Abstract
Heat shock proteins (HSPs) are proteins that are expressed more strongly when the cells are exposed to physiological and stressful conditions. In this study, the full-length cDNAs of heat shock proteins 40 (MjHSP40), 70 (MjHSP70) and 90 (MjHSP90) were cloned from kuruma shrimp Marsupenaeus japonicus. The open reading frames (ORFs) of the cDNA clones have lengths of 1,191, 1,959 and 2,172 bp and encode 396, 652 and 723 amino acid residues, respectively. The predicted MjHSP40 amino acid sequence contains a J domain, a glycine/phenylalanine-rich region, and a central domain containing four repeats of a CxxCxGxG motif, indicating that it is a type I HSP40 homolog. The signature sequences of the HSP70 and HSP90 gene families are conserved in the MjHSP70 and MjHSP90 amino acid sequences. The deduced amino acid sequences of MjHSP70 and MjHSP90 share high identity with previously reported shrimp HSP70s and HSP90s, respectively. The expression of MjHSP90 mRNA increased at 32°C. Additionally, the expressions of MjHSP40, MjHSP70 and MjHSP90 mRNAs increased in defense-related tissues (i.e., hemocytes and lymphoid organ) when the shrimp were challenged with white spot syndrome virus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cultured shrimp, like most commercially cultured aquatic species, are constantly exposed to stresses, such as elevated temperatures, high pressure, or the presence of toxic compounds, in their rearing environment. These stresses ultimately affect the various biological processes in shrimp, and how they respond to them is crucial to shrimp health. Heat shock proteins (HSPs) can help shrimp and other aquatic animals cope with various stresses in their environment.
Heat shock proteins are found in both eukaryotes and prokaryotes, where they function as molecular chaperones that are known to participate in protein folding, transport and assembly. They are synthesized in response to a variety of stresses, including extremes of temperature, cellular energy depletion, and extreme concentrations of ions, other osmolytes, gases and various toxic substances [1]. HSPs have been classified according to their sequence homology and molecular weight into several families, such as HSP110, HSP100, HSP90, HSP70, HSP60, HSP40 and HSP20 [2].
HSP40s (also called DnaJs) have been conserved throughout evolution and are important for protein homeostasis, where they stimulate the ATPase activity of the HSP70 proteins that are involved in protein translation, folding, unfolding, translocation, and degradation [3]. The HSP40 family is large. For example, the genomes of Saccharomyces cerevisiae and Homo sapiens encode 20 and 44 members, respectively [4–6]. All HSP40s contain a 70-amino acid-long J domain that is responsible for interactions with HSP70 [7, 8]. HSP40s can be classified into three subtypes [5]. Type I HSP40s have a glycine and phenylalanine (G/F)-rich region and a cysteine-rich, zinc finger-like region (ZFLR). Type II HSP40s have a G/F-rich region, while type III HSP40s have neither a G/F-rich region nor a ZFLR. In yeast, the J domain alone is sufficient to enable HSP70 to perform its essential cellular function [9]. Recent studies suggest a mechanism where the substrate appears to be released from HSP40 and transferred to HSP70 [10–12]. Furthermore, HSP40 proteins can regulate the activities of other chaperones, such as HSP90 [13].
HSP70s play a role in protein synthesis under normal cellular conditions, fixing denatured proteins and preventing the misfolding or aggregation of proteins [14–16]. The HSP70 family is composed of several members, including heat-inducible HSP70, constitutively expressed heat shock cognate 70 (HSC70), glucose-regulated protein (GRP78), and others [17]. HSC70s, unlike HSP70s, have no introns. However, they share common structural features, including a 44-kDa N-terminal ATPase domain, an 18-kDa peptide-binding domain, and a 10-kDa C-terminal substrate-binding domain [17, 18].
HSP90 participates in the folding, maintenance of structural integrity, and the proper regulation of a subset of cytosolic proteins [19], and accounts for 1% of the soluble protein in most tissues, even in the absence of stress [20]. HSP90 also functions as a specialized chaperone for a set of signaling proteins, including several protein kinases and transcription factors [21]. HSP90 has roles in cell growth and differentiation, apoptosis, signal transduction and cell–cell communication. Eukaryotic HSP90 proteins consist of three domains: a 25-kDa N-terminal ATP-binding domain, a 40-kDa middle domain, and a 12-kDa C-terminal dimerization domain. The N-terminal ATP-binding domain is connected to the middle domain by a “linker” of variable length, and the C-terminal dimerization domain provides the binding site for a set of co-chaperone molecules that function with HSP90 as part of a multi-chaperone complex [21].
In recent years, there has been increasing interest in shrimp HSPs (mostly HSP70 and HSP90), because of their roles in shrimp immune response. HSP70 genes and their expressions have been studied in Chinese shrimp Fenneropenaeus chinensis [22], Pacific white shrimp Litopenaeus vannamei [23, 24], and black tiger shrimp Penaeus monodon [25, 26], while the expressions of HSP90 genes have been studied in F. chinensis [27], P. monodon [28], and greasyback shrimp Metapenaeus ensis [29]. In contrast, we were unable to find reports on HSP40 in shrimp. In the present study, HSP40, HSP70 and HSP90 were cloned from kuruma shrimp (Marsupenaeus japonicus), and their expressions were examined after heat shock and being challenged with white spot syndrome virus (WSSV).
Materials and methods
Shrimp
The kuruma shrimp used in this study were purchased from a commercial shrimp farm in Miyazaki, Japan. The shrimp were analyzed for signs of infectious diseases, kept in artificial seawater maintained at 25°C and 30–32 ppt, and fed daily with commercial shrimp feed prior to all experimental procedures.
Cloning of MjHSP40, MjHSP70 and MjHSP90
The MjHSP40, MjHSP70 and MjHSP90 cDNAs were amplified from a normal kuruma shrimp hepatopancreas cDNA library prepared in our laboratory. The pairs of specific primers designed based on the partial cDNA sequences of MjHSP40, MjHSP70 and MjHSP90 previously identified in our laboratory were used for PCR amplification (Table 1). Moreover, the pair of primers used to amplify MjHSP70 were also designed based on the cDNA sequence of kuruma shrimp HSP70 (GenBank accession no. ABK76338). The PCR reaction was performed as follows: an initial denaturation at 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min, and a final extension at 72°C for 5 min. After electrophoresis on 1% agarose gel, the PCR products were subsequently purified. The purified DNAs of the PCR products were ligated into the pGEM-T easy vector (Promega, USA) and were then transformed into Escherichia coli strain JM109. The positive clones were screened by colony PCR with M13 forward and reverse primers. The subsequent PCR products were directly sequenced with an ABI 3130xl capillary sequencer using BigDye chemistry (Applied Biosystems, USA).
Sequence data analysis
Nucleotide and amino acid sequences were analyzed using GENETYX WIN (v.7.0.3) software. Homology analysis and cleavage site prediction were accomplished with BLASTP (see http://ncbi.nlm.nih.gov/). Conceptual translation was performed and the characteristics of the protein were predicted using the ExPASy web server (http://www.expasy.ch). The motifs were predicted using SMART (http://smart.embl-heidelberg.de/) [30], and subcellular localization predictions were performed on the PSORT II sever (http://psort.hgc.jp/form2.html). Multiple sequence alignments were created using ClustalW [31]. The neighbor-jointing phylogenetic trees were then generated by MEGA4 software [32].
Expression analysis after heat shock and white spot syndrome virus (WSSV) challenge experiments
In order to study the MjHSP40, MjHSP70 and MjHSP90 expression after heat shock treatment, apparently healthy kuruma shrimp, each weighing about 10 g, were acclimated to a salinity of 30–32 ppt at 25°C for 7 days before experimentation, and then heat shock treatment was performed at 32°C for 3 h. Gills were dissected from three shrimp sampled before the heat shock treatment as an initial control, and at 1 and 3 h after heat shock. After this, MjHSP40, MjHSP70 and MjHSP90 mRNA expression levels were examined by quantitative real-time PCR. Total RNAs were extracted using the RNAiso reagent (TaKaRa Bio Inc., Japan), as described in the manufacturer’s protocol. The first-strand cDNAs were synthesized from 2 μg of the total RNAs using Moloney murine leukemia virus reverse transcriptase (Invitrogen, USA). Primer sets for MjHSP40, MjHSP70 and MjHSP90 were designed with ABI Primer Express Software v.3.0 (Applied Biosystems, USA), as shown in Table 1. Quantitative real-time PCR assays were done in a 20 μl reaction volume consisting of 5 μl template cDNA (2 μg/ml), 0.4 μl of both forward and reverse primers (10 pM), 10 μl Power SYBR Green Master Mix (Applied Biosystems), and 4.2 μl distilled water. Real-time PCR analysis was performed on an ABI7300 real-time PCR system (Applied Biosystems), following the manufacturer’s protocol. Expression levels were measured by the \(2^{{ - \rm\Updelta \rm\Updelta \it{C}_{{\text T}} }} \) method [33]. Elongation factor-1 α (EF-1α) mRNA levels were used as an internal control. Differences in expression were measured by one-way analysis of variance followed by the Tukey significant difference test using SPSS 16.0 software. P values of <0.05 were considered significant.
In addition, the MjHSP40, MjHSP70 and MjHSP90 mRNA expression levels were examined after the WSSV challenge experiment. Apparently healthy kuruma shrimp, each weighing about 10 g, were acclimated to laboratory conditions for 7 days and then injected with 50 μl of 106 × WSSV stock suspension. This virus stock dilution was used based on an earlier in vivo viral titration assay which suggested that this particular dilution is optimal for use in challenge experiments. Hemocytes, lymphoid organ and hepatopancreas were collected from three shrimp sampled before the WSSV challenge experiment as an initial control and at 1, 3 and 5 days post-WSSV injection, and total RNA was extracted and pooled together at each sampling time. Then the first-strand cDNA was synthesized as described above. RT-PCR was performed to analyze the expression profiles of MjHSP40, MjHSP70 and MjHSP90 using MjHSP40s-F, MjHSP40s-R, MjHSP70s-F, MjHSP70s-R, MjHSP90s-F and MjHSP90s-R (see Table 1). EF-1α was amplified as an internal control using EFs-F and EFs-R (see Table 1). One microliter of the first-strand cDNA was used as the template in the PCR amplification. The RT-PCR reaction was conducted with an initial predenaturation step performed at 95°C for 5 min followed by 25 and 30 cycles of 95°C for 30 s, 55°C for 30 s and 72°C for 30 s, and a final extension at 72°C for 5 min. Ten microliters of the amplified products were separated by electrophoresis with a 1% agarose gel and visualized with ethidium bromide. The mRNA bands were semi-quantitatively assessed for their relative expression following the method described by Lindenstrøm et al. [34] using ImageJ software to measure light intensity [35].
Results
Characterization of MjHSP40, MjHSP70 and MjHSP90 genes in kuruma shrimp
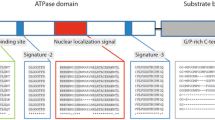
The full-length MjHSP40, MjHSP70 and MjHSP90 cDNA sequences (GenBank accession no. AB520825, AB520826 and AB520827, respectively) from kuruma shrimp were obtained by PCR amplification. For MjHSP40, the sequence consists of 1,191 nucleotides of an open reading frame (ORF) encoding 396 amino acids with a calculated molecular weight of 44.42 kDa and a theoretical pI of 6.62. The deduced amino acid sequence of MjHSP40 contains an N-terminal conserved domain (J domain, aa 5–60), a glycine/phenylalanine region (G/F domain, aa 67–96), a central domain containing four highly conserved cysteine-rich repeats with a consensus sequence of CxxCxGxG where x is any amino acid (CRR domain, aa 122–207), and a C-terminal domain (C domain, aa 220–344) (Fig. 1). In a GenBank BLASTP search, the deduced amino acid sequence of MjHSP40 showed high homology with those of other invertebrates: domestic silkworm Bombyx mori (NP_001040292, 69% identity), red flour beetle Tribolium castaneum (XP_971446, 64%), and jewel wasp Nasonia vitripennis (XP_001607240, 64%). MjHSP40 also had high similarities to DnaJ (Hsp40) homolog subfamily A member 1 from human Homo sapiens (NP_001530, 66%), heat shock protein 40 from American alligator Alligator mississippiensis (BAF94139, 65%), and DnaJ-like subfamily A member 4 from zebrafish Danio rerio (XP_689328, 62%) (Fig. 2). The deduced amino acid sequence of MjHSP70 was found to be very similar to M. japonicus HSP70s in the GenBank (accession nos. ABF83607 and ABK76338). Since both submissions were unpublished, we first verified the sequences of these genes.
Multiple alignment of the deduced amino acid sequences of HSP40 from kuruma shrimp and other animals. The amino acid positions are shown on the right. The N-terminal conserved domain (J domain) is boxed with a solid line, the glycine/phenylalanine-rich region (G/F domain) is boxed with a dashed line, and the C-terminal domain (CTD domain) is boxed with a dotted line. The central domain containing four repeats of a CxxCxGxG motif (CRR domain) is shown with a gray background. The HPD motif is marked with asterisks. Mj-HSP40 M. japonicus HSP40, Tc-HSP40h Tribolium castaneus HSP40 homolog (XP_971446), Tc-HSP40 T. castaneus HSP40 (XP_966855), Bm-HSP40h Bombyx mori DnaJ homolog subfamily A member 1 (NP_001040292), Bm-HSP40 B. mori HSP40 (BAD90846), Nv-HSP40h Nasonia vitripennis DnaJ homolog subfamily A member 1 (XP_001607240), Nv-HSP40 N. vitripennis HSP40 (XP_001601548), Cq-HSP40 Culex quinquefasciatus HSP40 (DnaJ chaperone) (XP_001844792), and Aa-HSP40 Aedes aegypti HSP40 (DnaJ chaperone) (ABF18277)
Phylogenetic analysis of MjHSP40. The neighbor joining (NJ) was done with MEGA4 software. A bootstrap test of the NJ analysis was conducted to evaluate the reliability of each branch in the tree. Numbers at the branches indicate the percentage of bootstrap values estimated from 1000 bootstrap replicates. Mj-HSP40 M. japonicus HSP40, Tc-HSP40h T. castaneus HSP40 homolog (XP_971446), Tc-HSP40 T. castaneus HSP40 (XP_966855), Bm-HSP40h B. mori DnaJ homolog subfamily A member 1 (NP_001040292), Bm-HSP40 B. mori HSP40 (BAD90846), Nv-HSP40h N. vitripennis DnaJ homolog subfamily A member 1 (XP_001607240), Nv-HSP40 N. vitripennis HSP40 (XP_001601548), Cq-HSP40 C. quinquefasciatus HSP40 (DnaJ chaperone) (XP_001844792), Aa-HSP40 A. aegypti HSP40 (DnaJ chaperone) (ABF18277), Dr-HSP40h Danio rerio DnaJ (HSP40) homolog subfamily A member 1 (NP_955956), Xl-HSP40h Xenopus laevis DnaJ (HSP40) homolog subfamily A member 1 (NP_00108365), Hs-HSP40h Homo sapiens DnaJ (HSP40) homolog subfamily A member 1 (NP_001530), Hs-HSP40 H. sapiens HSP740 (BAA12819), Mm-HSP40h Mus musculus DnaJ (HSP40) homolog subfamily A member 1 (NP_032324), Mm-HSP40 M. musculus HSP40 (BAA95672), Am-HSP40 Alligator mississippiensis HSP40 (BAF94139), and Gg-HSP40h Gallus gallus DnaJ (HSP40) homolog subfamily A member 1 (NP_001012963)
The deduced amino acid sequences of MjHSP70 also contain HSP70 family motifs and signatures, including an adenosine triphosphate/guanosine triphosphate (ATP/GTP)-binding site, a bi-partite nuclear localization signal, a non-organellar motif, and a conserved EEVD motif. MjHSP70 displayed very high homology with shrimp HSP70s from L. vannamei (AAT46566, 99%), P. monodon (AAQ05768, 99%), and M. ensis (ABF20530, 97%). Furthermore, MjHSP70 showed high similarities to HSP70 from American lobster Homarus americanus (ABA02165, 96%), marbled crab Pachygrapsus marmoratus (ABA02164, 94%), and pearl oyster Pteria penguin (ABJ97377, 86%).
The MjHSP90 cDNA contains a 2,172-bp ORF that encodes 723 amino acids with a calculated molecular weight of 83.6 kDa and a theoretical pI of 4.92. The deduced amino acid sequence of MjHSP90 also contains HSP90 family motifs and signatures, including five signatures, a typical histidine kinase-like ATPase (HATPase) domain, a GxxGxG motif, and a conserved MEEVD motif. MjHSP90 exhibited high homology with HSP90s of other invertebrates: P. monodon (ABM54577, 96%), M. enesis (ABR66910, 94%), Chinese mitten crab Eriocheir sinensis (ACJ01642, 90%), honey bee Apis mellifera (NP_001153536, 85%); and even with vertebrates: cattle Bos taurus (NP_001012688, 84%), African clawed frog Xenopus laevis (NP_001086624, 81%), and chicken Gallus gallus (NP_996842, 81%).
Expression profiles of MJHSP40, MjHSP70 and MjHSP90 after heat shock and white spot syndrome virus (WSSV) challenge experiments
In a preliminary experiment, we conducted a heat shock experiment at 35°C, and all shrimp died within 10 min. Thereafter, we tried it at 33°C, and we observed that most of the shrimp became moribund. We therefore conducted a heat shock experiment at 32°C. The expression profiles of MjHSP40, MjHSP70 and MjHSP90 mRNA after the shrimp had experienced the heat shock treatment are shown in Fig. 3. At 3 h post heat shock, the expression of MjHSP90 was significantly increased (fourfold) compared with that in shrimp that had not experienced heat shock (P < 0.05), whereas the expression of MjHSP40 and MjHSP70 showed no change. Furthermore, MjHSP40, MjHSP70 and MjHSP90 exhibited increased expression in the hemocytes and lymphoid organ after WSSV challenge compared with the initial control (Fig. 4). Both tissues play important roles in the innate immune response in shrimp. However, the gene expressions of the 3 MjHSPs in the hepatopancreas, another major defense-related tissue, showed no noticeable change.
Relative expressions of MjHSP40, MjHSP70 and MjHSP90 in gills of kuruma shrimp at 0, 1 and 3 h after heat shock treatment, as examined by quantitative real-time PCR. Each group at each time point consists of three samples pooled together. EF-1α mRNA levels were used as an internal control. Data are presented as the mean ± SD of triplicate measurements. Column bars with asterisks indicate values that are significantly different from those at time 0 (P < 0.05)
Expression profiles of MjHSP40, MjHSP70 and MjHSP90 mRNAs in defense-related tissues (i.e., hemocytes, lymphoid organ and hepatopancreas) of kuruma shrimp at 0, 1, 3 and 5 days post-WSSV injection (d.p.i.), as determined by RT-PCR (a), and a subsequent semi-quantitative analysis of their expression relative to EF-1α (b). Three samples were analyzed independently for each sampling period. EF-1α was used as an internal control. For the semi-quantitative analysis, mean values of the three measurements of specific gene expression relative to the corresponding EF-1α expression are presented. Bars represent standard deviations
Discussion
Recently, a number of HSPs were identified while extensively annotating the GenBank databases established from many crustaceans. HSPs function prominently in stress tolerance and promoting cell survival, and are especially involved in refolding proteins and preventing their denaturation [1, 19]. They also participate in a variety of normal cellular processes, including protein trafficking, signal transduction, DNA replication, and protein synthesis [14]. HSP genes such as HSP100, HSP90, HSP70, HSP60, HSP40, and small heat shock protein families consist of stress-inducible and constitutively expressed genes. Inducible genes are expressed at low levels under non-stress conditions, but are expressed increasingly in response to stress such as elevated temperatures, nutritional deficiencies, viral infection, ischemia–reperfusion injury, and exposure to oxidative stress, ultraviolet radiation, chemicals, and ethanol [36–42]. In the present study, the full-length cDNA sequences of HSP40, HSP70, and HSP90 were isolated from kuruma shrimp. Structural analysis shows that MjHSP40 contains an N-terminal conserved domain, a G/F domain, a central domain containing a CRR domain, and a C-terminal domain (Fig. 1). All DnaJ/HSP40 proteins contain the J domain, through which they bind to HSP70s [43–46], and they can be categorized into three groups depending on the presence of other domains [3]. This domain consists of four helices and a loop region between helices II and III that contains a highly conserved tripeptide of histidine, proline and aspartic acid (the HPD motif, aa 34–36) [8] (Fig. 1). Because the amino acid sequence of MjHSP40 is present in most type I HSP40 homologs [5], we classified it as a type I HSP40 homolog. Type I and type II HSP40s form chaperone pairs with cytosolic HSP70 that fold proteins with different efficiencies and carry out specific cellular functions [47, 48]. Type I DnaJ/HSP40 homolog or HSP40 and type II HSP40 differ in terms of structure, such that the former contains a central domain made up of four highly conserved cysteine-rich repeats with a consensus sequence of CxxCxGxG, and the latter contains only a J and G/F domain.
Our experiments also show that the expression of MjHSP40, MjHSP70 and MjHSP90 respond to heat shock and WSSV infection. In the heat shock treatment, the expression of MjHSP90 was increased (fourfold) compared with that in shrimp without heat shock at 3 h post heat shock (Fig. 3). Recently, although the HSP40 expression after heat shock treatment was not studied in shrimp, in the larvae of the midge Chironomus riparius, the expressions of HSP40 and HSP90 were upregulated within 1 h of heat shock treatment [49]. Similarly, in F. chinensis, P. monodon and freshwater shrimp, Macrobrachium rosenbergii, HSP70s and HSP90s appear to be transcriptionally up-regulated when the temperature is increased [22, 27, 28, 50]. Our results show that only the expression of MjHSP90 was increased after heat shock treatment, suggesting that MjHSP90 plays a role during and after thermal stresses by refolding and repairing denatured proteins. Hence, HSP90 can be used as a molecular marker gene for temperature stress conditions in shrimp. Moreover, the expressions of MjHSP40, MjHSP70 and MjHSP90 after WSSV challenge were increased in the hemocytes and lymphoid organ (Fig. 4). The expression profiles of F. chinensis HSP70 and HSP90 were also upregulated after infection with WSSV [51]. Therefore, the expressions of MjHSP40, MjHSP70 and MjHSP90 after WSSV challenge suggest that these proteins may play important roles in the immune function of shrimp against viruses. However, the expressions of these three MJHPS did not show any apparent change in the hepatopancreas (Fig. 4). EST analysis of cDNA libraries derived from shrimp hemocytes and hepatopancreas showed differences in the overall patterns of gene expression, including those of HSPs, suggesting that they have different functional roles in shrimp [52], which could explain why the expression patterns of MJHSPs in the two tissues are different.
In conclusion, HSP40 (MjHSP40), HSP70 (MjHSP70) and HSP90 (MjHSP90) from kuruma shrimp were characterized. A J domain, a G/F-rich region, and a central domain containing four repeats of a CxxCxGxG motif were found in the predicted MjHSP40 amino acid sequence. The conserved signature sequences of the HSP70 and HSP90 gene families were also found in the MjHSP70 and MjHSP90 amino acid sequences, respectively. The expression profiles of MjHSP40, MjHSP70 and MjHSP90 mRNAs were also examined after heat shock and WSSV challenge treatment. Our results show that only MjHSP90 mRNA levels were significantly induced by heat shock at 32°C, and that the expressions of MjHSP40, MjHSP70 and MjHSP90 increased after WSSV injection.
References
Feder ME, Hofmann GE (1999) Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol 61:243–282
Nover L, Scharf KD (1997) Heat stress proteins and transcription factors. Cell Mol Life Sci 53:80–103
Qiu XB, Shao YM, Miao S, Wang L (2006) The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci 63:2560–2570
Cyr DM, Langer T, Douglas MG (1994) DnaJ-like proteins: molecular chaperones and specific regulators of Hsp70. Trends Biochem Sci 19:176–181
Cheetham ME, Caplan AJ (1998) Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones 3:28–36
Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C (2001) The sequence of the human genome. Science 291:1304–1351
Hill RB, Flanagan JM, Prestegard JH (1995) 1H and 15 N magnetic resonance assignments, secondary structure, and tertiary fold of Escherichia coli DnaJ(1–78). Biochemistry 34:5587–5596
Qian YQ, Patel D, Hartl FU, McColl DJ (1996) Nuclear magnetic resonance solution structure of the human Hsp40 (HDJ-1) J-domain. J Mol Biol 260:224–235
Sahi C, Craig EA (2007) Network of general and specialty J protein chaperones of the yeast cytosol. Proc Natl Acad Sci USA 104:7163–7168
Jin Y, Awad W, Petrova K, Hendershot LM (2008) Regulated release of ERdj3 from unfolded proteins by BiP. EMBO J 27:2873–2882
Petrova K, Oyadomari S, Hendershot LM, Ron D (2008) Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. EMBO J 27:2862–2872
Summers DW, Douglas PM, Ramos CH, Cyr DM (2009) Polypeptide transfer from Hsp40 to Hsp70 molecular chaperones. Trends Biochem Sci 34:230–233
Brychzy A, Rein T, Winklhofer KF, Hartl FU, Young JC, Obermann WM (2003) Cofactor Tpr2 combines two TPR domains and a J domain to regulate the Hsp70/Hsp90 chaperone system. EMBO J 22:3613–3623
Hartl FU (1996) Molecular chaperones in cellular protein folding. Nature 381:571–579
Bukau B, Horwich AL (1998) The Hsp70 and Hsp60 chaperone machines. Cell 92:351–366
Kregel KC (2002) Heat shock proteins: modifying factors in physiological stress responses and acquired thermotolerance. J Appl Physiol 92:2177–2186
Kiang JG, Tsokos GC (1998) Heat shock protein 70 kDa: molecular biology, biochemistry, and physiology. Pharmacol Ther 80:183–201
Flaherty KM, DeLuca-Flaherty C, McKay DB (1990) Three-dimensional structure of the ATPase fragment of a 70 K heat-shock cognate protein. Nature 346:623–628
Picard D (2002) Heat-shock protein 90, a chaperone for folding and regulation. Cell Mol Life Sci 59:1640–1648
Buchner J (1999) Hsp90 & Co.—a holding for folding. Trends Biochem Sci 24:136–141
Pratt WB, Toft DO (2003) Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med (Maywood) 228:111–133
Luan W, Li F, Zhang J, Wen R, Li Y, Xiang J (2010) Identification of a novel inducible cytosolic Hsp70 gene in Chinese shrimp Fenneropenaeus chinensis and comparison of its expression with the cognate Hsc70 under different stresses. Cell Stress Chaperones 5:83–93
Cesar JR, Yang J (2007) Expression patterns of ubiquitin, heat shock protein 70, α-actin and β-actin over the molt cycle in the abdominal muscle of marine shrimp Litopenaeus vannamei. Mol Reprod Dev 74:554–559
Wu R, Sun L, Lei M, Xie ST (2008) Molecular identification and expression of heat shock cognate 70 (HSC70) in the Pacific white shrimp Litopenaeus vannamei. Mol Biol (Mosk) 42:265–274
Lo WY, Liu KF, Liao IC, Song YL (2004) Cloning and molecular characterization of heat shock cognate 70 from tiger shrimp (Penaeus monodon). Cell Stress Chaperones 9:332–343
Chuang KH, Ho SH, Song YL (2007) Cloning and expression analysis of heat shock cognate 70 gene promoter in tiger shrimp (Penaeus monodon). Gene 405:10–18
Li F, Luan W, Zhang C, Zhang J, Wang B, Xie Y, Li S, Xiang J (2009) Cloning of cytoplasmic heat shock protein 90 (FcHSP90) from Fenneropenaeus chinensis and its expression response to heat shock and hypoxia. Cell Stress Chaperones 14:161–172
Jiang S, Qiu L, Zhou F, Huang J, Guo Y, Yang K (2009) Molecular cloning and expression analysis of a heat shock protein (Hsp90) gene from black tiger shrimp (Penaeus monodon). Mol Biol Rep 36:127–134
Wu LT, Chu KH (2008) Characterization of heat shock protein 90 in the shrimp Metapenaeus ensis: evidence for its role in the regulation of vitellogenin synthesis. Mol Reprod Dev 75:952–959
Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P (2006) SMART 5: domains in the context of genomes and networks. Nucleic Acids Res 34:D257–D260
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the \(2^{{ - \rm\Updelta \rm\Updelta \it{C}_{{\text T}} }} \) method. Methods 25:402–408
Lindenstrøm T, Secombes CJ, Buchmann K (2004) Expression of immune response genes in rainbow trout skin induced by Gyrodactylus derjavini infections. Vet Immunol Immunopathol 97:137–148
Abramoff MD, Magelhaes PJ, Ran SJ (2004) Image processing with ImageJ. Biophoto Int 11:36–42
Welch WJ (1993) How cells respond to stress. Sci Am 268:56–64
Chouchane L, Bowers S, Sawasdikosol S, Simpson RM, Kindt TJ (1994) Heat shock proteins expressed on the surface of human T cell leukemia virus type I-infected cell lines induce autoantibodies in rabbits. J Infect Dis 169:253–259
Lathangue NB, Latchman DS (1988) A cellular protein related to heat shock protein 90 accumulates during herpes-simplex-virus infection and is overexpressed in transformed cells. Exp Cell Res 178:169–179
Donati YRA, Slosman DO, Polla BS (1990) Oxidative injury and the heat shock response. Biochem Pharmacol 40:2571–2577
Fincato G, Polentarutti N, Sica A, Mantovab A, Collotti F (1991) Expression of a heat inducible gene of the Hsp70 family in human myelomonocytic cells: regulation by bacterial products and cytokines. Blood 77:579–586
Morimoto RI (1998) Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Develop 12:3788–3796
Parsell DA, Lindquist S (1993) The function of heat shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Ann Rev Genetics 27:437–496
Szyperski T, Pellecchia M, Wall D, Georgopoulos C, Wuthrich K (1994) NMR structure determination of the Escherichia coli DnaJ molecular chaperone: secondary structure and backbone fold of the N-terminal region (residues 2–108) containing the highly conserved J domain. Proc Natl Acad Sci USA 91:11343–11347
Corsi AK, Schekman R (1997) The lumenal domain of Sec63p stimulates the ATPase activity of BiP and mediates BiP recruitment to the translocon in Saccharomyces cerevisiae. J Cell Biol 137:1483–1493
Young JC, Agashe VR, Siegers K, Hartl FU (2004) Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol 5:781–791
Hennessy F, Nicoll WS, Zimmermann R, Cheetham ME, Blatch GL (2005) Not all J domains are created equal: implications for the specificity of Hsp40–Hsp70 interactions. Protein Sci 14:1697–1709
Rudiger S, Scneider-Mergener J, Bukau B (2001) Its substrate specificity characterizes the DnaJ co-chaperones as a scanning factor for the DnaK chaperone. EMBO 20:1042–1050
Fan C-Y, Lee S, Ren H-Y, Cyr DM (2004) Exchangeable chaperone modules contribute to specification of type I and type II Hsp40 cellular function. Mol Biol Cell 15:761–773
Park K, Kwak IS (2008) Characterization of heat shock protein 40 and 90 in Chironomus riparinus larvae: effects of di(2-ethylhexayl) phthalate exposure on gene expression. Chemosphere 74:89–95
Liu J, Yang W-J, Zhu X-J, Karouna-Renier NK, Rao RK (2004) Molecular cloning and expression of HSP70 genes in the prawn, Macrobrachium rosenbergii. Cell Stress Chaperones 9:313–323
Wang B, Li F, Dong B, Zhang X, Xiang J (2006) Discovery of the genes in response to white spot syndrome virus (WSSV) infection in Fenneropenaeus chinensis through cDNA microarray. Mar Biotechnol (NY) 8:491–500
Gross PS, Bartlett TC, Browdy CL, Chapman RW, Warr GW (2001) Immune gene discovery by expressed sequence tag analysis of hemocytes and hepatopancreas in the Pacific White Shrimp, Litopenaeus vannamei, and the Atlantic White Shrimp, L. setiferus. Dev Comp Immunol 25:565–577
Acknowledgments
This study was supported in part by grants from the Ministry of Agriculture, Forestry, and Fisheries of Japan and Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the JSPS-NRCT Asian Core University program.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Danwattananusorn, T., Fagutao, F.F., Shitara, A. et al. Molecular characterization and expression analysis of heat shock proteins 40, 70 and 90 from kuruma shrimp Marsupenaeus japonicus . Fish Sci 77, 929–937 (2011). https://doi.org/10.1007/s12562-011-0394-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12562-011-0394-z