Abstract
Melanocortin 4 receptor: (MC4R) and Myostatin (MSTN) are two important growth trait-related genes in animals. In this study, we showed that two SNPs, MC4R-719A>G and MSTN-519C>T, found in the promoters of the MC4R and MSTN genes, respectively, are both associated with growth traits in Spinibarbus hollandi. Furthermore, we observed that there were significant associations between the expression levels of the MC4R and MSTN genes and these two growth trait-related SNPs. The expression level of MC4R gene in brain was lower in GG genotype fish with extremely high growth performance than that in AA genotype fish with extremely low growth performance. Expression level of the MSTN gene in muscle was lower in TT genotype fish with extremely high growth performance than that in CC and CT genotype fish with lower growth performance. The results indicated that these SNPs located in the promoters of MC4R and MSTN are associated with growth-related traits through modification of gene expression levels. The MSTN and MC4R SNPs may have useful application in effective marker-assisted selection aimed to increase output in S. hollandi.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Melanocortin-4 receptor (MC4R) is a G-protein-coupled receptor expressed in the appetite-regulating area of brain, which is associated with feed intake regulation and energy balance (Fan et al. 1997; Meidtner et al. 2010). Huszar et al. (1997) observed that knockout of MC4R induced symptoms of polyphagia, obesity, and higher levels of insulin secretion in mice. Moreover, mutations of the MC4R gene have been found to be the most common cause of hereditary obesity in humans (Carroll et al. 2005). Since MC4R links to the control of body weight via regulation of energy homeostasis, it has been considered as a potentially valuable gene for improving growth-related traits in animals.
Myostatin (MSTN) is a negative regulator of muscle development through regulating both the number and growth of muscle fibers (Lee and Mcpherron 1999). Mutations of MSTN leading to non-functional proteins have been reported to cause the “double-muscling” phenotype in cattle (Mcpherron and Lee 1997; Kambadur et al. 1997). Similarly, MSTN gene knockout was shown to cause a significant increase in muscle mass in mice, as a consequence of muscle cell hypertrophy and hyperplasia (McPherron et al. 1997). Additionally, knockdown of the MSTN gene has been shown to give rise to muscular hypertrophy in zebra fish (Lee et al. 2009). This gene has therefore been considered a potential candidate gene for identification of genetic markers and improving growth and meat quality traits in livestock and fish.
Single-nucleotide polymorphism (SNP) is a common form of variation in genes, promoters, and regulatory regions, it is widely distributed throughout genomes. Therefore, some SNPs may affect biological phenotypes through modifying the expression of genes. SNPs have become the focuses of intense research in the fields of fish genetics and breeding (Houston et al. 2012; Poćwierz-Kotus et al. 2014; Yang et al. 2016). SNPs of MSTN associated with production traits have been widely reported, both in livestock, such as sheep (Hickford et al. 2010), pig (Jiang et al. 2002), and cattle (Sellick et al. 2007), and in fish, such Atlantic salmon (Salmo salar; Peñaloza et al. 2013), common carp (Cyprinus carpio; Sun et al. 2012), spotted halibut (Verasper variegatus; Li et al. 2012), and bighead carp (Aristichthys nobilis; Liu et al. 2012). Although polymorphisms located in the MC4R gene have been widely demonstrated to be associated with growth traits (Zeng et al. 2014; Kim et al. 2000; Cai et al. 2015; Lee et al. 2013; Fontanesi et al. 2013), to date there have been few reports regarding associations between MC4R gene polymorphisms and growth quality traits in fish.
The cyprinid fish S. hollandi (Cyprinidae: Cypriniformes) is widely distributed in the south of China, including the provinces of Guangxi, Guangdong, Fujian, and Anhui. As S. hollandi has high nutritional and medicinal value, it produces high economic benefit (Cai et al. 2007). In recent years, increasing demand for S. hollandi has stimulated considerable research on this species. Most previous studies on S. hollandi have focused on genetic resources (Shu et al. 2015), rearing conditions (Lv et al. 2008), ethology (Li et al. 2011), and morphology (Wang 2013), whereas there is considerably less attention paid to analysis of marker-assisted selection.
Therefore, the aims of this study were to: (1) identify SNPs of the MSTN and MC4R genes and investigate the relationships between MSTN and MC4R polymorphisms and growth traits in S. hollandi, (2) analyze the relationships between expression levels of the MC4R and MSTN genes, their SNPs and growth traits in S. hollandi.
Materials and methods
Materials and phenotypic data collection
The experimental fish were obtained from Shaoguan Fisheries Research Institute in Guangdong, China. The parent fish of the experimental population used for association analysis were selected from the first generation of wild stock collected from the Beijiang River. All fish were hatched at the same time and cultured under the same rearing and management conditions. At the age of 1 year, 235 S. hollandi with an average weight of 134.75 ± 38.66 g were randomly selected without differentiating sexes. Five growth traits, including body weight (BWT), body length (BL), total length (TL), body depth (BD), and body width (BWH), were measured in each fish for association analysis. The part of caudal fin of each fish was collected and preserved in 95% ethanol. After completing measurements, each fish was marked and maintained with feeding under pre-measurement conditions in another pool.
PCR amplification and SNP identification
To detect MSTN and MC4R polymorphisms, two primer pairs, MSTNPF, 5′- ACAGATACGTGAATATTATC-3′, MSTNPR, 5′-TGCGCCGTTATATCTCCATG-3′, and MC4RPF, 5′-TCTTTATGAGTGAATTACTG-3′, MC4RPR, 5′-CAAAGCAGGTGCTGTGTGAG-3′, were designed using Primer Premier 5.0 software (Premier Biosoft International, Palo Alto, CA) based on the DNA sequences of S. hollandi MSTN (GenBank: KY853657) and MC4R (GenBank: KY022411) genes. PCR amplification was performed in a reaction volume of 50 µL, containing 25 µL 2 × Taq Master Mix (Dye Plus, Vazyme Biotech, Nanjing, China), 20 µL double-distilled water, 3 µL DNA solution, and 1 µL of each primer (10 µM). The PCR amplification reactions were performed using the following thermo cycle program: 94 °C for 10 min, followed by 35 cycles of 94 °C for 45 s, 55 °C for 30 s, and 72 °C for 1 min, with a final extension at 72 °C for 10 min. Amplification results were verified by 2% agarose gel electrophoresis and PCR fragments of the predicted size were purified using an agarose gel DNA Extraction kit (Generay Biotech Co., Ltd, Shanghai, China).
SNP identification, genotyping, and association analysis
Sequencing of the amplified DNA fragments was performed by Generay Biotech Co., Ltd using an ABI 3730XL sequencer (Applied Biosystems, USA). Differences of gene sequences between individuals were detected using SeqMan version 7.1.0 (DNASTAR Inc., Madison, WI, USA). SNPs were detected and genotyped through observing and comparing chromatogram files using Chromas version 2.33 (Technelysium Pty Ltd., South Brisbane, Australia). Association analyses between genotypes of the MSTN gene and the MC4R gene and the five selected growth traits were performed using post hoc multiple comparisons (the Duncan method) with SPSS 19.0 software (IBM, Armonk, NY, USA).
Association between gene expression levels and growth trait-related SNPs
On the basis of the results of association analyses, for each genotype of the growth trait-related SNPs, 12 fish were randomly collected from marked fish to analyze the association between gene expression levels and the growth trait-related SNPs. Sampled fish were anesthetized by immersion in 0.1% eugenol for 1–2 min, and these fish were then dissected using stainless steel scissors. Brain or/and muscle samples were frozen immediately in liquid nitrogen and stored at − 80 °C for RNA isolation. The brains were used to analyze the association between expression levels and the growth trait-related SNP in MC4R, whereas both muscle and brains were used for the same analysis in MSTN.
RNA isolation and the real-time PCR
Total RNA was isolated from frozen samples using RNA Isolater Total RNA Extraction Reagent (Vazyme Biotech, Nanjing, China) according to the manufacturer’s instructions. RNA concentrations were determined at 260 nm. The 260/280 ratio was used to verify the quality of the RNA in each sample. RNA samples were dissolved in diethylpyrocarbonate-treated water and stored at − 80 °C.
Real-time PCR was performed using an ABI 7000 thermal cycler in 20-µL reaction volumes containing the following components: 100 ng of cDNA, 10 µL Power SYBR Green PCR Master Mix (Vazyme Biotech Co., Ltd, Nanjing, China), 0.3 µL of each primer (10 µmolL−1), and 7.4 µL double-distilled water. The primer pairs, QMSTNF, 5′-ATGACCATGGCCACAGAGCCTG-3′, QMSTNR, 5′-CCGGTCTCAGATGAACCCAGAGC-3′, and QMC4RF, 5′-AGCCGTAGCAGACTTGTTGGTC-3′, QMC4RR, 5′-TGTTCTTGATGATGCTCTCGCG-3′, were used for RT-PCR amplification of MSTN and MC4R, respectively. All samples were analyzed in triplicate and the mean value was used to calculate mRNA expression levels. β-actin was amplified as the internal control gene with the primers F (5′-CAGCCATCCTTCTTGGGTATG-3′) and R (5′-TCTGCATACGGTCAGCAATGC-3′). The relative mRNA expression levels of each genotype were analyzed using the \({2^{ - \Delta \Delta {C_{\text{t}}}}}\) method.
Statistical analysis
Genetic analyses, including Hardy–Weinberg equilibrium (HWE), expected heterozygosity (He), and observed heterozygosity (Ho), were calculated using Haploview software (Broad Institute, America). Polymorphism information content (PIC) was calculated using PIC CALC version 0.6 (Yellow Sea Fisheries Research Institute, Qingdao, China). The effects of different SNP genotypes on the five selected growth traits were analyzed by one-way ANOVA using SPSS 19.0 software. The genotypes of SNPs significantly associated with the growth traits of S. hollandi were analyzed through post hoc multiple comparisons (the Duncan method). Analysis of the association between gene expression levels and growth trait-related SNPs was performed using Student’s t-test. The following statistical model was applied:
where Y is the phenotypic value of each trait, u is the population mean value of each growth trait, G is the fixed genotypic effect of each SNP, and e is the random error effect.
Results
SNP identification and genotyping
Two SNPs (MC4R-719A>G and MSTN-519C>T) were detected in the promoters of MC4R and MSTN, respectively. Chi square tests revealed that MC4R -719A>G was in HWE (p > 0.05), whereas MSTN-519C > T deviated from the HWE (p < 0.05), The He, Ho, PIC, and Hardy–Weinberg p-values of these two SNPs are shown in Table 1. The two SNPs were classified as being moderately polymorphic loci based on the following criteria: loci with PIC > 0.5 are highly polymorphic; loci with 0.5 > PIC > 0.25 are moderately polymorphic; and loci with PIC < 0.25 have low polymorphism (Vaiman et al. 1994). The allele and genotype frequencies of the SNPs are shown in Table 2.
Analysis of associations between SNPs and growth traits
The results of association analyses between different SNP genotypes and the five selected growth traits are shown in Table 3. For the MC4R-719A>G SNP, the five growth traits of the GG genotype fish were significantly higher than those of the AA and AG genotype fish. Measurements of the five growth traits of the AG genotype fish were also higher than those of the AA genotype fish, although the differences were not significant. For the MSTN-519C>T SNP, the BWT of the TT genotype fish was significant higher than that of the CC and CT genotype fish, and the BD and BWH of the TT genotype fish were significantly higher than those of CC genotype fish. Measurements of the five growth traits were not significantly different between CC and CT genotype fish.
Association between gene expression levels and growth trait-related SNPs
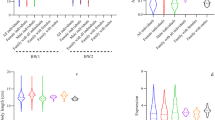
Expression levels of the MC4R gene in brain were significantly higher in the AA genotype fish than in the AG and GG genotype fish (p < 0.05) and slightly higher in the AG genotype fish than in the GG genotype fish (Fig. 1). The GG genotype fish, which had the highest BWT, had lowest expression of the MC4R gene in the brain. The AA genotype fish, which had the lowest BWT, had the highest expression of MC4R in the brain.
Body weight and MSTN mRNA expressions in the brain were shown in Fig. 2. No significant difference was detected in the MSTN expression levels in the brain among the different genotype groups of MSTN-519C>T (p > 0.05).
The expression levels of the MSTN gene in muscle were significantly lower in TT genotype fish than in the CC and CT genotype fish (p < 0.05). The BWT of TT genotype fish was significant higher than that of CC and CT genotype fish (p < 0.05), whereas there were no significant differences in BTW and expression levels of the MSTN gene between CC and CT genotype fish (Fig. 3).
Discussion
In this experiment, we detected two mutations (MC4R-719A>G and MSTN-519C>T) in the promoters of the MC4R and MSTN genes of S. hollandi, respectively. Both SNPs were moderately polymorphic loci. MC4R-719A>G was in HWE, whereas MSTN-519C>T deviated from the HWE, it is possible that the MSTN-519C>T, or other variation closely linked to the SNP, was subjected to natural selection pressure. This phenomenon was also found in Argopecten irradians (Meng et al. 2017). The two SNPs are both related with growth traits in this fish. The expression levels of the MC4R gene in the brain are associated with the growth trait-related SNP, MC4R-719A>G, whereas the SNP, MSTN-519C>T, is associated with expression levels of the MSTN gene in muscle, but not in the brain.
Our results showed that the MC4R-719A>G is associated with all five growth traits of S. hollandi examined in the present study, with the GG genotype being predominant among the three genotypes. As an important candidate gene affecting growth ratio, numerous association studies have focused on the detection of MC4R gene polymorphisms associated with economic traits in domestic animals. Zhang et al. (2006) reported a missense mutation in MC4R that was significantly associated with birth weight and average daily gain in cattle. Kim et al. (2000) observed that an SNP, Asp298Asn, located in MC4R was significantly associated with growth traits in pigs, and Song et al. (2012) detected four SNPs in the 3′-UTR of MC4R that were significantly associated with birth weight in sheep. However, there have been few studies that have focused on the association between MC4R SNPs and growth quality traits in fish. Although several SNPs have previously been identified in the MC4R gene of Oreochromis niloticus (tilapia) and Takifugu rubripes, no growth trait-related SNPs were identified in the two species (Liu et al. 2009; Zhang et al. 2012). The growth trait-related SNP identified in the MC4R gene in the present study will therefore make a potentially important contribution to the analysis of molecular markers in fish.
In the present study, we observed that the MC4R-719A>G is associated with expression levels of the MC4R gene in the brain of S. hollandi. The GG allele has the lowest expression level and the GG genotype fish have the most predominant growth traits. These observations are consistent with previous observations indicating that inactivation or decreased activity of MC4R can result in an increase in body weight (Huszar et al. 1997).We hypothesized that an SNP located in the 5′-flanking region might have an impact on MC4R expression and growth performance, by affecting MC4R promoter activity. However, we cannot exclude the possibility that the MC4R SNP identified in the present study is in linkage disequilibrium with an unidentified causal mutation for MC4R expression and growth performance.
In association studies, the MSTN-519C>T was also found to be associated with growth traits of S. hollandi, with the TT genotype being predominant. Significant associations between MSTN polymorphisms and production traits have been widely reported in aquacultural species, including bighead carp (Aristichthys nobilis; Liu et al. 2012), yellow catfish (Pelteobagrus fulvidraco; Zhu et al. 2012), spotted halibut (Verasper variegatus; Li et al. 2012), common carp (Cyprinus carpio; Sun et al. 2012), Atlantic bay scallop (Argopecten irradians; Guo et al. 2011), and gilthead seabream (Sparus aurata; Sánchez-Ramos et al. 2012). Moreover, the MSTN-519C>T departed from Hardy–Weinberg equilibrium, and thus it is possible that this SNP is linked to unidentified genes that are affected by natural selection.
The MSTN-519C>T is associated with expression levels of the MSTN gene in the muscles of S. hollandi. This result indicated that MSTN is a negative regulator of growth in muscle. Knocking down MSTN using antisense morpholinos resulted in the increase of length and width of somites in juvenile zebra fish (Amali et al. 2004). Muscle hyperplasia and hypertrophy were also found in Myostatin dsRNA-microinjected zebrafish (Acosta et al. 2005). No significant association between expression level of the MSTN gene in brain and its SNP was found, suggesting that MSTN expressed in brain may plays another role at the stage of development in S. hollandi. Further investigation of this possibility, including RNA-Seq after MSTN knockdown and other functional studies, will provide important insights into MSTN function. The possibility same as above, the SNP might have a direct impact on MSTN expression, as well as is in linkage disequilibrium with unidentified loci that impact on MSTN expression.
In conclusion, the findings of this study indicate that SNPs located in the MSTN and MC4R promoters have effects on growth-related traits via modifications of gene expression. Information obtained in this study on the MC4R and MSTN SNPs may have potential applications in effective marker-assisted selection to increase body weight and lean meat percentage in S. hollandi.
References
Acosta J, Carpio Y, Borroto I, González O, Estrada MP (2005) Myostatin gene silenced by RNAi show a zebrafish giant phenotype. J Biotechnol 119:324–331
Amali AA, Lin CJF, Chen YH, Wang WL, Gong HY, Lee CY, Ko YL, Lu JK, Her GM, Chen TT (2004) Up-regulation of muscle-specific transcription factors during embryonic somitogenesis of zebrafish (Danio rerio) by knock-down of Myostatin-1. Dev Dynam 229:847–856
Cai ZD, Lin G, Ni JY, Ye Z, Tang ZB, Zhang S (2007) Study on the reproductive biology of Spinibabus calddwellis. Guangxi Agric Sci 2:24–28 (Chinese with English abstract)
Cai X, Mipam TD, Zhao FF, Sun L (2015) SNPs detected in the yak MC4R gene and their association with growth traits. Animal 9:1097–1103
Carroll L, Voisey J, Van DA (2005) Gene polymorphisms and their effects in the melanocortin system. Peptides 26:1871–1885
Fan W, Boston BA, Kesterson RA, Hruby VJ, Cone RD (1997) Role of melanocortinergic neurons in feeding and the agouti obesity syndrome. Nature 385:165–168
Fontanesi L, Scotti E, Cisarova K, Di BP, Dall’Olio S, Fornasini D (2013) A missense mutation in the rabbit melanocortin 4 receptor (MC4R) gene is associated with finishing weight in a meat rabbit line. Anim Biotechnol 24:268–277
Guo L, Li L, Zhang S, Guo X, Zhang G (2011) Novel polymorphisms in the Myostatin, gene and their association with growth traits in a variety of bay scallop, Argopecten irradians. Anim Genet 42:339–340
Hickford JGH, Forrest RH, Zhou H, Fang Q, Han J, Frampton CM, Horrell AL (2010) Polymorphisms in the ovine Myostatin gene (MSTN) and their association with growth and carcass traits in New Zealand Romney sheep. Anim Genet 41:64–72
Houston DD, Elzinga DB, Maughan PJ, Smith SM, Kauwe JS, Evans RP (2012) Single nucleotide polymorphism discovery in cutthroat trout subspecies using genome reduction, barcoding, and 454 pyro-sequencing. BMC Genom 13:724
Huszar D, Lynch CA, Fairchildhuntress V, Dunmore JH, Fang Q, Berkemeier LR (1997) Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell 88:131–141
Jiang YL, Li N, Fan XZ, Xiao LR, Xiang RL, Hu XX (2002) Associations of t leads to a mutation in the promoter region of Myostatin gene with birth weight in yorkshire pigs. Asian Austral J Anim 15:1543–1545
Kambadur R, Sharma M, Smith TP, Bass JJ (1997) Mutations in Myostatin (GDF8) in double-muscled belgian blue and piedmontese cattle. Genome Res 7:910–915
Kim KS, Larsen N, Short T, Plastow G, Rothschild MF (2000) A missense variant of the porcine melanocortin-4 receptor (MC4R) gene is associated with fatness, growth, and feed intake traits. Mamm Genome 11:131–135
Lee SJ, Mcpherron AC (1999) Myostatin and the control of skeletal muscle mass: commentary. Curr Opin Genet Dev 9:604–607
Lee CY, Hu SY, Gong HY, Chen MH, Lu JK, Wu JL (2009) Suppression of Myostatin with vector-based RNA interference causes a double-muscle effect in transgenic zebrafish. Biochem Bioph Res Commun 387:766–771
Lee Y, Park S, Kim H, Lee SK, Kim JW, Lee HK, Lee SJ (2013) A C1069G SNP of the MC4R gene and its association with economic traits in Korean native cattle (brown, brindle, and black). Electron J Biotechnol 16:14–14
Li W, Chen Q, Huang Y (2011) Laboratory study on ethology of Spinibarbus hollandi. Acta Ecol Sin 31:1404–1411
Li H, Fan J, Liu S, Yang Q, Mu G, He C (2012) Characterization of a Myostatin gene (MSTN1) from spotted halibut (Verasper variegatus) and association between its promoter polymorphism and individual growth performance. Comp Biochem Phys B 161:315–322
Liu FP, Bai JJ, Xing YE, Li SJ, Li XH, Yu LY (2009) Cloning of MC4R gene and study on the association between SNPs of MC4R and growth trait in tilapia Oreochromis niloticus. J Fish Sci China 16:816–823 (Chinese with English abstract)
Liu L, Yu X, Tong J (2012) Molecular characterization of Myostatin (MSTN) gene and association analysis with growth traits in the bighead carp (Aristichthys nobilis). Mol Biol Rep 39:9211–9221
Lv YJ, Zhang S, Lin G, Ni JY, Ye Z (2008) Research on the high-yielding Spininbabus hollandi oshima cultured in cage. J Anhui Agric Sci 36:15522–15523 (Chinese with English abstract)
Mcpherron AC, Lee SJ (1997) Double muscling in cattle due to mutations in the Myostatin gene. Proc Natl Acad Sci USA 94:12457–12461
McPherron AC, Lawler AM, Lee SJ (1997) Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 387:83–90
Meidtner K, Wermter AK, Hinney A, Remschmidt H, Hebebrand J, Fries R (2010) Association of the melanocortin 4 receptor with feed intake and daily gain in F2 mangalitsa × pietrain pigs. Anim Genet 37:245–247
Meng X, Wang H, Qiu X, Liu H, Wang X (2017) SNPs of myostatin (MSTN) gene and their association with growth traits in three bay scallop (Argopecten irradians) populations. Aquac Res 48:531–536
Peñaloza C, Hamilton A, Guy DR, Bishop SC, Houston RD (2013) A SNP in the 5′ flanking region of the Myostatin-1b gene is associated with harvest traits in Atlantic salmon (Salmo salar). BMC Genom 14:112
Poćwierz-Kotus A, Bernaś R, Dębowski P, Kent MP, Lien S, Kesler M, Wenne R (2014) Genetic differentiation of southeast Baltic populations of sea trout inferred from single nucleotide polymorphisms. Anim Genet 45:96–104
Sánchez-Ramos I, Cross I, Mácha J, Martossa-Rodríguez G, Krylov V, Rebordinos L (2012) Assessment of tools for marker-assisted selection in a marine commercial species: Significant association between MSTN-1 gene polymorphism and growth traits. Sci World J 2012: 369802
Sellick GS, Pitchford WS, Morris CA, Cullen NG, Crawford AM, Raadsma HW (2007) Effect of Myostatin F94L on carcass yield in cattle. Anim Genet 38:440–446
Shu H, Yue L, Li Q, Lan ZJ, Gu PH, Wang HJ, Wu TJ (2015) Genetic variability of mtDNA COI genes in Spinibarbus hollandi. Oceanol Limnol Sin 46:1509–1515 (Chinese with English abstract)
Song XM, Jiang JF, Zhang GZ, Shi FX, Jiang YQ (2012) DNA polymorphisms of the hu sheep melanocortin-4 receptor gene associated with birth weight and 45-day weaning weight. Genet Mol Res 11:4432–4441
Sun Y, Yu X, Tong J (2012) Polymorphisms in Myostatin gene and associations with growth traits in the common carp (Cyprinus carpio L.). Int J Mol Sci 13:14956–14961
Vaiman D, Mercier D, Moazami-Goudarzi K, Eggen A, Ciampolini R, Lépingle A (1994) A set of 99 cattle microsatellites: characterization, synteny mapping, and polymorphism. Mamm Genome 5:288–297
Wang FQ (2013) Morphological variation analysis of different Spinibarbus hollandi Groups. J Anhui Agric Sci 41:2464–2465 (Chinese with English abstract)
Yang J, Liu Z, Shi H, Zhang J, Wang J, Huang J (2016) Association between MHC II beta chain gene polymorphisms and resistance to infectious haematopoietic necrosis virus in rainbow trout (Oncorhynchus mykiss, walbaum, 1792). Aquac Res 47:570–578
Zeng R, Zhang Y, Du P (2014) SNPs of melanocortin 4 receptor (MC4R) associated with body weight in beagle dogs. Exp Anim 63:73–78
Zhang CL, Chen H, Wang YH, Zhang L, Zhang AL, Zhang RF (2006) Association of a missense mutation of the MC4R gene with growth traits in cattle (brief report). Arch Tierz 49:515–516
Zhang L, Qiu XM, Wang J, Jiang ZQ, Liu Y, Wang XL (2012) Polymorphism analysis on melanocortin-4 receptor (MC4R) gene in Takifugu rubripes. Biotechn Bull 33:97–102 (Chinese with English abstract)
Zhu YY, Liang HW, Li Z, Luo XZ, Li L, Zhang ZW (2012) Polymorphism of MSTN gene and its association with growth traits in yellow catfish (Pelteobagruse fulvidraco). Hereditas 34:72–78
Acknowledgements
This work is supported by the National public industry major projects of China (No. 201303048) and the Guangdong Provincial Science and Technology Program (No. 2014A020208145).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Yang Yang declares that he does not have conflict of interest. Zhaojun LAN declares that he does not have conflict of interest. Hu Shu declares that he does not have conflict of interest. Huiqiang Zhou declares that he does not have conflict of interest. Xiaolu Jiang declares that she does not have conflict of interest. Liping Hou declares that she does not have conflict of interest. Pinghua Gu declares that he does not have conflict of interest.
Research involving human and animal rights
All animal experiments throughout the study were conducted in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals for the Science and Technology Bureau of China.
Rights and permissions
About this article
Cite this article
Yang, Y., Lan, Z., Shu, H. et al. Association between expression levels and growth trait-related SNPs located in promoters of the MC4R and MSTN genes in Spinibarbus hollandi. Genes Genom 40, 1119–1125 (2018). https://doi.org/10.1007/s13258-018-0666-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-018-0666-4