Abstract
Investigating the microbial communities associated with invasive plant species can provide insights into how these species establish and thrive in new environments. Here, we explored the fungal species associated with the roots of the invasive species Anthemis cotula L. at 12 sites with varying elevations in the Kashmir Himalaya. Illumina MiSeq platform was used to identify the species composition, diversity, and guild structure of these root-associated fungi. The study found a total of 706 fungal operational taxonomic units (OTUs) belonging to 8 phyla, 20 classes, 53 orders, 109 families, and 160 genera associated with roots of A. cotula, with the most common genus being Funneliformis. Arbuscular mycorrhizal fungi (AMF) constituted the largest guild at higher elevations. The study also revealed that out of the 12 OTUs comprising the core mycobiome, 4 OTUs constituted the stable component while the remaining 8 OTUs comprised the dynamic component. While α-diversity did not vary across sites, significant variation was noted in β-diversity. The study confirmed the facilitative role of the microbiome through a greenhouse trial in which a significant effect of soil microbiome on height, shoot biomass, root biomass, number of flower heads, and internal CO2 concentration of the host plant was observed. The study indicates that diverse fungal mutualists get associated with this invasive alien species even in nutrient-rich ruderal habitats and may be contributing to its spread into higher elevations. This study highlights the importance of understanding the role of root-associated fungi in invasion dynamics and the potential use of mycobiome management strategies to control invasive species.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Root-associated fungi (RAF) play a crucial role in the growth, development, and fitness of host plants (Bergelson et al. 2019; Nguyen et al. 2020; Abrego et al. 2020). The diversity of RAF is influenced by a range of factors, such as host plant species, root exudates, soil fertility, temperature, and other biotic and abiotic factors (Oh and Lim 2018; Goldmann et al. 2020; Abrego et al. 2020; Hu et al. 2021, 2022). Recent advances in metagenomics have enabled the study of the diversity and guild structure of RAF associated with a wide range of plants under different environmental conditions (Orellana 2013). The mycorrhizal fungal guild has received more attention (Rai and Agarkar 2016) but it is important to document the entire guild structure of these fungi associated with the roots of host plants to gain deeper insights into the community structure and assembly of RAF. Studies have revealed that the interactions of RAF with host plants range from mutualism to parasitism (Ahmad and Zaib 2020). Understanding the role of RAF in the growth and development of host plants is critical for developing sustainable agricultural practices and for conserving plant diversity in natural ecosystems (Maciá-Vicente 2022; Tondera et al. 2023).
Recent studies have started to shed light on the diversity of belowground microbial diversity in various ecosystems, including those impacted by invasive species. For example, Li et al. (2022) explored the effect of invasive bamboo species, namely Phyllostachys edulis, on the microbial community structure and function(s) of bulk soil, rhizosphere, and roots of a dominant native tree species (Cyclobalanopsis glauca) in a subtropical evergreen forest. Similarly, Řezáčová et al. (2022) investigated the root-associated and soil-dwelling arbuscular mycorrhizal fungi (AMF) of 11 invasive plant species in their native and exotic ranges and found that root-associated AMF assemblages were simplified, and many AMF genera were not associated with the invasive species in the exotic ranges. However, the dominant fungal genera were the same in both native and exotic ranges.
Notwithstanding these studies, role of root-associated microbes in plant invasiveness has been less studied compared to other factors, such as abiotic tolerance (Alpert et al. 2000; Levine et al. 2004), biotic resistance (Case 1990; Levine et al. 2004; Bogdziewicz et al. 2019), and propagule pressure (Lockwood et al. 2005; Simberloff 2009). But recent research has suggested that these microbes can have a significant impact on the performance and spread of invasive plant species (Byun et al. 2018; Paolucci et al. 2021). For example, studies have shown that invasive plants often have a greater abundance and diversity of root-associated microbes compared to non-invasive plants (Byun et al. 2018; Ramirez et al. 2019). These microbes can promote plant growth and increase nutrient uptake, allowing invasive species to outcompete native species for resources (Callaway and Aschehoug 2000; Kamutando et al. 2019). Additionally, some studies have found that root-associated microbes can help invasive plants to tolerate environmental stressors, such as drought or high salinity (Hierro et al. 2005). This could allow invasive species to expand into new areas where native species may not be able to survive (Vorstenbosch et al. 2020).
The study of these interactions in relation to abiotic factors, such as elevation, is an important area of research that could enhance our understanding of how plant–microbe interactions are influenced by environmental conditions (Zhang et al. 2021). Elevation can have a significant impact on the composition and abundance of microbial communities associated with plant roots (Merino-Martín et al. 2023). As elevation increases, environmental factors, such as temperature, precipitation, and soil type, can change, which in turn can influence the types of microbes that are able to survive and thrive in a given location (Midolo and Wellstein 2020). Additionally, elevation can affect the physiology of plants, which can also impact their ability to interact with microbes (Rudgers et al. 2020). Studying the interaction between resident microbes and non-resident host plants in relation to elevation could shed light on the mechanisms by which environmental factors shape plant–microbe interactions. There is a growing body of research suggesting that RAFs may play a facilitative role in the upward expansion of alien (non-native) species in nature (Pauchard et al. 2016). This is particularly relevant in the context of climate change, where it is believed that the upward expansion of alien plant species facilitated by microbes could increase significantly (Dainese et al. 2017). The study by Petitpierre et al. (2016) suggests that climate change could lead to the range expansion of non-native plant species through increased interactions with beneficial microbes, including RAFs. Another recent study by Ullah et al. (2022) also suggests that climate change could increase the impact of RAFs on the upward expansion of non-native plant species.
There is currently limited research on the specific relationship between the microbiome and the invasiveness of Anthemis cotula L. (mayweed or stinking chamomile) in Kashmir Himalaya. Given the well-known impact of microbiome on the growth and survival of plants, including invasive species, it is possible that it may be playing a role in the invasiveness of A. cotula in Kashmir Himalaya, but further research is needed to fully understand the extent of this relationship. Thus, we aimed to investigate how the composition and diversity of fungi associated with the roots of A. cotula change in relation to elevation. We assumed a very limited diversity of RAF with this invasive alien species, particularly in the recently invaded higher elevations. We also hypothesized that fungi, particularly arbuscular fungi, may not be associated with A. cotula in ruderal habitats that are disturbed and nutrient-rich (Ward et al. 2020) and the host species may not entail carbon costs for such a mutualistic association in these habitats (Jin et al. 2017). By examining the composition of root-associated fungi at different elevations, the study is expected to provide insights into how environmental factors that change with elevation influence fungal communities. Additionally, the study may help to identify fungal species that are adapted to specific elevations, which could have implications for management of this highly invasive species. The research questions addressed in the present study are:
-
a.
How rich is the assemblage of fungi associated with the roots of A. cotula in Kashmir Himalaya and whether they form a stable core mycobiome or tend to show opportunistic relationship across elevations?
-
b.
Does the guild structure of the root-associated fungi show any elevation-specific pattern?
-
c.
Is there any effect of microbiome on the traits that contribute to the invasiveness of A. cotula in Kashmir Himalaya?
Material and methods
Study species
Anthemis cotula L., commonly known as mayweed or stinking chamomile, is an annual monocarpic species and belongs to the family Asteraceae (Reshi et al. 2011). It is known by several other synonyms, including Chamaemelum cotula, Maruta cotula, and Matricaria cotula. Native to Eurasia and parts of northern Africa, A. cotula is believed to have been introduced outside its native range through trade as a contaminant of crop seeds or propagules (Erneberg 1999). Due to its ruderal life history strategy, which is characterized by its ability to colonize disturbed habitats, the species grows as a common weed in arable land, farmyards, roadsides, moist meadows, and overgrazed pastures (Adhikari et al. 2020). It is highly invasive in Kashmir Himalaya and several traits that contribute to its invasiveness in the Kashmir Himalayan region include synchronous germination of its achenes with favourable climatic conditions (Rashid et al. 2007), over-compensatory growth upon herbivory (Shah et al. 2012), allelopathic potential (Allaie et al. 2006; Rashid and Reshi 2012), widespread mycorrhizal association (Shah and Reshi 2007), and high reproductive output (Rashid and Reshi 2012). These traits allow the species to quickly spread and compete with native plant species, posing a threat to native biodiversity. This threat is likely to increase due to anthropogenic activities and global climate change.
Study sites
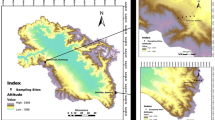
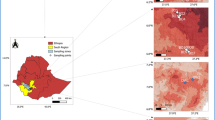
Extensive field surveys were conducted in Kashmir Himalaya, India, during the summer (April–July) of 2018 which revealed that A. cotula L. grows in ruderal habitats over an elevation range of 1600 m amsl to 3700 m amsl. To cover this entire elevation range, twelve sites (Fig. 1), having an average area of around 0.25 km2, were selected for the collection of root samples. The survey sites were grouped into lower (1550–1700 m amsl), middle (1701–2100 m amsl), and higher (2101–3700 m amsl) elevation sites and various features of these sites are given in Table S1.
Root and soil sample collection
Fifty individuals of A. cotula, at least 5 m apart from each other, were collected from each of the 12 sites within the first 2 weeks of April 2019. It was ensured that all the collected individuals were at the same developmental stage (spring cohort rosettes). The samples were processed immediately in the laboratory for separation of roots from rest of the plant. The root samples were washed with tap water and pooled to form a composite sample. In all, we had 12 composite root samples (four from each elevation) which were stored at − 20 °C until further use. Additionally, soil samples were collected from all the 12 sites for nutrient analysis. Five soil samples were collected from randomly selected places at each site by inserting a corer (2.25 cm diameter) vertically into the soil up to a depth of 15 cm. These sub-samples were thoroughly mixed and collected in plastic bags. The resulting composite soils were carried in polythene bags to the laboratory where these were gently crushed manually, air-dried, and sieved through a 2-mm sieve to eliminate debris.
DNA extraction
DNA was extracted from the 12 composite root samples using the CTAB method (Doyle 1991). DNA quality was checked on 0.8% (w/v) agarose gel. The quantity of extracted DNA was determined using a NanoDrop 2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). DNA was diluted to 50 ng μL−1 and stored at − 20 °C. For amplification of fungal DNA, a reaction volume of 25μL was prepared using 10μL 2 × reaction buffer (4 mM MgCl2, 0.4 mM of each dNTP, Taq polymerase (0.05 U/µL) (Thermo Scientific, USA), 0.5μL of forward (ITS1F) (White et al. 1990) and 0.5 μl reverse (ITS2) (Gardes and Bruns 1993) primer (10 μM), 13μL of sterilized distilled water, and 1.0μL of template DNA (50 ng). The amplification was carried out in Applied Biosystems Thermocycler, USA, and the following PCR conditions were followed: 94 °C 1 min; 94 °C 30 s; primer specific temperature (Tm) 30 s; 68 °C 30 s, 35 cycles; and a final extension at 68 °C 10 min and 4 °C hold. The resulting PCR products were separated on 0.75% (w/v) Ultrapure™ agarose (Invitrogen, USA) and stained with ethidium bromide using 1 × TAE (Tris–acetate EDTA) as running buffer. Amplicon library preparation and sequencing on an Illumina MiSeq, version 2, 2 × 250-bp paired-end chemistry were outsourced to Bionivid Technology (A GENOME “IT” COMPANY), Bangalore. Data were deposited with the National Centre for Biotechnology Information (NCBI) short read archive (SRA) under the bio-project accession number PRJNA613184.
Soil analysis
The soil samples were processed for analysis at the Soil Science Division, Department of Quality Control, Sher-e-Kashmir University of Agricultural Sciences and Technology, Shalimar, Srinagar. The pH of the soil samples (soil and distilled water in the ratio of 1:5) was measured by a portable pH meter (SYSTRONICS Model: MKVI). The soil organic carbon was estimated using the Walkley and Black (1934) rapid titration method. Olsen’s method was used for measuring phosphorus concentration (Olsen et al. 1954). The flame photometry procedure outlined by Jackson (1973) was employed to estimate total potassium (K) content. Sulphur in the soil samples was estimated by the calcium chloride method (Chesnin and Yien 1951) and calcium was measured in ammonium acetate extracts of soil by titration with EDTA (Cheng et al. 1953). Data on these soil variables is presented in Table 1.
Greenhouse experimental setup
The greenhouse experiment consisted of two treatments (sterilized soil and unsterilized soil) with 10 replications. The earthenware pots of 20 × 12 cm dimensions were used for the experiment. Soil (pH = 7.5 and organic carbon = 1.6%) was collected from an A. cotula invaded site (to mimic the natural rhizospheric conditions) and 10 pots were filled with this soil and pre-washed sand in the ratio of 1:1. Half of the soil and sand mixture was autoclaved thrice at 120 °C for 45 min with a 12-h interval between autoclaving (Shah et al. 2008a) and 10 pots were filled with this autoclaved soil and sand mixture to serve as control.
Seeds (achenes) collected in the fall of 2019 were germinated under natural light conditions in the greenhouse during April 2020 on plastic mesh plates containing vermiculite and peat. Three A. cotula seedlings after attaining a height of almost 4 cm were transplanted into each pot containing sterilized and unsterilized soil and sand mixture as described above. Finally, only 1 seedling of more or less similar size was retained per pot. The pots were arranged in a completely randomized design (in the greenhouse with natural light conditions and an average daily temperature of 28˚C). Pots were re-randomized every week to ensure randomization of all posts during the entire duration of the experiment. Pots were watered daily to keep the moisture at water-holding capacity and no additional nutrients were added to the pots. The experiment lasted from April 2020 to September 2020.
Data collection
Morphological and reproductive traits
Plants were harvested after the completion of experiment and their height (cm) and number of lateral branches (NLB) were recorded. Subsequently, the harvested plants were separated into belowground (root) and aboveground (shoot) parts. After thoroughly washing with tap water to remove any adhered soil particles, roots and shoots were dried in a hot air oven at 55 °C for 48 h to constant weight and expressed as root and shoot dry mass (g/plant). Since it was difficult to count the exact number of flowers or achenes per plant owing to high flower and achene number in capitula, the number of flower heads (capitula) (NFH) per plant was used as a surrogate for fitness.
Measurement of gas exchange parameters
Various gas exchange parameters, including photosynthetic rate/PN (µmolCO2 m−2 s−1) stomatal conductance/gs (mmol H2O m−2 s−1), internal leaf carbon-dioxide concentration (µmol/mol), and transpiration rates/E (mmol H2O m−2 s−1) of both treatment and control plants, were recorded just before their harvest using a Portable LI-6400 XT infrared gas analyser (Li-Cor, USA).
Bioinformatic and data analyses
The PIPITS v.2.3 bioinformatic pipeline was used to analyse the fungal internal transcribed spacer (ITS) sequence data from the Illumina MiSeq sequencing platform (Gweon et al. 2015). Using default parameters, preliminary processing of the data included de-multiplexing and quality filtering with a minimum quality score of 30. ITS2 region was extracted by PIPITS FUNITS. Sequences less than 100 bp and singletons were removed. Sequences were clustered into operational taxonomical units (OTUs) using the VSEARCH with an identity threshold of 97% by using the tools of QIIME v.1.9.1 (Rognes et al. 2016; Edgar 2018; Zou et al. 2018). The identified clusters were subjected to chimera removal. Chimera-free representatives were taxonomically assigned using the RDP classifier against the UNITE fungal ITS reference dataset version 8.2 (released on 04.02.2020).
To reduce sample variability across elevation zones, OTU table was rarefied to an equal sequencing depth with the rarefy_even_depth command in phyloseq package (McMurdie and Holmes 2013). Rarefaction curves were plotted using the modified function ggrare of ranacapa package (Kandlikar et al. 2018). The rarefied OTU data were used to compute alpha diversity indices, namely “Observed”, “ACE”, “Shannon”, “Simpson”, “InvSimpson”, “Fisher” using Phyloseq package in R environment and visualized as boxplots, and means of alpha diversity indices were statistically compared across elevation zones using the Wilcoxon test.
The variations in the composition of fungal communities across elevation zones were visualized utilizing the non-metric multidiomensional scaling (NMDS) based on Bray–Curtis dissimilarity using ordinate function of phyloseq package in R. Permutational multivariate analysis of variance (PERMANOVA) was used to determine if fungal communities differed significantly between the elevation zones, and adonis2 in Vegan package in R was used for these analyses. Finally, the taxon composition of samples was elucidated and reflected in the form of stacked bar plots in Phyloseq package using ggplot2.
Differential abundance of taxa across sites and elevation zones were computed using microeco package and plotted using ggalluvial and also as a heatmap. Venn and petal plots showing number of shared taxa were also prepared with microeco package.
Core mycobiome analysis covering unique and shared taxa for each site based upon a robust 75% prevalence threshold with at least 1% detection threshold was identified using the core function in microbiome R package version 1.5.28 (Lahti and Sudarshan 2017). We also created a Venn diagram using an online tool, InteractiveVenn, to define stable and dynamic components of the core mycobiome (Bardou et al. 2014). Functional guilds and trophic modes were assigned to fungal taxa using FUNGuild (Nguyen et al. 2016; Martínez-Diz et al. 2019) and the results were plotted as PieDonut plots using webr package in R. One-way anova was used to test the influence of elevation zones on guild structure and trophic modes. t-test was employed to compare the mean values of different traits of A. cotula between the plants grown in sterilized and unsterilized soil: sand medium. All data analyses were carried out in R (version 4.2.2; R Core Team 2022).
Results
In all, we obtained 2,367,266 reads after processing the 12 composite root samples, but only 2,242,792 reads remained after quality filtering that were resolved into 8 phyla, 20 classes, 53 orders, 109 families, 160 genera, and 706 OTUs. The average number of sequences across the samples ranged from 925 to 53,517 and the average number of counts per sample was 7888. In view of large difference in the sequence depth across sites, rarefaction of the data was performed which allowed us to resolve the reads into a total of 521 OTUs (Table S2). Furthermore, rarefaction analysis indicated that the sequencing was thorough in capturing the fungal alpha diversity across elevation zones (Fig. 2). Out of 521 OTUs, 119, 102, and 79 OTUs were unique to lower, middle, and higher elevations, respectively. Sixty-eight OTUs were shared by lower and middle elevations and 37 by middle and higher while only 18 by higher and lower elevations. Only 98 (19%) OTUs were common to all the elevations (Fig. S1). Moreover, the percent abundance of OTUs of root-associated fungi of A. cotula across the sites is presented in Fig. 3. Only rarefied data was used for further downstream analyses.
The present study revealed that phylum Ascomycota was the most abundant taxon with a relative abundance of 71.5% in the samples, followed by Basidiomycota, Glomeromycota, and Olpidiomycota with relative abundance of 12.6%, 9%, and 4%, respectively. Overrepresentation of Ascomycota and Basidiomycota and relatively low incidence of Glomeromycota can be attributed to the primer (ITS1F and ITS2) selection. The above-mentioned taxa together comprise about 97% of reads in all the samples. The remaining 3% reads were represented by Mortierellomycota, Rozellomycota, Chytridiomycota, Mucoromycota, and some unidentified fungal groups. Elevation-specific abundance of the fungal groups revealed that Ascomycota was the most abundant phylum at lower and middle elevations followed by Basidiomycota, Glomeromycota, and others. Though Ascomycota was also most abundant at higher elevations, the next most abundant phyla at this elevation zone, however, was Glomeromycota followed by Olpidiomycota, Basidiomycota, and others (Fig. 4). Across elevation zones, the most common classes were Dothideomycetes, Sordariomycetes, Agaricomycetes, and Glomeromycetes with relative abundance of 40%, 20.3%, 10%, and 9%, respectively.
The most common orders, across all the sites spanning three elevation zones, were Pleosporales (38%), Hypocreales (9.3%), and Glomerales (8.5%) followed by Agaricales (5%). Elevation-specific abundance of different orders showed that Pleosporales was the most abundant order at all three elevation zones with relative abundance of 36%, 53%, and 24% at lower, middle, and higher elevation zones, respectively. At lower elevation zone, the next orders in terms of their relative abundance were Agaricales (13%), Cantharellales (11%), and Glomerales (6%). At middle elevation zone, the next common fungal orders after Pleosporales were Hypocreales (53%), Sordariales (10%), and Glomerales (4.1%). In the higher elevation zone, on the other hand, Glomerales (16%) was the second most abundant order followed by Hypocreales (11%) and Olpidiales (6.5%).
The most abundant fungal families were Didymellaceae (10%), Nectriaceae (9%), Glomeraceae (8.1%), and Ceratobasidiaceae (4%) across all the elevation zones (Fig. 5). Lower elevations zone had the dominance of Ceratobasidiaceae (11%), Tricholomataceae (9.7%), Didymellaceae (8%), Nectriaceae (6%), and Glomeraceae (5%). At middle elevation zone, Didymellaceae (19%), Nectriaceae (9%), Lasiosphariaceae (4.4%), and Glomeraceae (4%) were more abundant. On the other hand, Glomeraceae (15%) was the most abundant family followed by Nectriaceae (11%), Olpidiaceae (6.5%), and Didymellaceae (4%) at the higher elevation zone.
At the genus level, the most abundant fungal genera were Funneliformis (7.6%), Olpidium (4%), Vishniacozyma (2.2%), and Pyrenochaeptopsis (1.2%) across all the elevation zones (Fig. 6). Funneliformis (5%), Olpidium (3%), and Vishniacozyma (2.7%) were the most common genera at lower elevation zone. Vishniacozyma (3.7%), Funneliformis (3.5%), and Mortierella (2%) were the most abundant genera at middle elevation zone. At the higher elevation zone, the most commonly occurring genera were Funneliformis (15%), Olpidium (6.5%), and Ophiosphaerella (2.7%). The most abundant taxa at lower, middle, and higher elevation zones at species level are shown in differential heat trees (Fig. S2).
Alpha diversity
Alpha diversity based on various indices is presented in Fig. 7. Except ACE, all other indices of alpha diversity revealed higher values in middle elevation zone and lower values in higher elevation zone (Table S3). For instance, observed species richness was 127 in middle elevation zone and 96 in higher elevation zone. The values of other alpha diversity indices including Shannon, Simpson, Inverse Simpson, and Fisher followed a pattern similar to observed species richness across the three elevational zones. However, none of these indices varied significantly across elevations (p > 0.05) (Table 2).
β-Diversity
In order to visualize the dissimilarities in root-associated fungal community composition between the samples, NMDS analysis based on the Bray–Curtis dissimilarity index was undertaken. The NMDS ordination had a stress value of 0.102, and the visual interpretation of the NMDS plot (Fig. 8) indicated some overlap between root-associated fungal communities of various elevation zones. However, PERMANOVA analysis revealed significant difference between the groups (p < 0.05) Table 3.
Relationship between the fungal community composition and edaphic factors
Redundancy analysis (RDA) was used to explore the influence of edaphic factors (used as explanatory variables) on fungal community composition in roots of A. cotula. The first two axes explained 67.5% of the total variance (Fig. 9). This analysis indicated that organic carbon (OC) was the major determinant that explained most of the variations in the fungal community composition of the three elevation zones. While the relative abundance of Funneliformis at higher elevations had strong correlation with OC content, it was inversely related to nitrogen (N), pH, and calcium (Ca). The abundance of genus Olpidium, Vishniacozyma, and Neoascochyta was related with lower levels of OC and potassium (K) but higher pH, N, and Ca. The abundance of Tetracladium was strongly related to K but negatively related to sulphur (S). Likewise, the abundance of genera, including Ophioshaerella, Calyptella, Podospora, Mortirella, and Pyrenochaetopsis, is determined more by S than by P, Ca, K, N, and pH.
Redundancy analysis (RDA) showing the influence of soil variables on the root-associated fungal community composition in roots of A. cotula. Red arrows represent edaphic factors; grey arrows represent fungal genera; each dot represents a sample. For clarity, only the twenty most abundant taxa are shown
Core mycobiome
The majority of the OTUs had elevation-specific distribution. Overall, a total of 12 different OTUs across elevations comprised the core mycobiome. Many reproducible and other non-reproducible OTUs which did not follow the criteria of being core comprised the opportunistic mycobiome. The number of OTUs constituting core mycobiome at the three elevation zones was 9, 7, and 8 for lower, middle, and higher elevation zones, respectively. A small proportion of these core OTUs was reproducibly detected only at one or two elevation zones. For instance, 1 OTU was exclusively present at lower elevation zone, 1 at middle elevation zone, and 2 at higher elevation zone (Fig. 10). A considerable fraction of core OTUs were consistently present at only two of the three elevation zones (e.g. 2 OTUs were constantly present at middle and lower elevation zones, 2 at the higher and lower elevation zones, and none at higher and middle elevation zones). However, only 4 OTUs were invariably present across all the elevation zones. Thus, the core mycobiome comprising 12 OTUs can be categorized into a stable component comprising only 4 OTUs (Fig. 11) and a dynamic component with 8 OTUs (occurring in either one or two of the elevation zones).
Guild structure and trophic modes
A total of 19 guilds were recorded at lower elevation zone and 17 each at middle and higher elevation zones. These guild types were either unique to an elevation zone or were shared between two or all three elevation zones. At lower elevation zone, the most abundant guilds were plant pathogens, AMF, and endophytes with relative abundance of 7%, 5%, and 4%, respectively. Likewise, middle elevation zones showed predominance of plant pathogens (7%), endophytes (5%), animal pathogen (5%), and AMF (3%). However, at higher elevation zone, AMF and plant pathogens formed the most abundant guilds with relative abundance of 5% each, followed by endophytes (3.5%) and animal pathogen (3%) (Fig. 12). Besides the above-mentioned guild types, some guilds were unique to specific elevations. For instance, orchid mycorrhizal guild occurred exclusively at lower elevation zone, whereas ericoid mycorrhizal guild existed at middle and higher elevation zones. Likewise, plant parasite and bryophyte parasite guilds occurred only at lower and higher elevation zones (Table S4). One-way analysis of variance revealed insignificant relationship between elevation zone and guild structure (F2,42 = 0.182, p = 0.834).
All the identified species were successfully assigned to different trophic modes (Fig. 13). A total of three trophic modes, including saprotrophs, symbiotrophs, and pathotrophs, were recorded from each elevation zone. Saprotrophs constituted the most abundant trophic mode across elevations. While symbiotrophs were relatively more abundant at lower elevation zone (12%) and higher elevation zones (10%); saprotrophs and pathotrophs both showed higher abundance at middle elevation zone in comparison to lower and higher elevation zones (Table S5). Abundance of trophic modes did not vary significantly across elevation zones (F2,6 = 0.684, p = 0.54).
Effect of microbiome on host plant traits
The effect of entire soil microbiome including the root-associated fungi on various morphological and reproductive traits of A. cotula is presented in Table 4. Among the morphological traits, height (cm) (t = 3.99; p = 0.003) and shoot biomass (g) (t = 5.8; p = 0.0003) showed a significant increase in unsterilized soil over control. On the contrary, root biomass (g) declined significantly in unsterilized soil as compared to control (t = 10.8; p = 0.0001). The number of lateral branches (NLB), however, did not vary significantly between sterilized and unsterilized soil treatments. The number of flower heads (NFH) varied significantly between the two treatments (t = 4.5; p = 0.001). Among the physiological attributes, only leaf internal CO2 concentration differed significantly between the sterilized and unsterilized soil treatments (t = 3.09; p = 0.01).
Discussion
The study presents an assessment of the root-associated fungal species and guilds of A. cotula in various elevation zones in Kashmir Himalaya. It was revealed that the roots of A. cotula host a diverse group of fungal species, with a higher representation of Ascomycota. Several other studies have also reported high fungal species diversity associated with invasive plants, such as Phragmites australis (Van Bael 2020) and Berberis thunbergii (Coats and Rumpho 2014), and also the predominance of Ascomycota. This pattern may be due to several factors, including the ability of Ascomycota to form associations with a broad range of host plants and contribute to invasiveness by providing the hosts with a diverse functions, such as nutrient acquisition, pathogen defence, and stress tolerance (James et al. 2006; Challacombe et al. 2019; Sudová et al. 2020). It is also worth noting that the predominance of Ascomycota could be due to their higher abundance in the roots at the time of the investigation or the choice of primers used in the study (ITS1F and ITS2). It is important to note that the relationship between invasive plants and their fungal associates is complex and can vary depending on the specific plant-fungal interactions involved, as well as the environment in which they occur. Therefore, the results of the present study may not be generalizable to other plant species or regions. However, the findings highlight the importance of understanding the diversity of fungal species associated with plant roots, as these fungi play important roles in nutrient acquisition and plant growth. Further research is needed to fully understand the mechanisms underlying the association between A. cotula and its root-associated fungi, and how these interactions contribute to invasiveness of A. cotula in Kashmir Himalaya.
We also found that most of the fungal species associated with A. cotula were opportunistic, meaning they were generalists and not specifically adapted to this plant. This was expected, as A. cotula is not native to the area and therefore does not share a long-term evolutionary association with its root-associated fungi. The lack of static core mycobiome may have contributed to the success of A. cotula in the area, as it can benefit from different root-associated fungi in different elevations. In fact, the role of resident soil microbiota in the success of alien plant species has been well documented (Klironomos 2002). Mycorrhizal mutualists, for example, can aid the alien plant species in overcoming biotic resistance in invaded ranges and reduce the incidence of insect herbivory, promoting successful invasion (Yin et al. 2020).
Our results also suggest that there is a shift in the prevalence of different fungal guilds from lower to higher elevations, with arbuscular mycorrhizal fungi (AMF) being predominant at higher elevations and plant pathogens being dominant at lower and middle elevations. This trend is supported by previous research which suggests that climatic factors play a significant role in determining the prevalence of fungal guilds in different habitat types (Tedersoo et al. 2014; Veach et al. 2018).
The present study also brings out the inconsistent pattern in the number and abundance of different fungal genera at different elevations, except for AMF genus Funneliformis, which was found associated with A. cotula roots across all elevations, with the highest relative abundance at higher elevations. This indicates that Funneliformis may be contributing to the establishment and expansion of A. cotula at higher elevations. The possible explanations for the increase in dominance of arbuscular mycorrhizal fungi (AMF) and decrease in abundance of saprotrophic fungi with elevation are as follows: (1) soil nutrient availability which decreases as elevation increases. AMF may dominate in nutrient-poor soils found at higher elevations because these are specialized in forming symbiotic relationships with plant roots to obtain nutrients, particularly phosphorus, in exchange for carbohydrates. In contrast, saprotrophic fungi rely on decaying organic matter as a source of nutrients, which may be less abundant in higher elevation soils; (2) carbon availability in soil may decrease with elevation due to lower temperatures and slower decomposition rates. As a result, saprotrophic fungi may have less access to carbon sources in higher elevation soils, leading to their decreased abundance. AMF may be better adapted to survive in low carbon availability environments because they get the carbon from the host plants; (3) plant communities can vary with elevation, with different plant species dominating at different elevations. Some plant species prefer forming relationships with AMF, while others prefer saprotrophic fungi. If the dominant plant species at higher elevations have a greater preference for AMF, then this could contribute to the dominance of AMF at these elevations, and (4) soil pH and moisture content can also vary with elevation, and these factors can influence fungal community composition. AMF may be better adapted to survive in acidic soils found at higher elevations, while saprotrophic fungi may be better adapted to more neutral or alkaline soils. Additionally, moisture content can affect fungal community composition, with AMF generally preferring drier soils compared to saprotrophic fungi. In fact, studies have reported AMF colonization and diverse spore types in high-altitude regions, such as the world’s highest summits (Pan et al. 2013) and coldest places (Upson et al. 2008; Bueno De Mesquita et al. 2018). These findings indicate that AMF symbiosis can occur in environments previously believed to be unsuitable for it. Contrary to our results, a study of alpine plant communities in the Rocky Mountains revealed that dominance of arbuscular mycorrhizal fungi (AMF) decreased with elevation, while the abundance of saprotrophic fungi increased (Li et al. 2020). This pattern was attributed to the lower soil nutrient availability at higher elevations, which favours saprotrophic fungi that can decompose organic matter and release nutrients into the soil (Oehl and Körner 2014; Kotilínek et al. 2017). Similarly, a study of tropical montane forests in Costa Rica found that the abundance of ectomycorrhizal fungi (EMF) increased with elevation, while AMF became less abundant (Desai et al. 2016). This pattern was attributed to changes in the dominant tree species along the elevation gradient, with EMF being more associated with certain tree species at higher elevations. Therefore, it is likely that the guild structure of root-associated fungi does show elevation-specific patterns, but the specific patterns can vary depending on the environmental conditions and plant communities present at different elevations.
The consistent occurrence of genus Funneliformis across all elevation zones also confirms an earlier report by Shah et al. (2008b) of the occurrence of Glomus in the rhizospheric soil of A. cotula and presumed its association with the host. However, Shah et al. (2008b) did not screen the roots, which is necessary to authenticate the association of a rhizospheric AMF with a host. Additionally, we found that Claroideoglomus was also associated with the roots of A. cotula. This is consistent with the widespread occurrence of Claroideoglomus in the rhizosphere of plants found in disturbed sites (Zubek et al. 2012) and river valleys (Nobis et al. 2015). Glomus and Claroideoglomus genera have been frequently reported from Rudbeckia laciniata and Solidago gigantea invaded sites (Majewska et al. 2017), and these two plant species and A. cotula belong to the same family, Asteraceae. This suggests that there may be some similarities in the fungal communities associated with invasive Asteraceae species in different regions, and that A. cotula may be benefiting from a similar fungal community as these other invasive species. Another taxon, Mortierella, which was found to be quite predominant in the roots of A. cotula during the present study may be contributing towards its establishment and invasiveness. Studies have shown that Mortierella species can enhance the access of plants to bioavailable forms of iron and phosphorus in the soil (Shi et al. 2014), and can also help the host plants synthesize important phytohormones and enzyme like 1-aminocyclopropane-1-carboxylate (ACC) deaminase (Ozimek et al. 2018). Additionally, Mortierella species have been reported to play a role in protecting agricultural plants from pathogenic organisms (Ozimek et al. 2020). However, the precise role of Mortierella and other such taxa in the invasiveness of A. cotula in the Kashmir Himalaya needs further detailed investigations.
We also observed variations in α-diversity and species richness across the sampling sites, but the variations were insignificant. This is consistent with the findings of Li et al. (2014) who also did not find significant differences in OTU richness of AMF species along an elevational gradient. However, other studies have shown that fungal diversity decreases with an increase in elevation due to detrimental climate, resource scarcity, and adverse soil conditions (Bahram et al. 2012; Gai et al. 2012; Dar et al. 2020; Liu et al. 2021). Interestingly, some studies have reported peaks in fungal and bacterial abundance and diversity at mid-elevations (Jarvis et al. 2015), which have been attributed to the mid-domain effect (Miyamoto et al. 2014; Guo et al. 2020) and greater availability of organic carbon, nitrogen, and mineral nutrients (Siles et al. 2016). Therefore, the relationship between elevation and fungal diversity is not straightforward, and it varies depending on the location and environmental conditions.
Significant variation in root-associated fungal species turnover (β-diversity) of A. cotula across sites varying in elevation points towards a possible role of elevation in structuring the composition of belowground microbiota (Fig. 14).
Furthermore, elevation-based clustering of sites into separate groups is also suggestive of elevational influence on root-associated fungi of A. cotula. These results are in agreement with the findings of some previous investigations which have advocated the role of elevation and environmental variables that co-vary with elevation in determining the microbial community structure and species turnover composition (Veach et al. 2018). Redundancy analysis (RDA), used to explore the influence of soil variables on the fungal community composition in roots of A. cotula, revealed that the two axes explained about 67.40% variability. It is well-established that the relationship between fungal species composition and elevation is complex and can vary across different studies. This may be due to a variety of factors, such as differences in study design, sample size, geographic location, and environmental conditions. In some cases, elevation may play a significant role in shaping fungal communities, as observed in the studies by Gorzelak et al. (2012) and Li et al. (2014). Other studies, such as those by Ruotsalainen et al. (2002) and Kivlin et al. (2017), suggest that elevation may have a lesser or no effect on fungal communities. The study by Li et al. (2014) also suggests that the composition of mycorrhizal fungi may shift along elevational gradients, with different types of mycorrhizal fungi dominating at different elevations.
The findings from the preliminary greenhouse pot trial suggest that the soil microbiome may be playing a facilitative role in the spread of this invasive alien plant species. The positive effects observed on various morphological, physiological, and fitness parameters suggest that the microbiome may be enhancing the plant’s ability to establish and thrive in new environments. The facilitative role of soil microbes in invasive plant species has been supported by recent research, such as the study by Beals et al. (2022) on the invasive plant species Solidago canadensis. These findings suggest that soil microbial communities may be contributing to the success of invasive plant species by promoting their growth and competitive ability (Day et al. 2016). However, it is important to note that the facilitative role of soil microbes in invasive plant species is not universal and may depend on the specific plant species, the composition of the soil microbial community, and other factors such as climate, soil characteristics, and competition with native plant species.
Conclusion
The present study suggests that A. cotula roots host a diverse assemblage of fungal species. The dominance of the AMF guild at higher elevations and the representation of the core mycobiome by only 12 OTUs indicate that there are various factors which shape the association of RAF with A. cotula and predominance of Funneliformis, particularly at higher elevations may be driving its upward expansion in the study region. The significant beta diversity variations across elevations further confirm the existence of elevation-specific association of fungi with the roots of A. cotula.
Based on these findings, it is concluded that future studies investigating the upward spread of this invasive species should focus on interactions between the host plant and below-ground soil microbial community particularly in the context of recent elevation-dependent climate change.
Overall, this study highlights the importance of understanding the ecological interactions between invasive species and their associated fungal communities in predicting their potential spread and impact on the ecosystems.
Data availability
The OTU datasets generated during the present study are available in the Bio Project at the NCBI (SRA) as PRJNA613184. Other data can be obtained from the corresponding author on a reasonable request.
References
Abrego N, Crosier B, Somervuo P et al (2020) Fungal communities decline with urbanization—more in air than in soil. ISME J 14:2806–2815. https://doi.org/10.1038/S41396-020-0732-1
Adhikari S, Burke IC, Eigenbrode SD (2020) Mayweed chamomile (Anthemis cotula L.) biology and management—A review of an emerging global invader. Weed Res 60:313–322. https://doi.org/10.1111/wre.12426
Ahmad I, Zaib S (2020) Mighty microbes: Plant growth promoting microbes in soil health and sustainable agriculture. Soil Health 243–264. https://doi.org/10.1007/978-3-030-44364-1_14
Allaie RR, Reshi Z, Rashid I, Wafai BA (2006) Effect of aqueous leaf leachate of Anthemis cotula - An alien invasive species on germination behaviour of some field crops. J Agron Crop Sci 192:186–191. https://doi.org/10.1111/J.1439-037X.2006.00205.X
Alpert P, Bone E, Holzapfel C (2000) Invasiveness, invasibility and the role of environmental stress in the spread of non-native plants. Perspect Plant Ecol Evol Syst 3:52–66. https://doi.org/10.1078/1433-8319-00004
Bahram M, Põlme S, Kõljalg U et al (2012) Regional and local patterns of ectomycorrhizal fungal diversity and community structure along an altitudinal gradient in the Hyrcanian forests of northern Iran. New Phytol 193:465–473. https://doi.org/10.1111/j.1469-8137.2011.03927.x
Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C (2014) Jvenn: An interactive Venn diagram viewer. BMC Bioinformatics 15(1):1–7. https://doi.org/10.1186/1471-2105-15-293
Beals KK, Lebeis SL, Bailey JK, Schweitzer JA (2022) Conditionality of soil microbial mediation of Solidago plant phenotype: indicator taxa within complex microbiomes influence some, but not all Solidago traits. Plant Soil. https://doi.org/10.1007/S11104-022-05828-0
Bergelson J, Mittelstrass J, Horton MW (2019) Characterizing both bacteria and fungi improves understanding of the Arabidopsis root microbiome. Sci Rep 9:1–11
Bogdziewicz M, Lichti NI, Zwolak R (2019) Consumer-mediated indirect interaction with a native plant lowers the fitness of an invasive competitor. J Ecol 107:12–22. https://doi.org/10.1111/1365-2745.13023
Bueno De Mesquita CP, Sartwell SA, Ordemann EV et al (2018) Patterns of root colonization by arbuscular mycorrhizal fungi and dark septate endophytes across a mostly-unvegetated, high-elevation landscape. Elsevier. https://doi.org/10.1016/j.funeco.2018.07.009
Byun C, de Blois S, Brisson J (2018) Management of invasive plants through ecological resistance. Biol Invasions 20:13–27. https://doi.org/10.1007/S10530-017-1529-7
Callaway RM, Aschehoug ET (2000) Invasive plants versus their new and old neighbors: A mechanism for exotic invasion. Science 290(5491):290:521–523. https://doi.org/10.1126/SCIENCE.290.5491.521
Case TJ (1990) Invasion resistance arises in strongly interacting species-rich model competition communities. Proc Natl Acad Sci U S A 87:9610–9614. https://doi.org/10.1073/PNAS.87.24.9610
Challacombe JF, Hesse CN, Bramer LM et al (2019) Genomes and secretomes of Ascomycota fungi reveal diverse functions in plant biomass decomposition and pathogenesis. BMC Genomics 20(1):1–27. https://doi.org/10.1186/S12864-019-6358-X
Cheng KL, Melsted SW, Bray RH (1953) Removing interfering metals in the versenate determination of calcium and magnesium. Soil Sci 75:37–40. https://doi.org/10.1097/00010694-195301000-00004
Chesnin L, Yien CH (1951) Turbidimetric Determination of Available Sulfates. Soil Sci Soc Am J 15:149–151. https://doi.org/10.2136/SSSAJ1951.036159950015000C0032X
Coats VC, Rumpho ME (2014) The rhizosphere microbiota of plant invaders: An overview of recent advances in the microbiomics of invasive plants. Front Microbiol 5:368. https://doi.org/10.3389/fmicb.2014.00368
R Core Team (2022). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Dainese M, Aikio S, Hulme PE et al (2017) Human disturbance and upward expansion of plants in a warming climate. Nat Clim Chang 7:577–580. https://doi.org/10.1038/NCLIMATE3337
Dar MA, Afshana ,Sheikh AH et al (2020) Dynamics of Mycorrhizal Mutual- ism in Relation to Plant Invasion Along an Altitudinal Gradient in Kashmir Himalaya. Bot Rev 86:1–38. https://doi.org/10.1007/s12229-020-09221-3
Day NJ, Dunfield KE, Antunes PM (2016) Fungi from a non-native invasive plant increase its growth but have different growth effects on native plants. Biol Invasions 18:231–243. https://doi.org/10.1007/S10530-015-1004-2
Desai NS, Wilson AW, Powers JS et al (2016) Ectomycorrhizal diversity and community structure in stands of Quercus oleoides in the seasonally dry tropical forests of Costa Rica. Environ Res Lett 11(12):125007. https://doi.org/10.1088/1748-9326/11/12/125007
Doyle J (1991) DNA Protocols for Plants. Mol Tech Taxon. Springer Berlin 283–293. https://doi.org/10.1007/978-3-642-83962-7_18
Edgar RC (2018) Accuracy of taxonomy prediction for 16S rRNA and fungal ITS sequences. PeerJ 6:e4652. https://doi.org/10.7717/PEERJ.4652
Erneberg M (1999) Effects of herbivory and competition on an introduced plant in decline. Oecologia 118:203–209. https://doi.org/10.1007/S004420050719
Gai JP, Tian H, Yang FY et al (2012) Arbuscular mycorrhizal fungal diversity along a Tibetan elevation gradient. Pedobiologia (jena) 55:145–151. https://doi.org/10.1016/J.PEDOBI.2011.12.004
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118. https://doi.org/10.1111/J.1365-294X.1993.TB00005.X
Goldmann K, Ammerschubert S, Pena R et al (2020) Early stage root- associated fungi show a high temporal turnover, but are independ- ent of beech progeny. Microorganisms 8(2):210. https://doi.org/10.3390/microorganisms8020210
Gorzelak MA, Hambleton S, Massicotte HB (2012) Community structure of ericoid mycorrhizas and root-associated fungi of Vaccinium membranaceum across an elevation gradient in the Canadian Rocky Mountains. Fungal Ecol 5:36–45. https://doi.org/10.1016/j.funeco.2011.08.008
Guo Y, Ren C, Yi J et al (2020) Contrasting responses of Rhizosphere bacteria, fungi and arbuscular mycorrhizal fungi along an Ele-vational gradient in a temperate montane forest of China. Front Microbiol 11:2042. https://doi.org/10.3389/FMICB.2020.02042/FULL
Gweon H, Oliver A, Taylor J et al (2015) PIPITS: An automated pipeline for analyses of fungal ITS sequences from the Illumina sequencing platform. Researchgate net 6:973–980. https://doi.org/10.1111/2041-210X.12399
Hierro JL, Maron JL, Callaway RM (2005) A biogeographical approach to plant invasions: The importance of studying exotics in their introduced and native range. J Ecol 93:5–15
Hu W, Ran J, Dong L et al (2021) (2021) Aridity-driven shift in biodiversity–soil multifunctionality relationships. Nat Commun 121(12):1–15. https://doi.org/10.1038/s41467-021-25641-0
Hu W, Hou Q, Delgado-Baquerizo M et al (2022) Continental-scale niche differentiation of dominant topsoil archaea in drylands. Environ Microbiol 24:5483–5497. https://doi.org/10.1111/1462-2920.16099
Jackson ML (1973) Soil chemical analysis, pentice hall of India Pvt. Ltd, New Delhi, India 498:151–154
James TY, Kauff F, Schoch CL et al (2006) Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443:818–822. https://doi.org/10.1038/NATURE05110
Jarvis SG, Woodward S, Taylor AFS (2015) Strong altitudinal partitioning in the distributions of ectomycorrhizal fungi along a short (300 m) elevation gradient. New Phytol 206:1145–1155. https://doi.org/10.1111/nph.13315
Jin L, Wang Q, Wang Q et al (2017) Mycorrhizal-induced growth depression in plants. Symbiosis 72:81–88. https://doi.org/10.1007/S13199-016-0444-5
Kamutando CN, Vikram S, Kamgan-Nkuekam G et al (2019) The Functional Potential of the Rhizospheric Microbiome of an Invasive Tree Species, Acacia dealbata. Microb Ecol 77:191–200. https://doi.org/10.1007/S00248-018-1214-0
Kandlikar GS, Gold ZJ, Cowen MC et al (2018) Ranacapa: An R pack- age and shiny web app to explore environmental DNA data with exploratory statistics and interactive visualizations. F1000Re- search 7:1734. https://doi.org/10.12688/F1000RESEARCH.16680.1
Kivlin SN, Lynn JS, Kazenel MR et al (2017) Biogeography of plant-associated fungal symbionts in mountain ecosystems: A meta-analysis. Wiley Online Libr 23:1067–1077. https://doi.org/10.1111/ddi.12595
Klironomos JN (2002) Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature 417:67–70. https://doi.org/10.1038/417067a
Kotilínek M, Hiiesalu I, Košnar J et al (2017) Fungal root symbionts of high-altitude vascular plants in the Himalayas. Sci Rep 7(1):1–14. https://doi.org/10.1038/s41598-017-06938-x
Lahti L, Shetty S, Blake T, Salojarvi J (2017) Tools for microbiome analysis in R. R package version 1.5.28
Levine JM, Adler PB, Yelenik SG (2004) A meta-analysis of biotic resistance to exotic plant invasions. Ecol Lett 7:975–989
Li X, Gai J, Cai X et al (2014) Molecular diversity of arbuscular mycorrhizal fungi associated with two co-occurring perennial plant species on a Tibetan altitudinal gradient. Mycorrhiza 24:95–107. https://doi.org/10.1007/S00572-013-0518-7
Li X, Xu M, Li X et al (2020) Linkages between changes in plant and mycorrhizal fungal community composition at high versus low elevation in alpine ecosystems. Environ Microbiol Rep 12:229–240. https://doi.org/10.1111/1758-2229.12827
Li S, Xie D, Ge X, Dong W, Luan J (2022) Altered diversity and functioning of soil and root-associated microbiomes by an invasive native plant. Plant Soil 473(1–2):235–249
Liu G, Liu RL, Zhang WG et al (2021) Arbuscular mycorrhizal colonization rate of an exotic plant, Galinsoga quadriradiata, in mountain ranges changes with altitude. Mycorrhiza 31:161–171. https://doi.org/10.1007/S00572-020-01009-Y
Lockwood JL, Cassey P, Blackburn T (2005) The role of propagule pressure in explaining species invasions. Trends Ecol Evol 20:223–228. https://doi.org/10.1016/J.TREE.2005.02.004
Maciá-Vicente JG (2022) Fungi living in plant roots have low habi-tat and host specificities, but highly restricted distributions. Bull Ecol Soc Am 103(1):1–6. https://doi.org/10.1002/BES2.1951
Majewska ML, Rola K, Zubek S (2017) The growth and phosphorus acquisition of invasive plants Rudbeckia laciniata and Solidago gigantea are enhanced by arbuscular mycorrhizal fungi. Mycorrhiza 27:83–94. https://doi.org/10.1007/S00572-016-0729-9
Martínez-Diz M del P, Andrés-Sodupe M, Bujanda R, Díaz-Losada E, Eichmeier A, Gramaje D (2019) Soil-plant compartments affect fungal microbiome diversity and composition in grapevine. Fungal Ecol 41:234–244
McMurdie PJ, Holmes S (2013) Phyloseq: An R Package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8(4): e61217. https://doi.org/10.1371/JOURNAL.PONE.0061217
Merino-Martín L, Hernández-Cáceres D, Reverchon F et al (2023) Habitat partitioning of soil microbial communities along an elevation gradient: from plant root to landscape scale. Oikos 2023(1):e09034. https://doi.org/10.1111/OIK.09034
Midolo G, Wellstein C (2020) Plant performance and survival across transplant experiments depend upon temperature and precipitation change along elevation. J Ecol 108:2107–2120. https://doi.org/10.1111/1365-2745.13387
Miyamoto Y, Nakano T, Hattori M, Nara K (2014) The mid-domain effect in ectomycorrhizal fungi: Range overlap along an elevation gradient on Mount Fuji, Japan. ISME J 8:1739–1746. https://doi.org/10.1038/ISMEJ.2014.34
Nguyen NH, Song Z, Bates ST, Branco S, Tedersoo L, Menke J, Schilling JS, Kennedy PG (2016) FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol 20:241–248
Nguyen DQ, Schneider D, Brinkmann N et al (2020) Soil and root nutrient chemistry structure root-associated fungal assemblages in temperate forests. Wiley Online Libr 22:3081–3095. https://doi.org/10.1111/1462-2920.15037
Nobis A, Błaszkowski J, Zubek S (2015) Arbuscular mycorrhizal fungi associations of vascular plants confined to river valleys: towards understanding the river corridor plant distribution. J Plant Res 128:127–137. https://doi.org/10.1007/S10265-014-0680-9
Oehl F, Körner C (2014) Multiple mycorrhization at the coldest place known for Angiosperm plant life. Alp Bot 124:193–198. https://doi.org/10.1007/S00035-014-0138-7
Oh SY, Lim YW (2018) Root-associated bacteria influencing mycelial growth of Tricholoma matsutake (pine mushroom). J Microbiol 56:399–407. https://doi.org/10.1007/s12275-018-7491-y
Olsen SR, Watanabe FS, Cosper HR et al (1954) Residual phosphorus availability in long-time rotations on calcareous soils. Soil Sci 78:141–151. https://doi.org/10.1097/00010694-195408000-00008
Orellana, SC (2013) Assessment of Fungal Diversity in the Environment using Metagenomics:a Decade in Review. Fungal Genomics Biol 03: https://doi.org/10.4172/2165-8056.1000110
Ozimek E, Hanaka A (2021) Mortierella species as the plant growth-promoting fungi present in the agricultural soils. Agric 11:1–18. https://doi.org/10.3390/agriculture11010007
Ozimek E, Jaroszuk-ściseł J, Bohacz J, et al (2018) Synthesis of indoleacetic acid, gibberellic acid and acc-deaminase by Mortierella strains promote winter wheat seedlings growth under different conditions. Int J Mol Sci 19. https://doi.org/10.3390/ijms19103218
Pan J, Liu Y, He X et al (2013) Arbuscular mycorrhizal and dark septate endophytic fungi at 5,500 m on a glacier forefront in the Qinghai-Tibet Plateau, China. Symbiosis 60:101–105. https://doi.org/10.1007/S13199-013-0245-Z
Paolucci A, Rauschert ES, Carrino-Kyker S, Burke D (2021) Root fungal communities associated with better performance of an invasive spring ephemeral. Biol Invasions 23:181–192
Pauchard A, Milbau A, Albihn A et al (2016) Non-native and native organisms moving into high elevation and high latitude ecosystems in an era of climate change: new challenges for ecology and conservation. Biol Invasions 18:345–353. https://doi.org/10.1007/S10530-015-1025-X/FIGURES/1
Petitpierre B, McDougall K, Seipel T et al (2016) Will climate change increase the risk of plant invasions into mountains? Ecol Appl 26:530–544. https://doi.org/10.1890/14-1871
Rai M, Agarkar G (2016) Plant-fungal interactions: What triggers the fungi to switch among lifestyles? Crit Rev Microbiol 42:428–438
Ramirez KS, Snoek LB, Koorem K et al (2019) Range-expansion effects on the belowground plant microbiome. Nat Ecol Evol 3:604–611. https://doi.org/10.1038/S41559-019-0828-Z
Rashid I, Reshi ZA (2012) Allelopathic interaction of an alien invasive specie Anthemis cotula on its neighbours Conyza canadensis and Galinsoga parviflora. Allelopath J 29:77–92
Rashid I, Reshi Z, Allaie RR, Wafai BA (2007) Germination ecology of invasive alien Anthemis cotula helps it synchronise its successful recruitment with favourable habitat conditions. Ann Appl Biol 150:361–369. https://doi.org/10.1111/J.1744-7348.2007.00136.X
Reshi ZA, Shah MA, Rashid I, Rasool N (2012) Anthemis cotula L.: A highly invasive species in the Kashmir Himalaya, India. Invasive Alien Plants An Ecol Apprais Indian Subcont 108–125. Wallingford UK: CABI. https://doi.org/10.1079/9781845939076.0108
Řezáčová V, Michalová T, Řezáč M, Gryndler M, Duell EB, Wilson GW, Heneberg P (2022) The root-associated arbuscular mycorrhizal fungal assemblages of exotic alien plants are simplified in invaded distribution ranges, but dominant species are retained: A trans-continental perspective. Environ Microbiol Rep 14(5):732–741
Rognes T, Flouri T, Nichols B et al (2016) VSEARCH: A versatile open source tool for metagenomics. PeerJ 4:e2584. https://doi.org/10.7717/PEERJ.2584
Rudgers JA, Afkhami ME, Bell-Dereske L et al (2020) Climate Disruption of Plant-Microbe Interactions 51:561–586. https://doi.org/10.1146/ANNUREV-ECOLSYS-011720-090819
Ruotsalainen AL, Tuomi J, Väre H (2002) A model for optimal mycorrhizal colonization along altitudinal gradients. Silva Fenn 36:681–694. https://doi.org/10.14214/SF.533
Shah MA, Reshi Z (2007) Invasion by alien Anthemis cotula L. in a biodiversity hotspot: Release from native foes or relief from alien friends? Curr Sci 92:21–22
Shah MA, Reshi Z, Rashid I (2008) Mycorrhizosphere mediated Mayweed Chamomile invasion in the Kashmir Himalaya, India. Plant Soil 312:219–225. https://doi.org/10.1007/S11104-008-9706-1
Shah MA, Reshi Z, Rashid I (2008) Mycorrhizal source and neighbour identity differently influence Anthemis cotula L. invasion in the Kashmir Himalaya. India Appl Soil Ecol 40:330–337. https://doi.org/10.1016/j.apsoil.2008.06.002
Shah MA, Reshi ZA, Rashid I (2012) Synergistic effect of herbivory and mycorrhizal interactions on plant invasiveness. African J Microbiol Res 6:4107–4112. https://doi.org/10.5897/AJMR11.1278
Shi LL, Mortimer PE, Ferry Slik JW et al (2014) Variation in forest soil fungal diversity along a latitudinal gradient. Fungal Divers 64:305–315. https://doi.org/10.1007/S13225-013-0270-5
Siles JA, Cajthaml T, Minerbi S, Margesin R (2016) Effect of altitude and season on microbial activity, abundance and community structure in Alpine forest soils. FEMS Microbiol Ecol 92:. https://doi.org/10.1093/FEMSEC/FIW008
Simberloff D (2009) The role of propagule pressure in biological invasions. Annu Rev Ecol Evol Syst 40:81–102. https://doi.org/10.1146/ANNUREV.ECOLSYS.110308.120304
Sudová R, Kohout P, Rydlová J et al (2020) Diverse fungal commu- nities associated with the roots of isoetid plants are structured by host plant identity. Fungal Ecol 45:100914. https://doi.org/10.1016/J.FUNECO.2020.100914
Tedersoo L, Bahram M, Põlme S et al (2014) Global diversity and geography of soil fungi. Science 346(6213):1256688. https://doi.org/10.1126/SCIENCE.1256688
Tondera K, Chazarenc F, Brisson J, Chagnon P-L (2023) Structure and impact of root-associated fungi in treatment wetland mesocosms. Sci Total Environ 858:159958. https://doi.org/10.1016/J.SCITOTENV.2022.159958
Ullah R, Khan N, Ali K (2022) Which factor explains the life history of Xanthium strumarium L., an aggressive alien invasive plant species, along its altitudinal gradient? Plant Direct 6(1):e375. https://doi.org/10.1002/pld3.375
Upson R, Newsham KK, Read DJ (2008) Root-fungal associations of Colobanthus quitensis and Deschampsia antarctica in the maritime and subantarctic. Arctic, Antarct Alp Res 40:592–599. https://doi.org/10.1657/1523-0430(07-057)[UPSON]2.0.CO;2
Van Bael SA (2020) Fungal Diversity Diversity 12:1–2. https://doi.org/10.3390/D12110437
Veach AM, Stokes CE, Knoepp J et al (2018) Fungal Communities and Functional Guilds Shift Along an Elevational Gradient in the Southern Appalachian Mountains. Microb Ecol 76:156–168. https://doi.org/10.1007/s00248-017-1116-6
Vorstenbosch T, Essl F, Lenzner B (2020) An uphill battle? The elevational distribution of alien plant species along rivers and roads in the Austrian Alps. NeoBiota 63:1–24. https://doi.org/10.3897/neobiota.63.55096
Walkley A, Black IA (1934) An examination of the degtjareff method for determining soil organic matter, and a proposed modification of the chromic acid titration method. Soil Sci 37:29–38. https://doi.org/10.1097/00010694-193401000-00003
Ward EB, Pregitzer CC, Kuebbing SE, Bradford MA (2020) Invasive lianas are drivers of and passengers to altered soil nutrient availability in urban forests. Biol Invasions 22:935–955. https://doi.org/10.1007/S10530-019-02134-2
White T, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18(1):315–322
Yin L, Liu B, Wang H et al (2020) The rhizosphere microbiome of Mikania micrantha provides insight into adaptation and invasion. Front Microbiol 11:1462. https://doi.org/10.3389/FMICB.2020.01462/FULL
Zhang M, Shi Z, Yang M et al (2021) Molecular diversity and distribution of arbuscular mycorrhizal fungi at different elevations in Mt. Taibai of Qinling Mountain. Front Microbiol 12: 609386. https://doi.org/10.3389/FMICB.2021.609386/FULL
Zou Q, Lin G, Jiang X et al (2018) Sequence clustering in bioinformatics: An empirical study. Brief Bioinform 21:1–10. https://doi.org/10.1093/BIB/BBY090
Zubek S, Błaszkowski J, Buchwald W (2012) Fungal root endophyte associations of medicinal plants. Nov Hedwigia 94:525–540. https://doi.org/10.1127/0029-5035/2012/0024
Acknowledgements
We thank Head, Department of Botany, University of Kashmir, for providing laboratory facilities. Award of fellowship by the UGC-CSIR, India, to the Afshana and the support under the CPEPA by the UGC, New Delhi, to the University of Kashmir are also gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
Afshana and ZAR conceived the idea and designed the study together with MAS and IR. Afshana did sampling, generated data, and wrote the first draft of the manuscript. ZAR and RAM performed data analysis and ZAR revised and corrected the manuscript. All authors have read and agreed with the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Afshana, Reshi, Z.A., Shah, M.A. et al. Species composition of root-associated mycobiome of ruderal invasive Anthemis cotula L. varies with elevation in Kashmir Himalaya. Int Microbiol 26, 1053–1071 (2023). https://doi.org/10.1007/s10123-023-00359-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10123-023-00359-9