Abstract
Injection of shrimp with non-specific double-stranded RNA (dsRNA) of diverse lengths, sequences, and base compositions is known to induce non-specific immunity and protect against lethal disease, although the mechanisms are unclear. Previous shrimp studies examined the effects of non-specific RNA on particular pathways, while their global effects have not been examined. To understand the global effects of non-specific RNA in shrimp, we injected kuruma shrimp (Marsupenaeus japonicus) with a dsRNA and a small interfering RNA (siRNA) that is not specific to any gene in the shrimp genome and then examined global gene expression at 24 and 48 h with a microarray. For the non-specific RNA, we chose double-stranded green fluorescent protein (dsGFP) and siGFP because they are commonly used as mock controls and their effects on shrimp have not yet been studied. Injection of PBS was used as a control. The microarray results showed that many genes were up-regulated and some were down-regulated by dsGFP. In addition, dsGFP injection increased survival following WSSV challenge. The changes in expression for several genes were confirmed by quantitative PCR. The up-regulated genes included genes for eight immune-related proteins: c-type lectin 2, hemocyte homeostasis-associated protein, viral responsive protein, fibrinogen-related protein 1, sid-1 like protein, argonaute 2, Dicer 2, and heat shock protein 90. These results show that injection of shrimp with non-specific dsRNA hinders viral accumulation and prevents significant mortalities.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Shrimp farming has become a vital source of export income for many countries since the 1970s (Flegel and Sritunyalucksana 2011). From small-scale operations, it has become a vibrant industry that approaches the production level of capture fisheries. However, further growth has been impeded by the spread of viral pandemics, and production is still being impacted by viral pathogens such white spot syndrome virus (WSSV). There has been a rapid increase in the work on shrimp viral responses since 2000 (Flegel and Sritunyalucksana 2011). A notable discovery in the study of shrimp immune response is the presence of an RNA interference (RNAi) mechanism that was initially identified as a natural antiviral immunity in plants and arthropods (Wang et al. 2006a, b). In RNAi, double-stranded RNA (dsRNA) molecules suppress the expression of genes with homologous sequences (Robalino et al. 2005). Injecting shrimp with dsRNA specific to viral genes has had considerable success in blocking disease progression (Robalino et al. 2007). RNAi had been used as an antiviral therapy for Taura syndrome virus (TSV), WSSV, and yellowhead virus (YHV) because these viruses form dsRNAs that have the potential to engage the shrimp RNAi pathway, leading to effective antiviral responses. However, another interesting RNAi phenomenon is that injection of dsRNA of any length, sequence, or base composition can provide animals with partial protection from viral challenge.
The first study to exhibit such results was done by Robalino et al. (2004) where the injection of duck IgƲ dsRNA reduced cumulative mortality of Litopenaeus vannamei after TSV and WSSV challenge. It was speculated that this phenomenon is an example of a conserved antiviral mechanism of vertebrate and invertebrate innate immunity. Subsequently, other studies looked for novel molecular mechanisms of innate immunity in crustaceans. Because RNAi components are capable of dsRNA binding and recognition, some of them have been speculated to be involved in the observed protection against viral infection after dsRNA introduction. Clear mechanisms for such a response are yet to be established, and thus, numerous studies have examined the direct effect of dsRNA or viral DNA/RNA on the transcriptional expression of some RNAi components. Several lines of evidence suggest a functional overlap between non-specific activation of antiviral immunity and some aspects of the RNAi mechanism: (1) up-regulation of RNAi components such as Dicer (Su et al. 2008; Chen et al. 2011) and toll-like receptors (Labreuche et al. 2010) after viral infection; (2) increased WSSV replication after knockdown of RNAi components, trans-activation response RNA-binding protein (TRBP), and eukaryotic initiation factor 6 (eIF6) (Wang et al. 2012; Chendrimada et al. 2007); and (3) increased mRNA expression of argonaute and sid-1 (proteins that serve as channels for systemic spread of dsRNA) after injection of non-specific dsRNA (Labreuche et al. 2010).
Although microarray analyses have provided important insights into the regulation of biodefense mechanisms of shrimp, they have not yet been applied to understanding the effects of non-specific RNA on injected shrimp. Using microarray technology, the current study aimed to determine the global expression of shrimp injected with non-specific dsRNA. The microarray data were then used to identify genes that become up-regulated in comparison with non-injected and mock-injected shrimp. The relationship of transcript expression in experimental shrimp and observed increase in survival after pathogen challenge of pre-injected shrimp will give important insights about the components of this induced non-specific immune response.
Methods
Shrimp Samples
Apparently healthy kuruma shrimp (Marsupenaeus japonicus) with an average body weight of 14 g were used in the experiments. Delivered farm shrimp were temporarily kept in large stocking tanks prior to experiments. They were acclimatized for 5 days in a recirculating water tank with maintained temperature of 22–25 °C, salinity of 30–35 ppt, and aeration. Shrimp were fed with commercial pellets once a day during the course of acclimatization and injection experiments.
dsRNA Preparation/Synthesis
dsRNAs were in vitro synthesized using T7 RiboMAX™ Express Large-Scale RNA Production System (Promega, USA). Primers specific to GFP were designed and incorporated with T7 promoter to produce sense and anti-sense templates, which were then column-purified using Amicon® Ultra-0.5 centrifugal filter devices (Millipore, USA). They were immediately checked by gel electrophoresis for the presence of a clean fragment with the expected size. The concentration of these purified products was quantified using a spectrophotometer. Single-stranded RNAs (ssRNAs) were transcribed by incubation at 37 °C for 30 min using T7 RNA polymerase. Equal amounts of sense and anti-sense transcripts were annealed by incubation at 70 °C for 10 min to produce double-stranded RNA. Freshly synthesized dsRNAs were purified by phenol/chloroform/isoamyl alcohol extraction and finally verified by gel electrophoresis.
Injection and Hemocyte Collection
After acclimatization, shrimp were intramuscularly injected with 100 μl of phosphate-buffered saline (PBS) solution, small interfering green fluorescent protein (siGFP), or double-stranded green fluorescent protein (dsGFP) at a concentration of 3 μg per gram body weight of shrimp. Hemolymph of treated kuruma shrimp was collected at designated time points such as 0, 24, and 48 h postinjection. Approximately 500 μl of hemolymph was drawn out from the ventral sinus using a 2.5-ml sterile syringe with 1 ml of filter sterilized and pre-cooled anticoagulant (0.82 % NaCl, 0.55 % citric acid, 1.98 % glucose, and 0.88 % sodium citrate dissolved in water and adjusted to pH 5.6). Hemocytes were immediately collected by centrifugation at 3000×g for 3 min, and the resulting pellet was washed with 1× PBS.
Challenge Experiments
Two days after injection of PBS or dsGFP, 20 kuruma shrimp were injected intramuscularly with WSSV inoculums, which were previously prepared from tissues of WSSV-infected shrimp. These inoculums were diluted to 10−5 in 100 μl of PBS prior to injection. Mortalities were monitored and recorded daily for 14 to 20 days. Survival analysis was done using the statistics and graphing software packaged in GraphPad Prism 6.0d (GraphPad Software, Inc., USA).
Microarray Analysis
Total RNA from hemocytes of control and shrimp injected with PBS, dsGFP, and siGFP was extracted using RNAiso reagent (Takara, Japan). The extracted total RNA was then purified by RNeasy Mini Kit (Qiagen, Germany), and the purity was checked by Agilent 2200 TapeStation System (Agilent Technologies, USA). Total RNA (200 ng) was reverse transcribed and labeled with Cy3 using a Low Input Quick Amp Labeling kit (Agilent Technologies) and then hybridized on the microarray in accordance to the protocol listed in one-color microarray-based expression analysis. The hemocyte transcriptome was analyzed by using the Agilent Gene Expression oligo microarray. An 8 × 15-k microarray format, which contains around 13,875 probes, was used (eight microarrays per slide). Hybridization was done for 17 h at 65 °C and then followed by washing in buffers 1 and 2 (Agilent Technologies, USA) for 1 min each at room temperature and 37 °C, respectively. The slides were eventually dried and scanned immediately with Sure Scan High-Resolution Scanner (Agilent Technologies, USA). Data were extracted from the scanned slides using Agilent Feature Extraction Software and imported into GeneSpring GX Software (Agilent Technologies, USA). After data were filtered on expression, flags, and error, all treatment groups were compared by one-way analysis of variance (ANOVA) with a corrected p value of p ≤ 0.05. Only differentially expressed genes with a 4-fold change based on pairwise analysis with the control groups were further analyzed and visualized by Gene Cluster 3.0 program (de Hoon et al. 2004) and Java TreeView (Saldanha 2004). Gene ontology annotation was done using BLAST2GO (Conesa et al. 2005) with default parameters in BLAST (E value cutoff 1 × 10−3) and other analyses. Gene expression data were uploaded to NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE61541.
Gene Expression Analysis
Following the manufacturer’s protocol with slight modification, total RNA was extracted from pelleted hemocytes using RNAiso reagent. Concentration and purity were checked by a spectrophotometer, and 1 μg of total RNA was added as a template in reverse transcription reactions using High-Capacity cDNA Reverse Transcription kits (Applied Biosystems, USA). Expression levels were checked by quantitative PCR (qPCR). Primers used were designed using Primer Express 3 (Applied Biosystems, USA) and are listed in Table 1. qPCR was carried out in an Applied Biosystems 7300 real-time PCR system using Thunderbird™ SYBR® qPCR Mix with the following protocol: 60 s at 95 °C, 40 cycles of 15 s at 95 °C, and 60 s at 60 °C, and a dissociation curve analysis. Each reaction in a MicroAmp Optical 96-well reaction plate (Applied Biosystems, USA) contained 1× qPCR mix, 0.3 μM each of gene specific forward and reverse primers, 0.4 μl ROX reference dye, and 2-μl complementary DNA (cDNA) template. Gene expression data were determined by 2-ΔΔCT method using shrimp elongation factor-1 alpha (EF-1α) as reference gene and presented as relative expression ratio of treated shrimp and untreated shrimp. Statistical significance was determined using the Holm-Sidak method, with alpha = 5.000 %, packaged in GraphPad Prism 6.0d (GraphPad Software, Inc., USA). Computations assumed that all rows were sampled from populations with different scatter (SD).
Gene Expression Analysis of WSSV Viral Protein 28 (vp28)
cDNA templates from gills of control and experimental shrimp were used in real-time quantitative PCR analysis as described previously. Expression values are presented as ratios relative to gene expression of 1 dpi of WSSV in treated groups with EF-1α as the reference gene.
Results
Challenge Experiments
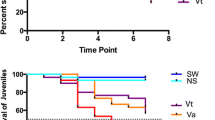
Shrimp were first injected with 3 μg of dsGFP and then injected with WSSV inoculum 2 days later. The results of challenge experiments are shown in Fig. 1. dsGFP injection prior to WSSV infection resulted to 60 % final cumulative survival of shrimp, which was significantly higher than the 5 % survival of the PBS control groups (chi-square and Gehan-Breslow-Wilcoxon tests). As shown in Fig. 1, most shrimp mortalities occurred during days 1–6.
Cumulative percent survival of shrimp challenged with white spot syndrome virus in relation to injection of PBS or dsGFP 2 days before the challenge. This is a representative of two experiments, each using 18 shrimp per treatment group. The survival curves are significantly different based on a chi-square and Gehan-Breslow-Wilcoxon test
Microarray Analysis
Out of 13,875 spotted genes on the microarray, 399 (14 %) genes were differentially expressed between treated and untreated groups based on 4-fold change significance analysis (Table 2). The 24 and 48-h dsGFP-treated groups had more up-regulated genes and fewer down-regulated genes than the siGFP and PBS groups. About 4 % (8) of the up-regulated genes and about 3 % (6) of the down-regulated genes had BLAST hits with E values less than 1 × 10−3. Cluster analysis of expression values based on expression and treatment is shown in Fig. 2. Four major groups were observed.
Hierarchical clustering analysis of 399 genes in hemocytes of shrimp injected with dsGFP, siGFP, and PBS at 24 and 48-h time points. Each row represents a single gene and each column is the average expression data of four samples in each experimental group. Genes were linked by the dendrogram shown on the left to illustrate similarity in expression pattern. Up-regulated genes are shown in red, down-regulated are in green, and genes without significant change from the control are in black
qPCR Analysis
At 5 dpi, expression of vp28 was much lower in the dsGFP-injected group than in the PBS-injected group (Fig. 3). qPCR was used to validate the microarray results for several genes from the shrimp innate immune response and components of the RNAi pathway. dsGFP injection significantly affected the expressions of several innate-immune-system-related genes, up-regulating eight of them (c-type lectin 2, hemocyte homeostasis-associated protein, viral responsive protein, fibrinogen-related protein, sid-1 like protein, argonaute 2, Dicer 2, and heat shock protein 90) and down-regulating one (serine proteinase inhibitor). The qPCR results for each of these genes (Fig. 4) were consistent with the microarray data.
Gene expression of WSSV viral protein 28 (vp28) at several time points after injection of PBS and dsGFP 2 days before viral challenge. The relative value was determined by 2-ΔΔCT method using shrimp elongation factor-1 alpha (EF-1α) as the reference gene of treated and untreated shrimp. The data were analyzed by multiple t tests, and statistical significance between PBS and dsGFP treatments (indicated by an asterisk) was determined using the Holm-Sidak method, with alpha = 5 %. Computations assume that all rows are sampled from populations with the same scatter (SD). Dotted line at the relative value of 1 represents no difference between control shrimp and experimental shrimp
Discussion
Microarray analyses have been shown to successfully elucidate the relationship of different sample conditions to the transcriptome response in many species (Calduch-Giner et al. 2014; Rebl et al. 2014; Mazurais et al. 2014; Limtipsuntorn et al. 2014; Kaitetzidou et al. 2012). In kuruma shrimp, recent transcriptome studies have shown that microarray technology is useful in studying the effects of gene-specific silencing on the expression of non-target genes. A link between different members of the shrimp immune system has been suggested because silencing transglutaminase, a protein responsible for blood coagulation, caused significant down-regulation of the gene expressions of other antimicrobial peptides like crustin and lysozyme (Fagutao et al. 2012). It is interesting to note that the number of down-regulated genes is higher than the number of up-regulated genes in global expression analyses of gene-specific silenced shrimp (Fagutao et al. 2009). In another microarray study of kuruma shrimp, peptidoglycan stimulation was found to increase the expressions of crustin, lysozyme, and a few antibacterial peptides (Fagutao et al. 2008). The antiviral ability of the shrimp immune system, however, is also not yet fully elucidated, and the mechanisms of immune stimulation by non-specific dsRNA remain unknown. Injecting dsRNA of diverse lengths, sequences, and base compositions has been demonstrated to protect shrimp from mortality induced by low doses of viruses, but the molecular mechanism is still unclear (Robalino et al. 2007). Subsequently, it was suggested that RNAi-related antiviral response and innate immune response may involve the same pathway in shrimp (Labreuche et al. 2010). These responses, however, required induction of sid-1 and Ago 2 by longer small RNA complexes like dsRNA. A similar result may be observed in this study because sid-1 and Ago 2 were up-regulated in the 24 and 48-h dsGFP-treated groups based on microarray and qPCR data. Additionally, the global expression patterns shown by microarray analysis indicated that only dsGFP induces an informative differential expression when the treatment groups are compared to the PBS-treated group.
In agreement with previous studies, our results show that injection of non-specific dsRNA like dsGFP significantly protected shrimp against WSSV infection (Fig. 1). During days 1 to 5, when most mortalities occurred, expression of WSSV vp28 was lower in the dsGFP-injected groups than in the control group (Fig. 3). At day 5, when the mortality of the control group reached 95 %, vp28 expression had been high for 4 days. On the other hand, the mortality of the dsGFP-injected group was 15 %, and vp28 expression was almost completely gone (RV = 0.28) at this time. In a study of Pacific white shrimp (Robalino et al. 2004), histological sections of non-specific dsRNA-treated shrimp at 3 dpi of WSSV showed no basophilic granular viral inclusions in the nuclei of gastric and cuticular epithelial tissues. Taken together, these results indicate that WSSV fails to accumulate significantly in non-specific dsRNA-treated shrimp. Our results also confirm that dsRNA induces an antiviral state in a sequence-independent manner, characteristic of the innate immunity.
Components of the RNAi mechanism that respond to non-specific dsRNA include Ago 2, Dicer 2, and sid-1-like protein (Labreuche et al. 2010). Consistent with these findings, Ago 2 and sid-1-like transcripts in kuruma shrimp were up-regulated by non-specific dsRNA. Up-regulation of RNAi components was expected because dsRNA is a known ligand of these molecules. The notable observation in this study, however, is that these genes are up-regulated not for silencing of a specific gene but for some other physiological response. We have observed in this study, as other studies also showed, that up-regulation of these genes caused a sufficient antiviral state that improves shrimp survival during WSSV infection. Subsequently, Ago 2 of Penaeus monodon was up-regulated by Staphylococcus aureus and WSSV, a clear indication that Ago 2 has an immune role (Yang et al. 2014). On the other hand, while the role of sid-1 in RNAi is already known, knockdown experiments reveal that sid-1 expression is a lethal phenotype. This indicated that the gene is involved in other essential functions (Labreuche et al. 2010). We speculate that one of these functions includes antiviral immunity. Dicer 2 is reported to be a host susceptibility locus in Drosophila (Galiana-Arnoux et al. 2006) and to be up-regulated in Chinese shrimp with acute WSSV infection (Li et al. 2010). Dicer 2 is up-regulated in Pacific white shrimp injected with poly-C/G and WSSV (Chen et al. 2011). Dicer 2 is also up-regulated in dsGFP-treated kuruma shrimp, which supports its role in non-specific activation of antiviral immunity. This activation of antiviral immunity may be observed because up-regulated transcript expression of Dicer 2 is reported to enhance RNAi potency and efficacy for antiviral action (Kim et al. 2005). Silencing of TRBP in kuruma shrimp increased viral replication, possibly as a result of destabilization in Dicer transcript expression (Labreuche et al. 2010). Dicer and TRBP directly interact with each other in the RNAi pathway, in which Dicer binds to dsRNA (Daniels et al. 2009). This finding supports the idea that Dicer has a more active role in preventing viral replication.
In addition to the analyzed RNAi components, many of the differentially expressed genes are known to have binding and catalytic activity as major molecular functions. The qPCR results (Fig. 4) confirm the up-regulation of many components of the innate immunity. In fact, dsGFP-injected groups have a considerably higher number of up-regulated genes than the siGFP- and PBS-injected groups (Table 1) confirming the results of other studies that non-specific long RNA complexes such as dsRNA can induce the expression of host genes. For example, the third highest gene in terms of expression at 2 dpi of dsGFP (based on microarray data) was identified as c-type lectin 2 gene (Online Resource 1). C-type lectins are pattern recognition proteins that recognize highly conserved pathogen-associated molecular patterns (PAMPs) on microbial surfaces. They were previously reported to agglutinate various bacteria but are also recently found to have antiviral functions (Xu et al. 2014). In this study, high up-regulation of c-type lectin 2 transcripts was observed at 1 and 2 dpi of dsGFP groups (Fig. 4), similar to what was found in studies analyzing c-type lectin expression after WSSV infection (Xu et al. 2014; Costa et al. 2011; Luo et al. 2007, 2003). Particularly, Xu et al. (2014) reported that kuruma shrimp c-type lectin 2 can inhibit the infection and replication of WSSV by binding to WSSV envelope protein vp28, thereby protecting the shrimp against WSSV infection. We suggest that up-regulation of c-type lectin 2 after injection of dsGFP (Fig. 4) led to inhibited vp28 expression (Fig. 3) which was then attributed to the observed increased survival in dsGFP-injected groups (Fig. 1). In addition, the transcripts of another pattern recognition receptor, fibrinogen-related protein (FREP) 1, were up-regulated in our study. Like c-type lectin 2, FREP 1 transcripts were also up-regulated in bacteria and WSSV-injected shrimp, and recombinant FREP 1 agglutinates several bacteria and binds peptidoglycan, LPS, bacteria, and vp28 of WSSV. These results confirm that dsGFP treatment induces many genes that are involved in binding of pathogens for an effective antiviral state.
Heat shock protein 90 (HSP90) transcripts were also found significantly up-regulated in the dsGFP groups as shown in both microarray and qPCR results. In other studies, this chaperone protein was up-regulated after WSSV challenge (Danwattananusorn et al. 2011; Wang et al. 2006a, b). However, it is not clear whether this up-regulation has a role in the host immune system or WSSV infectivity/pathogenicity. In this study, HSP90 was induced by non-specific RNA, which then supports that this protein has an immune role to protect the host from viral infection or stress caused by viral entry and not an infective factor.
We also analyzed the expression of novel viral responsive genes that were recently discovered. Both microarray and qPCR revealed their up-regulation in dsRNA-treated groups. For example, the hemocyte homeostasis-associated protein (HHAP) gene expression reached as high as 100-fold up-regulation in the dsGFP-injected groups at 1 and 2 dpi. HHAP is a viral responsive protein that is suggested to be important in hemocyte homeostasis. In WSSV-injected shrimp, it is speculated that the up-regulation of PmHHAP is a mechanism for maintaining the hemocyte level in the circulation by preventing too rapid hemocyte degradation and is also a way to control immune homeostasis in crustaceans during viral infection (Prapavorarat et al. 2010). Most likely in the same way, when shrimp was injected with dsGFP, the up-regulation of HHAP (Fig. 4) directed sufficient hemocyte persistence during viral challenge that perhaps led to increased shrimp survival. Meanwhile, our results also showed the up-regulation of a homolog of P. monodon viral responsive protein 15 (PmVRP15). This gene was the highest up-regulated viral responsive gene ever reported in shrimp (Vatanavicharn et al. 2014). Decreased WSSV propagation and shrimp mortality by infection after PmVRP15 knockdown indicate that this gene codes for a nuclear membrane protein that acts as a part of the WSSV propagation pathway. In this study, the gene homolog was significantly up-regulated. Because this gene is an infective factor, the up-regulation of this gene may be the reason why protection in dsGFP-injected groups is only partial and not as high as in experiments involving RNAi therapeutics such as dsVP28 (Sudhakaran et al. 2011). We speculate that this gene is triggered by the same receptors of WSSV in shrimp, and the up-regulation of other viral-defense-related genes has a probable compensatory mechanism to cancel out the effect of increased WSSV infectivity by the up-regulation of this gene. Likewise, it is also notable that MjVRP up-regulation is not as high as other immune-related genes tested (such as c-type lectin 2) or as reported by Vatanavicharn et al. (2014).
Expression of serine protease inhibitor (serpin) was slightly down-regulated after 1 dpi of dsGFP, but the change was not significant. In WSSV-challenged black tiger and Chinese shrimp, serpin was down-regulated (Homvises et al. 2010; Liu et al. 2009). In this period, the expression of Fc-serpin was strongly down-regulated, which might lead to the over-expression of serine proteinases and the disruption of the steady balance of serpin-serine proteinase in the immune system (Liu et al. 2009). More studies are needed to determine the effect of dsGFP-induced down-regulation of serpin on survival.
Our results demonstrate that dsGFP treatment induces the expression of about 180 genes related to the innate immune response or RNAi mechanism. Together, these genes hinder viral accumulation and prevent significant mortalities in infected shrimp.
References
Calduch-Giner JA, Echasseriau Y, Crespo D, Baron D, Planas JV, Prunet P, Pérez-Sánchez J (2014) Transcriptional assessment by microarray analysis and large-scale meta-analysis of the metabolic capacity of cardiac and skeletal muscle tissues to cope with reduced nutrient availability in gilthead sea bream (Sparus aurata L.). Mar Biotechnol 16(4):423–435
Chen YH, Jia XT, Zhao L, Li CZ, Zhang SA, Chen YG, Weng SP, He JG (2011) Identification and functional characterization of Dicer2 and five single VWC domain proteins of Litopenaeus vannamei. Dev Comp Immunol 35:661–671
Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA (2007) MicroRNA silencing through RISC recruitment of eIF6. Nature 447:823–828
Conesa A, Götz S, Garcia-Gomez JM, Terol J, Talon M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Costa F, Valença N, Silva A, Bezerra G, Cavada B, Rádis-Baptista G (2011) Cloning and molecular modeling of Litopenaeus vannamei (Penaeidae) C-type lectin homologs with mutated mannose binding domain-2. Genet Mol Res 10:650–664
Daniels SM, Melendez-Pena CE, Scarborough RJ, Daher A, Christensen HS et al (2009) Characterization of the TRBP domain required for dicer interaction and function in RNA interference. BMC Mol Biol 10:38
Danwattananusorn T, Fagutao FF, Shitara A, Kondo H, Aoki T, Nozaki R, Hirono I (2011) Molecular characterization and expression analysis of heat shock proteins 40, 70 and 90 from kuruma shrimp Marsupenaeus japonicus. Fish Sci 77:929–937
De Hoon MJL, Imoto S, Nolan J, Miyano S (2004) Open source clustering software. Bioinformatics 20(9):1453–1454
Fagutao FF, Koyama T, Kaizu A, Saito-Taki T, Kondo H, Aoki T, Hirono I (2009) Increased bacterial load in shrimp hemolymph in the absence of prophenoloxidase. FEBS J 276:5298–5306
Fagutao FF, Maningas MB, Kondo H, Aoki T, Hirono I (2012) Transglutaminase regulates immune-related genes in shrimp. Fish Shellfish Immunol 32:711–715
Fagutao FF, Yasuike M, Caipang CM, Kondo H, Hirono I, Takahashi Y (2008) Gene expression profile of hemocytes of kuruma shrimp, Marsupenaeus japonicus, following peptidoglycan stimulation. Mar Biotechnol 10:731–740
Flegel TW, Sritunyalucksana K (2011) Shrimp molecular responses to viral pathogens. Mar Biotechnol 13:587–607
Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL (2006) Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nature Immunol 7:590–597
Homvises T, Tassanakajon A, Somboonwiwat K (2010) Penaeus monodon SERPIN, PmSERPIN6, is implicated in the shrimp innate immunity. Fish Shellfish Immunol 29(5):890–898
Kaitetzidou E, Crespo D, Vraskou Y, Antonopoulou E, Planas JV (2012) Transcriptomic response of skeletal muscle to lipopolysaccharide in the gilthead seabream (Sparus aurata). Mar Biotechnol 14(5):605–619
Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ (2005) Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol 23:222–226
Labreuche Y, Veloso A, de la Vega E, Gross PS, Chapman RW, Browdy CL (2010) Non-specific activation of antiviral immunity and induction of RNA interference may engage the same pathway in the Pacific white leg shrimp Litopenaeus vannamei. Dev Comp Immunol 34:1209–1218
Li F, Wang D, Li S (2010) A Dorsal homolog (FcDorsal) in the Chinese shrimp Fenneropenaeus chinensis is responsive to both bacteria and WSSV challenge. Dev Comp Immunol 34(8):874–883
Limtipsuntorn U, Haga Y, Kondo H, Hirono I, Satoh S (2014) Microarray analysis of hepatic gene expression in juvenile Japanese flounder Paralichthys olivaceus fed diets supplemented with fish or vegetable oils. Mar Biotechnol 16(1):88–102
Liu Y, Li F, Wang B, Dong B, Zhang X, Xiang J (2009) A serpin from Chinese shrimp Fenneropenaeus chinensis is responsive to bacteria and WSSV challenge. Fish Shellfish Immunol 26(3):345–351
Luo T, Li F, Lei K, Xu X (2007) Genomic organization, promoter characterization and expression profiles of an antiviral gene PmAV from the shrimp Penaeus monodon. Mol Immunol 44:1516–1523
Luo T, Zhang X, Shao Z, Xu X (2003) PmAV, a novel gene involved in virus resistance of shrimp Penaeus monodon. FEBS Lett 551:53–57
Mazurais D, Ferraresso S, Gatta PP, Desbruyères E, Severe A, Corporeau C, Claireaux G, Bargelloni L, Zambonino-Infante JL (2014) Identification of hypoxia regulated genes in the liver of common sole (Solea solea) fed different dietary lipid contents. Mar Biotechnol 16(3):277–288
Prapavorarat A, Vatanavicharn T, Soderhall K, Tassanakajon A (2010) A novel viral responsive protein is involved in hemocyte homeostasis in the black tiger shrimp, Penaeus monodon. J Biol Chem 285:21467–21477
Rebl A, Korytář T, Köbis JM, Verleih M, Krasnov A, Jaros J, Kühn C, Köllner B, Goldammer T (2014) Transcriptome profiling reveals insight into distinct immune responses to Aeromonas salmonicida in gill of two rainbow trout strains. Mar Biotechnol 16(3):333–348
Robalino J, Bartlett T, Chapman R, Gross P, Browdy C, Warr G (2007) Double stranded RNA and antiviral immunity in marine shrimp: inducible host mechanisms and evidence for the evolution of viral counter-responses. Dev Comp Immunol 31:537–547
Robalino J, Bartlett T, Shepard E, Prior S, Jaramillo G, Scura E (2005) Double-stranded RNA induces sequence specific antiviral silencing in addition to nonspecific immunity in a marine shrimp: convergence of RNA interference and innate immunity in the invertebrate antiviral response? J Virol 79:13561–13571
Robalino J, Browdy CL, Prior S, Metz A, Parnell P, Gross P, Warr G (2004) Induction of antiviral immunity by double-stranded RNA in a marine invertebrate. J Virol 78:10442–10448
Saldanha AJ (2004) Java treeview-extensible visualization of microarray data. Bioinformatics 20:3246–3248
Sudhakaran R, Mekata T, Kono T, Inada M, Okugawa S, Yoshimine M, Yoshida T, Sakai M, Itami T (2011) Double-stranded RNA-mediated silencing of the white spot syndrome virus VP28 gene in kuruma shrimp, Marsupenaeus japonicus. Aquac Res 42:1153–1162
Su J, Oanh DTH, Lyons RE, Leeton L, van Hulten MCW, Tan S-H (2008) A key gene of the RNA interference pathway in the black tiger shrimp, Penaeus monodon: identification and functional characterization of Dicer-1. Fish Shellfish Immunol 24:223–233
Vatanavicharn T, Prapavorarat A, Jaree P, Somboonwiwat K, Tassanakajon A (2014) PmVRP15, a novel viral responsive protein from the black tiger shrimp, Penaeus monodon, promoted white spot syndrome virus replication. PLoS One 9(3):e91930
Wang B, Li F, Dong B, Zhang X, Zhang C, Xiang J (2006a) Discovery of the genes in response to white spot syndrome virus (WSSV) infection in Fenneropenaeus chinensis through cDNA microarray. Mar Biotechnol 8(5):491–500
Wang S, Chen AJ, Shi LJ, Zhao XF, Wang JX (2012) TRBP and eIF6 homologue in Marsupenaeus japonicus play crucial roles in antiviral response. PLoS One 7:e30057
Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R (2006b) RNA interference directs innate immunity against viruses in adult Drosophila. Science 312:452–454
Xu YH, Bi WJ, Wang XW, Zhao YR, Zhao XF, Wang JX (2014) Two novel C-type lectins with a low-density lipoprotein receptor class A domain have antiviral function in the shrimp Marsupenaeus japonicus. Dev Comp Immunol 42:323–332
Yang L, Li X, Jiang S, Qiu L, Zhou F, Liu W, Jiang S (2014) Characterization of Argonaute2 gene from black tiger shrimp (Penaeus monodon) and its responses to immune challenges. Fish Shellfish Immunol 36(1):261–269
Acknowledgments
This research was supported in part by grants-in-aid for scientific research from the Ministry of Education, Culture and Sports, Science and Technology of Japan.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(XLSX 25 kb)
Rights and permissions
About this article
Cite this article
Maralit, B.A., Komatsu, M., Hipolito, S.G. et al. Microarray Analysis of Immunity Against WSSV in Response to Injection of Non-specific Long dsRNA in Kuruma Shrimp, Marsupenaeus japonicus . Mar Biotechnol 17, 493–501 (2015). https://doi.org/10.1007/s10126-015-9637-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-015-9637-9