the Mediterranean diet pattern is low. Cardiovasc Diabetol 11:1–12
Qin Z, Li Y, Su B, Cheng Q, Ye Z, Perera DA, Fobes M, Shang M, Dunham RA (2016) Editing of the luteinizing hormone gene to sterilize channel catfish, Ictalurus punctatus, using a modified zinc finger nuclease technology with electroporation. Mar Biotechnol 18:255–263
Schjolden J, Schioth HB, Larhammar D, Winberg S, Larson ET (2009) Melanocortin peptides affect the motivation to feed in rainbow trout (Oncorhynchus mykiss). Gen Comp Endocrinol 160:134–138
Song Y, Cone RD (2007) Creation of a genetic model of obesity in a teleost. FASEB J 21:2042–2049
Tlusty M, Tyedmers P, Ziegler F, Jonell M, Henriksson P, Newton R, Little D, Fry JP, Love D, & Cao L (2018) Commentary: comparing efficiency in aquatic and terrestrial animal production systems. Environ Res Lett 13
Vaisse C, Clement K, Guy-Grand B, Froguel P (1998) A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nat Genet 20:113–114
Varshney GK, Pei W, LaFave MC, Idol J, Xu L, Gallardo V, Carrington B, Bishop K, Jones M, Li M, Harper U, Huang SC, Prakash A, Chen W, Sood R, Ledin J, Burgess SM (2015) High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res 25:1030–1042
Waltz E (2017) First genetically engineered salmon sold in Canada. Nature News 548:148
Wan Y, Zhang Y, Ji P, Li Y, Xu P, Sun X (2012) Molecular characterization of CART, AgRP, and MC4R genes and their expression with fasting and re-feeding in common carp (Cyprinus carpio). Mol Biol Rep 39:2215–2223
Xie SL, Bian WP, Wang C, Junaid M, Zou JX, Pei DS (2016) A novel technique based on in vitro oocyte injection to improve CRISPR/Cas9 gene editing in zebrafish. Sci Rep 6:34555
Yang Y, Lan Z, Shu H, Zhou H, Jiang X, Hou L, Gu P (2018) Association between expression levels and growth trait-related SNPs located in promoters of the MC4R and MSTN genes in Spinibarbus hollandi. Genes & Genomics 40:1119–1125
Yang Z, Liang XF, Li GL, Tao YX (2020) Biased signaling in fish melanocortin-4 receptors (MC4Rs): divergent pharmacology of four ligands on spotted scat (Scatophagus argus) and grass carp (Ctenopharyngodon idella) MC4Rs. Mol Cell Endocrinol 115:110929
Yeo GS, Farooqi IS, Aminian S, Halsall DJ, Stanhope RG, O’Rahilly S (1998) A frameshift mutation in MC4R associated with dominantly inherited human obesity. Nat Genet 20:111–112
Funding
This work was funded by the United States Department of Agriculture National Institute of Food and Agriculture award 2015–67015-23488 to Roger Cone and sub-award to Rex Dunham.
Author information
Authors and Affiliations
Contributions
Conceptualization: Michael Coogan, Rex Dunham, Karim Khalil, Ahmed Elaswad, Mohd Khan, Roger D. Cone; methodology: Michael Coogan, Rex Dunham, Karim Kalil, Ahmed Elaswad, Patrick Page-McCaw, Wenbiao Chen, Maximilian Michel, Mohd Khan; formal analysis and investigation: Michael Coogan; writing—original draft preparation: Michael Coogan; writing—review and editing: Michael Coogan, Veronica Alston, Baofeng Su, Rex Dunham, Ian Butts, Andrew Johnson, De Xing, Jinhai Wang, Shangjia Li, Rhoda M.C. Simora, Wenwen Wang, Darshika Hettiarachchi, Cuiyu Lu; funding acquisition: Rex Dunham, Roger D. Cone; Resources: Rex Dunham; supervision: Rex Dunham, Ian Butts; data curation: Andrew Johnson, De Xing, Jinhai Wang, Shangjia Li, Rhoda M.C. Simora, Wenwen Wang, Darshika Hettiarachchi, Cuiyu Lu, and Tasnuba Hasin.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Coogan, M., Alston, V., Su, B. et al. Improved Growth and High Inheritance of Melanocortin-4 Receptor (mc4r) Mutation in CRISPR/Cas-9 Gene-Edited Channel Catfish, Ictalurus punctatus. Mar Biotechnol 24, 843–855 (2022). https://doi.org/10.1007/s10126-022-10146-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-022-10146-8