Abstract
Grass pea (Lathyrus sativus L.) is widely cultivated for food and feed in some developing countries including Ethiopia. However, due to its overexaggerated neuro-lathyrism alkaloid causing paralysis of limbs, it failed to attract attention of the research community and is one of the most neglected orphan crops in the world. But, the crop is considered an insurance crop by resource-poor farmers due to its strong abiotic stress tolerance and ability to produce high yields when all other crops fail due to unfavorable environmental conditions. This study was aimed at screening rhizobial isolates of grass pea and evaluating their symbiotic nitrogen fixation efficiency and tolerance to abiotic stresses. Fifty rhizobial isolates collected from grass pea nodules were isolated, screened, and characterized based on standard microbiological methods. The rhizobial isolates showed diversity in nodulation, symbiotic nitrogen fixation, and nutrient utilization. The 16S rRNA gene sequencing of 14 rhizobial isolates showed that two of them were identified as Rhizobium leguminosarum and the remaining twelve as Rhizobium species. Based on their overall performance, strains AAUGR-9, AAUGR-11, and AAUGR-14 that performed top and identified as Rhizobium species were recommended for field trials. This study screened and identified effective and competitive rhizobial isolates enriched with high nitrogen-fixing and abiotic stress tolerant traits, which contributes much to the application of microbial inoculants as alternative to chemical fertilizers.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Grass pea is one of the cool-season legume crops within the tribe Viciae including the genera Vicia, Pisum, and Lens that fix inorganic nitrogen in association with root nodule bacteria known as rhizobia. Previous studies on Vicia, Pisum, Lathyrus, and Lens (Jordan 1984; Perret et al. 2000; Rivas et al. 2009) showed that the endosymbionts belong to Rhizobium leguminosarum biovar vicieae. However, recent reports revealed that nodules from Vicia faba, Pisum sativum, and Lens esculenta also harbor other groups of root nodule bacteria other than Rhizobium leguminosarum biovar vicieae (Santillana et al. 2008; Tena et al. 2017). These include Rhizobium leguminosarum biovar trifolli that nodulate trifolium spp. and Rhizobium etli that nodulate Phaseolus vulgaris (Beyene et al. 2004). Recently, Tena et al. (2017) made a polyphasic study on root nodule from lentil and identified three distinct sub-lineages as Rhizobium etli, Rhizobium leguminosarum, and Rhizobium spp.
Other studies also showed the different levels of symbiotic effectiveness of cross-nodulating rhizobia isolated from one host may not necessarily be effective on another host indicating diversity and specificity even among the Rhizobium leguminosarum var. viceae populations (Laguerre et al. 2003; Gebremariam and Assefa 2017). For several years now, extensive studies have been undertaken on phenotypic and symbiotic properties of the tribe Viceae in Ethiopia. Some of these studies were focused on faba bean (Mnalku et al. 2009; Keneni et al. 2010; Belay and Assefa 2011; Legesse and Assefa 2014), lentil (Jida and Assefa 2011; Setargie et al. 2015), and field pea (Amsalu et al. 2012; Baye et al. 2015), and cross-inoculation studies on the Viceae tribe (Gebremariam and Assefa 2017).
Grass pea is the fifth most important pulse crop in Ethiopia after faba bean, chickpea, field pea, and red haricot bean (CSA (Central Statistical Authority) 2017) and is used for food, feed, and fodder (Urga et al. 2005). The seed contains proteins, fats, and carbohydrates (about 35% of which is starch), and it also contains glucose, pentosans, phytin, lignin, albumin, prolamine, globulin, guttllin, and several essential amino acids and minerals (Demelash et al. 2015). The protein content of grass pea seeds collected from 15 major production areas of Ethiopia is 27–32% (Urga et al. 2005), which is higher than the average percentage of protein content (21–25%) in other legume seeds (Monsoor and Yusuf 2002). Grass pea also plays an important role as a legume crop in crop rotations, reportedly adding around 67 kg/ha of nitrogen to the soil from symbiosis with Rhizobium species in a single season and conferring yield and protein benefits on the subsequent non-legume crop (Jennifer 2003). Apart from biological nitrogen fixation, rhizobia have also plant growth-promoting activities including solubilization of nutrients and production of growth hormones, production of hydrolytic enzymes, production of siderophore, and production of HCN. Such activities directly or indirectly increase plant growth and productivity.
Despite its wide cultivation in Ethiopia due to its ability to grow on marginal lands and reduced moisture requirement, grass pea is one of the neglected orphan crops that failed to attract attention of the scientific research community. This inadvertent neglect of the crop is due to the negative perception of the crop for its alkaloid associated with lathyrism (Moges et al. 2004). Although extensive researches have been undertaken on other related pulse crops, little is known about grass pea contribution to soil fertility and plant health by its rhizobia. The limited studies showed that the root-nodulating rhizobia of grass pea were as diverse as other cross-nodulating hosts such as faba bean, field pea, and lentil (Drouin et al. 1996; Mahdavi et al. 2007; Aoki et al. 2010) contributing to diversity studies on Rhizobium leguminosarum var. viceae. The lack of due attention by the scientific community, as it was the case with other orphan crops, precludes the full exploitation of the fifth most important feed and cover crop in the country.
Rhizobial populations are known to vary in their tolerance to major environmental factors (Wei et al. 2008). Consequently, exploiting the enormous potential of symbiotic nitrogen fixation requires the selection and development of efficient inoculant strains compatible to the legume host and the target edaphic environment (Bhargava et al. 2016). To this end, studying the potential of the isolates for physiological versatility is important to select them for their competitiveness in the complex soil environment (Fitouri et al. 2012). Thus, screening and selecting symbiotically highly effective and most tolerant isolates with ecological adaptations that can be introduced as microbial inoculants is key not only for grass pea but also for the cross-nodulation crops such as faba bean, field pea, and lentil.
Materials and methods
Sample collection, isolation, and purification of isolates
Grass pea nodules were collected from the roots of selected standing crops grown in 50 sampling field sites (Table 1); 35 from some districts of South Wollo (Jamma, Woreilu, Tenta, Borena, Kalu, Tehuledere, and Kutaber), 3 from Oromia zone (Artumafursi), and 12 from Western Shoa (A/Gindeberet, Jeldu, and Dendi) (Fig. 1). The nodule samples were collected in vials containing a desiccant (silica gel) covered with 1 cm of cotton wool (Somasegaren and Hoben 1994) and brought to Addis Ababa University. The isolation and purification of the rhizobial isolates were carried out using Yeast Extract Mannitol Agar (YEMA) as described by Vincent (1970). The purified isolates were designated as AAUGR (Addis Ababa University Grasspea Rhizobia) with different numbers representing each isolate and kept at – 20 °C for further work. Colony characteristics including colony diameter, colony texture, acid base reaction on YEMA-BTB media, and mean doubling time were evaluated according to Somasegaren and Hoben (1994) and White (1995). Confirmatory tests that included gram reaction, Congo red absorption, and growth on peptone glucose agar (PGA) media were also carried out (Buck 1982; Somasegaren and Hoben 1994).
Authentication of the isolates under greenhouse conditions and determining their relative symbiotic effectiveness
All the rhizobial isolates were subjected to pot sand culture to authentication test (definitive test) for 60 days according to Somasegaren and Hoben (1994) (Fig. S1). “Wasse” grass pea variety which was obtained from Debrezeit Agricultural Research Centre was surface-sterilized using 95% ethanol (briefly), bleached with 3% sodium hypochlorite for 4 min, rinsed with five changes of sterile water, and germinated on water agar (0.75 w/v) for 3 days. Five germinated seeds were transferred into 3-kg capacity plastic pots, pre-cleaned with acid (95% H2SO4). After 7 days of growth, each seedling was inoculated with 1 ml (109cells/ml) of 72-h YEMB-grown culture. The experiment was statistically laid out in a complete random design (CRD) with three replicates in a greenhouse with a 12-h photoperiod and an average of 28 °C and 15 °C day and night temperature. The sampling included a negative control without treatment and inoculation and a positive control treated with 70 mg/l potassium nitrate (0.05 KNO3 (w/v)) solution every week. All pots were fertilized with quarter strength of Broughton and Dilworth N-free medium per week and watered every two days as described in Somasegaren and Hoben (1994).
Sixty days after planting (DAP), plants were uprooted to record nodule number, nodule dry weight, and shoot dry weight. The symbiotic effectiveness (SE) of the isolates was calculated using the formula (Mulongoy 2004):
where SDW = shoot dry weight, N = nitrogen, SE = symbiotic effectiveness
The rate of nitrogen-fixing effectiveness was evaluated as: highly effective > 80%, effective 50–80%, lowly effective 35–49%, and ineffective < 35%.
Tolerance to pH, temperature, and salt
Each isolate was grown on YEMB media adjusted at different pH (4.0, 4.5, and 5.0) and on YEMA media adjusted at pH 8.5, 9.0, 9.5, and 10.0 and incubated at 28 ± 2 °C for 3–5 days. Similarly, the isolates were grown at different salt concentrations of 1%, 2%, 3%, 4%, 5%, 6%, 7%, and 8%, of NaCl (Bernal and Graham 2001) and incubated at 28 ± 2 °C for 3–5 days. The isolates’ potential to grow at different incubation temperatures was also evaluated by incubating at 4 °C, 10 °C, 15 °C, 35 °C, 40 °C, and 45 °C for 3–5 days as indicated in Lupwayi and Haque (1994). Growth of bacterial colonies was recorded as an indication of resistance to these environmental stresses.
Inherent antibiotic resistance and heavy metal tolerance of the isolates
Inherent antibiotic resistance (IAR) of the rhizobial isolates was determined by inoculating (106cells ml−1) on solid YEMA medium containing filter-sterilized (0. 22 mm Millipore filters) antibiotics (μg/ml): ampicillin (30), chloramphinicol (40), erythromycin (30), nalidixic acid (20), neomycin (20), streptomycin (10), and tetracycline (30). The isolates (106 cells/ml) grown for 48 h were inoculated to the same medium containing filter-sterilized heavy metals (HM) at concentrations of (μg/ml) Hg (HgCl2) (5), Kr (K2Kr2O7) (50), Mn (MnCl2) (200), Ni (NiCl2) (200), Pb (Pb(CH3COO)2) (50), and Zn (ZnCl2) (100) culture (Mohamed et al. 2012). All plates were incubated at 28 ± 2 °C for 3–5 days. Growth of bacterial colonies was recorded as an indication of resistance.
Heterotrophic utilization of the isolates
Carbon utilization of isolates was determined on a basal medium (1 g of KH2PO4; 1 g K2HPO4; 0.01 g FeCl3·6H2O; 0.2 g MgSO4·7H2O; 0.1 g CaCl2; 15 g agar) containing each of the 12 carbohydrate starch, cellobiose, dextrin, lactose, sucrose, galactose, maltose, fructose, glucose, arabinose, xylose, and sorbitol as 10% (w/v) by reducing the yeast extract to 0.05 gl−1 liter following the method of Somasegaren and Hoben (1994). Similarly, the ability of isolates to utilize different nitrogen sources was tested on the same basal medium containing each of the 7 amino acids l-lysine, l-leucine, l-alanine, glycine, l-aspargine and l-cystine, and l-diphenylamine (Amargar et al. 1997). The plates were incubated at 28 ± 2 °C for 3–5 days. Growth of bacterial colonies was recorded as an indication of nutrient utilization.
Molecular characterization
Molecular characterization of some selected rhizobial isolates for the presence of 16S rRNA gene was conducted at the Institute for Agricultural Biosciences, Oklahoma State University, as previously described (Adal et al. 2018).
Colony PCR amplification of 16S rRNA
Fourteen rhizobial isolates that showed better performance in greenhouse and stress tolerance tests and for which amplicons were obtained were selected for 16S rRNA gene characterization. A single colony from a 48-h grown rhizobial culture was taken using a sterile pipette tip, added into PCR mix (10 μl 5× Phusion HF buffer, 1 μl 10 mM dNTPs, 2.5 μl 10 μM forward primer, 2.5 μl 10 μM reverse primer, 0.5 μl Phusion DNA polymerase, DNA template (small piece of single colony), and 33.5 μl Nuclease-Free Water), and well mixed through pipetting up and down for several times. PCR amplification of 16S rRNA genes was carried out using the primers 8.27F 5′ AGA GTT TGA TCC TGG CTC AG 3′ and rD1-5′ ACGGCTACCTTGTTA CGACTT 3′ (Weisburg et al. 1991; Eardly et al. 1996; and Bontemps et al. 2010). The PCR conditions were preheating at 98 °C for 30 s″, 35 cycles of 98 °C for 10 s″ denaturing, 56 °C for 30 s″ annealing, 72 °C for 30 s″ elongation with a final extension of 72 °C for 10 min′ (Weisburg et al. 1991).
Purification of PCR products and sequencing of 16S rRNA genes
The colony PCR products were run on 1% agarose gel, and appropriate bands were excised from the gel and purified using PCR purification kit (Qiagen, Chicago, IL, USA) following the standard protocol recommended by the manufacturer. Five microliters of each purified PCR product was used for Sanger sequencing at the Oklahoma State University Core Facility. PCR products were sequenced using the same primers used in the PCR amplification. Sequence reads were edited using Bioedit software and compared for homology with sequences of other organisms deposited in the National Center for Biotechnology Information (NCBI) GenBank database using the Basic Local Alignment Search Tool (BLAST) to determine their identity to known sequences (Shayne et al. 2003).
Clustering analysis
The isolates’s phenotypic variability was analyzed with a computer cluster analysis using similarity coefficient (Hammer et al. 2001). The ecophysiological and heterotrophic utilization that included resistance to temperature, pH, salt, antibiotics, and heavy metals and utilizations of carbohydrates and amino acids were used to construct a phenogram. Traits were coded 1 for positive and 0 for negative. The final matrix contained 22 isolates and 59 traits. A computer cluster analysis of 59 phenotypic variables was carried out using Ward’s linkage and Squared Euclidian distance as a measure of dissimilarity coefficient (Ward 1963), and a dendrogram was constructed PAST ver.2.17c (Hammer et al. 2001).
Data analysis
The data recoded were analyzed and interpreted using ANOVA. The experimental treatments were compared and contrasted against their control following Duncan’s multiple range test (DMRT). The correlation between different greenhouse data was evaluated by Pearson correlation coefficient using SPSS v.20.
Result
Isolation, presumptive, and cultural characterization of the isolates
A total of 50 legume-nodulating bacteria were isolated from grass pea–growing areas of South Wollo (35 isolates), Artumafursi (3 isolates), and West Shewa (12 isolates). The determination of the confirmatory tests and cultural characterization of the isolates are shown in Table 1. The confirmatory test result showed that all isolates but isolate AAUGR-47 were Gram-negative and failed to grow on PGA media and to absorb Congo red. Similarly, all the isolates except AAUGR-47 changed the YEMA-BTB to yellow and showed growth with doubling time ranging from 1.27 to 4.73 h. The isolates showed variations in colony size in which 7 (14 %) and 10 (20.4 %) displayed the highest and smallest colony diameter of 5. 5 mm and 2.0 mm. The colony diameter of the remaining isolates was in between this range. The isolates exhibited large watery (LW) (45%) and large mucoid (LM) (55%) colony textures. All these colony and growth features accompanied by acid production (yellow on YEMA-BTB) are characteristics of fast-growing and acid-producing rhizobia.
Symbiotic effectiveness of the isolates on sand culture under greenhouse conditions
The preliminary screening for symbiotic effectiveness of the isolates showed variations in nodulation and growth characters (Table 1, Figs. S2-S4). Accordingly, the rhizobia-induced nodules on the host plant with nodule number (NN) ranging from 17/plant (AAUGR-22) to 116/plant (AAUGR-15) and mean nodule dry weight (NDW) ranging from 0.011 g/p (AAUGR 22) to 0.098 g/p (AAUGR-15), indicating a 7-fold difference in nodule number and a 9-fold difference in nodule dry weight between the highest and lowest nodulating isolates.
Among the inoculated plants, all of them accumulated a higher shoot dry weight than the negative control and nearly half of them accumulated a higher shoot dry weight than the positive control plants (P < 0.05). The amount of shoot dry matter accumulated by the host plants was in the range of 0.27 g/p (AAUGR-39) to 1.15 g/p (AAUGR-15), suggesting that the most effective isolate helped to accumulate approximately 5-fold more biomass than the least effective isolate. The same host plants inoculated by the same isolates of AAUGR-15, AAUGR-14, AAUGR-11, AAUGR-6, and AAUGR-5 showed similar pattern in NN, NDW, and SDW, implying high correlation between nodule dry weight and shoot dry weight (r = 0.864 and 0.817). The data also showed a strong positive relationship at p < 0.01 among the parameters evaluated. The correlation between shoot dry weight and percent symbiotic effectiveness was found to form a relatively more direct and perfect linear relationship than the other parameters (r = 0.988). Based on the relative plant dry matter accumulation of the inoculated plants with uninoculated and nitrogen-fertilized control (positive control), 22 (45%) of the isolates were highly effective in nitrogen fixation with > 80% of shoot dry matter accumulation, whereas 20 (41%) of them accumulated 50–80% of shoot dry matter compared with the nitrogen-fertilized positive controls. In general, 86% of the isolates performed well in their symbiotic effectiveness.
Physiological tolerance of the isolates
All the isolates showed a significant difference in physiological tolerance to pH, salt, temperature, antibiotics, heavy metals, and utilization of carbohydrates and amino acids as substrates (Table 2). Nearly half of the isolates (45.5 %) tolerated both lower and higher pH and temperature ranges. Moreover, the isolates also showed variation in resistance to the tested antibiotics and toxic heavy metals. However, some exhibited similar resistance to both antibiotics and heavy metals.
Tolerance to salt, pH, and temperature
The potential of the isolates to tolerate a wide range of salt, pH, and temperature was determined (Table 2). All isolates displayed growth at 1–3% salt concentrations, 20–35 °C temperatures and a pH range of 6–9. Almost half of the isolates were tolerant to 40 °C, 6% NaCl, and pH of 4. However, fewer isolates were tolerant to 10 °C and 8% NaCl. Few isolates (18%) that included AAUGR-2, AAUGR-6, AAUGR-9, and AAUGR-24 were tolerant to a wide pH range of 4–10, whereas 9 isolates (40.9%) grew at a wide temperature range of 10–40 °C.
Intrinsic antibiotic and heavy metal resistance
The isolates also showed variations in IAR and HMR tolerance (Table 2). Many isolates were resistant to nalidixic acid, erythromycin, and neomycin, whereas fewer isolates were resistant to streptomycin and tetracycline. Among the isolates, AAUGR-6, AAUGR-9, AAUGR-24, AAUGR-2, AAUGR-11, AAUGR-14, AAUGR-15, AAUGR-19, and AAUGR-30 grew and tolerated the majority of the tested antibiotics. The isolates were resistant to manganese (96%), lead (82%), and Zn (77%) and fairly resistant to nickel (68%) and chromium (55%), but sensitive to mercury (23%), and 40% of the isolates including AAUGR-9, AAUGR-50, AAUGR-5, AAUGR-6, AAUGR-11, AAUGR-14, AAUGR-24, and AAUGR-46 were highly tolerant to many of the tested heavy metals. Isolates AAUGR-2, AAUGR-6, AAUGR-9, AAUGR-11, AAUGR-14, and AAUGR-24 showed the highest resistance to all or most of the tested antibiotics and heavy metals.
Pattern of carbohydrate and amino acid utilization of isolates
The potential of the isolates to utilize a wide range of carbohydrates and amino acids is shown in Table 2. All isolates utilized most of the monosaccharides and disaccharides except trehalose (63.6) and Na-citrate (41%) that were utilized by fewer isolates. Isolates AAUGR-2, AAUGR-9, AAUGR-11, AAUGR-12, AAUGR-24, and AAUGR-42 utilized all the tested carbohydrates, and the other isolates catabolized 80–90% of the carbon sources, indicating the versatility of the isolate to utilize carbohydrate sources. Isolates also showed versatility (46–100%) in utilizing different amino acid sources (Table 2), and 18 % of the isolates including AAUGR-2, AAUGR-5, AAUGR-11, and AAUGR-24 catabolized all nitrogen sources. The amino acids utilized the least were leucine (77.3%) and lysine (81.8%).
Clustering analysis
The numerical analysis conducted based on 59 phenotypic features showed the grouping of the 22 isolates into three distinct clusters at 25% dissimilarity level (Fig. 2). Cluster I was further divided into two sub-clusters comprising of three isolates in sub-clusters IA (AAUGR-37, AAUGR-42, and AAUGR-16) and four isolates in sub-cluster IB (AAUGR-3, AAUGR-19, AAUGR-17, and AAUGR-46). Cluster II was also further divided into two sub-clusters with four isolates consisting of AAUGR-7, AAUGR-8, and AAUGR-26 in sub-cluster IIA and AAUGR-12 in sub-cluster IIB. Cluster III was further classified into three sub-clusters comprising of eleven isolates that included isolates AAUGR-2, AAUGR-5, AAUGR-6, AAUGR-9, AAUGR-11, AAUGR-14, AAUGR-15, AAUGR-20, AAUGR-24, AAUGR-30, and AAUGR-50. Clustering decreased below 5% level of dissimilarity, indicating their phenotypic diversity. Taken together, the clusters indicated high diversity among grass pea rhizobia.
16S rRNA gene sequence analysis
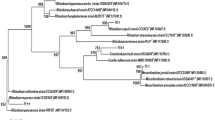
Among the rhizobial isolates characterized for 16S rRNA genes, only 2 (14.3%) of the sequenced isolates AAUGR-24 and AAUGR-42 matched with Rhizobium leguminosarum with 100% sequence similarity, whereas the majority 12 (85.7%) were grouped with Rhizobium species with 99.8% similarity (Table 3).
Discussion
The conventional presumptive and authentication tests together with cultural and growth characteristics showed that 98% of the isolates were authenticated as fast-growing root nodule bacteria (Vincent 1970). Thus, according to the plant host specificity for infection and nodulation as recorded in the cool-season legume crops of tribe Viceae, the isolates were categorized as Rhizobium leguminosarum bv. viciae (Berkum et al. 1995). The findings of this study were similar to the rhizobial phenotypic features recorded from rhizobia-nodulating cross-inoculating hosts: faba bean (Mnalku et al. 2009; Belay and Assefa 2011; Legesse and Assefa 2013), field pea (Amsalu et al. 2012; Baye et al. 2015), and lentil (Jida and Assefa 2011; Rashid et al. 2012; Setargie et al. 2015).
The isolates induced nodules on the host plant with nodule number (NN) ranging from 17 to 116/plant and mean nodule dry weight ranging from 0.010 to 0.098 g/plant, indicating 7- and 9-fold difference between the highest and lowest nodulating isolates, respectively. Likewise, other authors that worked on cross-inoculating group rhizobia also reported similar variations ranging from 2.5 to 9 times and from 2.5 to 15 times for nodule number/plant and nodule dry weight/plant, respectively (Belay and Assefa 2011; Mnalku et al. 2009; Argaw 2012a, b; Legesse and Assefa 2013; Tsegaye et al. 2015). This indicated the existence of variations in rhizobial effectiveness which accounts to low rhizobial density, incompatibility of the rhizobia, and difference in rhizobial strain and host plant type (Vincent 1970). The host plants inoculated with AAUGR (Rhizobium sp. AAUGR-15, AAUGR-14, AAUGR-11, and AAUGR-6) and isolate AAUGR-5 that displayed similar pattern in NN and NDW showed high correlation between nodule dry weight and shoot dry weight (r = 0.864 and 0.817).
The difference in shoot dry matter accumulated between the highest (1.15 g/p) and the least (0.27 g/p) inoculated plants was greater than fourfold which coincided with the findings of Argaw (2012a, b) and Belay and Assefa (2011) who reported 4.5 times and 5.8 times difference between the highest and least faba bean rhizobia-inoculated plants, indicating the existence of variations in rhizobial effectiveness (Vincent 1970). This could be due to difference in agro-ecological zones of Ethiopia (Berkum et al. 1995; Wolde-Meskel 2007) and variations in rhizobial strains and plant species (or cultivars) (Neelawan 2012). Among the isolates, 86% of them were found highly effective and effective in their symbiotic effectiveness similar to other reports in which authors reported 55–100% effective and highly effective rhizobial isolates compared with the positive control (Mnalku et al. 2009; Argaw 2012a, b; Legesse and Assefa 2013; Tsegaye et al. 2015). The correlation between shoot dry weight and percent symbiotic effectiveness showed a relatively more direct and perfect linear relationship than the other parameters (r = 0.988), suggesting that shoot dry matter is a good indicator of relative effectiveness revealing sound correlation between the nitrogen-fixing capacity of legumes and their shoot dry matter accumulation (Mulongoy 2004). Among the isolates, AAUGR-15, AAUGR-14, AAUGR-11, AAUGR-9, AAUGR-6, and AAUGR-5 promoted grass pea growth under greenhouse conditions, indicating their potential for application at field conditions.
Grass pea rhizobia showed variations in the tested physiological parameters. All isolates were tolerant to 6–9, whereas 5 (23 %) grew at pH 4 and 9 (40 %) grew on the medium adjusted at pH 10, indicating that the isolates were more tolerant to alkaline than acidic pH. Lebrazi and Benbrahim (2014) reported that rhizobia can grow at high pH since alkalinity is less harmful to the survival of rhizobia. Most isolates grew at higher salt concentrations similar to rhizobial isolates from Vicia faba, Lens culinaris, and Pisum sativum (Mohammad et al. 1991; Zahran et al. 2003; Keneni et al. 2010; Legesse and Assefa 2013; Setargie et al. 2015) showing that fast-growing rhizobia are more salt-tolerant than slow-growing rhizobia (Zahran 1999) and could be used as a remedy in areas where 40% of the world’s land surface is categorized as having potential salinity problems (Zahran 1999). The isolates of this study showed growth at a temperature range of 10–45 °C that was similar to the findings of Workalemahu (2009) and Legesse and Assefa (2013) who reported faba bean rhizobial isolates that showed growth at similar temperature range, implying that rhizobia are tolerant to heat (Michiels et al. 1994) and tolerant to freezing temperatures as low as – 10 °C (Cloutier et al. 1992). Among the isolates of this study, AAUGPR-6, AAUGPR-9, AAUGPR-11, AAUGPR-14, and AAUGPR-24 grew better in the tested tolerance parameters which could be considered potential candidates for use as inoculants in areas where pH, salinity, and temperature are persistent problems.
The isolates of this study showed variations in their tolerance and sensitivity to the tested antibiotics (Keneni et al. 2010; Zahran et al. 2012; Hewedy et al. 2014; Abera et al. 2010) with a pattern of ST > TE > CL > AM > NE > ER > NA which is adopted as a useful complementary tool for characterization and discrimination of rhizobial isolates (Zahran et al. 2012). Similarly, the isolates showed variability in heavy metal tolerance (Jida and Assefa 2011; Baye et al. 2015) with sensitivity pattern of HgCl2 > K2Cr2O7 > NiCl2 > ZnCl2 > Pb(CH3COO)2 > MnCl2. Most of the isolates of this study showed the ability to grow at high metal concentrations which is also found in many rhizospheric microorganisms (Lakzian et al. 2002) and may be the result of intrinsic or induced mechanisms (Giller et al. 1998). It can be concluded that no significant difference was observed in the ability of isolates to tolerate antibiotics and heavy metals. Isolates AAUGR-2, AAUGR-6, AAUGR-9, AAUGR-11, AAUGR-14, and AAUGR-24 showed the highest resistance both to the tested antibiotics and heavy metals, indicating correlation between metal and antibiotic tolerance in bacteria because of the likelihood that resistance genes to both antibiotics and heavy metals may be located closely together on the same plasmid in bacteria and are thus more likely to be transferred together in the environment (Elizabeth et al. 2012).
Grass pea rhizobia showed wide range of utilization to the tested carbohydrate and nitrogen sources as similarly reported by Maâtallah et al. 2002; Wielbo et al. 2012; Zahran et al. 2012; Legesse and Assefa 2013; Tena et al. 2016 which may be advantageous for survival in soil (Gauri et al. 2011) and be used as a marker for rhizobial classification (Zabaloy and Gomez 2005). The types of carbohydrates tested in this study showed variations in their degree of utilization by the isolates monosaccharides > disaccharides > polysaccharides similar with the findings of Jida and Assefa 2011; Zahran et al. 2012 reported a broad range carbohydrate utilization ability of fast-growing rhizobial isolates. Variability among the isolates in utilizing the amino acid sources was observed, indicating that utilization of amino acids is one of the phenotypic characteristics for possible taxonomic diversity of the study isolates (Zahran et al. 2012. The inconsistency observed in substrate utilization among rhizobial isolates may have ecological relevance which could be important to live as a saprophyte in soil or colonize plant roots and live symbiotically (Charabarti et al. 1981).
The ecological stresses and adaptations to nutritional utilization tests conducted showed that isolates AAUGR-2, AAUGR-5, AAUGR-6, AAUGR-9, AAUGR-11, AAUGR-14, AAUGR-15, AAUGR-24, and AAUGR-50 were the most tolerant and considered potential candidates to relieve environmental stresses including soil pH, salinity, and temperature that affect the survival, metabolism, and functioning of rhizobia (Abdullah 2002), which in turn limit crop productivity through affecting legume-rhizobia symbiosis (Andres et al. 2012). Consequently, isolating and screening rhizobial inoculants that are compatible to legume host and the target edaphic environment is important since rhizobial populations are known to vary in their tolerance to major environmental factors (Wei et al. 2008). These isolates that exhibited physiological versatility in all the tested physiological assays could be used as an important candidates of microbial inoculants in competitive and complex soil environments (Somasagaren and Hoben 1985).
In this study, isolates AAUGR-2, AAUGR-6, AAUGR-9, AAUGR-11, and AAUGR-24 showed better performances in the tested parameters which were followed by isolates AAUGR-5, AAUGR-14, AAUGR-15, AAUGR-19, AAUGR-20, AAUGR-30, and AAUGR-50 and were regarded as an important inoculants at soil where fluctuations in pH, nutrient availability, temperature, and salinity among other factors greatly influence the growth, survival, and metabolic activity of nitrogen-fixing bacteria and plants and their ability to enter into symbiotic interactions (Werner and Newton 2005). Screening isolates on the basis of their stress tolerance level in culture media is essential for the selection of microbial inoculants for field applications (Fitouri et al. 2012) to improve symbiotic efficiency and crop productivity in agricultural systems (Broughton et al. 2003). Isolates AAUGR-2, AAUGR-6, AAUGR-9, AAUGR-11, AAUGR-14, AAUGR-20, and AAUGR-30 showed similar diversity in both phenotypic functional traits and 16S rRNA sequence analysis, whereas the remaining isolates failed to show similarity which may be due to activity inconsistency at in vitro conditions.
The 16S rRNA gene sequencing result showed that out of the 14 rhizobial isolates, 12 (85.7%) of them were found to be closely related to Rhizobium species with 99.8% identity and 2 (14.3%) isolates to Rhizobium leguminosarum with100% identity which corroborated with the findings of (Berrada et al. 2012) who reported the identification of 2 Rhizobium leguminosarum and 12 Rhizobium species from bean plants with 98–100% identity. Rivas et al. (2009) reported Rhizobium leguminosarum bv. viciae is the specific microsymbiont of the legumes of the tribe Vicieae, which comprises the genera Vicia, Pisum, Lens, and Lathyrus. On the other hand, Aoki et al. (2010) reported 97–100% sequence identity of 16S rRNA gene of rhizobia isolated from Lathyrus japonicus nodules with 16S rRNA gene sequences of Rhizobium species, Rhizobium phaseoli, Rhizobium pisi, Rhizobium multihospitum, Rhizobium tropici, and Rhizobium radiobacter which is nearly similar with the findings of this study. Moreover, Drouin et al. (1996) reported 100% sequence identity of two Lathyrus japonicus and Lathyrus pratensis 16S rRNA gene with Rhizobium species, Rhizobium etli, Rhizobium meliloti, and Rhizobium leguminosarum bv. phaseoli. Similarly, Tena et al. (2017) also reported genotypically diverse rhizobia-nodulating lentil (Lens culinaris Medik.), implying that the phylogenetic positions of grass pea–nodulating rhizobia to Rhizobium species was not unexpected. Further, molecular identification using housekeeping genes such as recA, glnII, atpD, and ropB and other symbiotic genes needs to be conducted for these 12 Rhizobium species to specify as Rhizobium leguminosarum which are more informative for phylogenetic analysis and identification of closely related species than 16S rRNA (Rashid et al. 2012).
In a nutshell, the result of this study confirmed that grass pea rhizobia showed the potential of tolerating a wide range of abiotic stresses, broad range of nutrient versatility, and nitrogen fixation. This enhanced the screening of elite rhizobial strains having better symbiotic and ecophysiological potential that could help tolerate abiotic stresses, increase soil fertility, and thereby improve the growth and yield of rhizobia-associated crop plants. Some rhizobial isolates including AAUGR-5, AAUGR-6, AAUGR-9, AAUGR-11, and AAUGR-14 exhibited consistent performance in all the tested parameters indicating their effective and competitive ability. The 16S rRNA gene sequence analysis conducted on some selected rhizobial isolates showed the identification of Rhizobium species and Rhizobium leguminosarum with 99.8–99% and 100% identity, respectively. In general, the rhizobial strains AAUGR-6, AAUGR-9, AAUGR-11, AAUGR-14, and AAUGR-24 that were identified as Rhizobium species and Rhizobium leguminosarum and ranked top in all the tested symbiotic and stress tolerance parameters were recommended as microbial inoculants for field applications.
References
Abdullah MKA-F (2002) Factors affecting the efficiency of symbiotic nitrogen fixation by Rhizobium. Pak J Biol Sci 5(11):1277–1293
Abera T, Semu E, Debele T, Wegary D, Haekoo K (2010) Effects of faba bean break crop and N rates on subsequent grain yield and nitrogen use efficiency of highland maize varieties in Toke Kutaye, western Ethiopia. WJAS 11(5):311–324
Adal M, Tadege M, Assefa F (2018) Rhizospheric bacterial isolates of grass pea (Lathyrus sativus L.) endowed with multiple plant growth promoting traits. J Appl Microbiol 125(6):1–16
Amargar N, Macheret V, Aguerre G (1997) Rhizobum gallicum sp. Nov, and Rhizobum giardinii sp. Nov. from Phaseolus vulgaris nodules. Int J Syst Bacteriol 47:996–1006
Amsalu A, Assefa F, Hailemariam A (2012) Symbiotic and phenotypic characterization of Rhizobium isolates of field pea (Pisum sativum L.) Fabaceae, from central and southern Ethiopia. Ethiop J Biol Sci 11(2):163–179
Andres JA, Rovera M, Guiñazú LB, Pastor NA, Rosas SB (2012) Interactions between legumes and rhizobia under stress conditions. In: Dinesh KM (ed) Bacteria in agrobiology: stress management. Springer-Verlag, Berlin, pp 77–94. https://doi.org/10.1007/978-3-642-23465-1-5
Aoki S, Kondo T, Prévost D, Nakata S, Kajita T (2010) Genotypic and phenotypic diversity of rhizobia isolated from Lathyrus japonicus indigenous to Japan. Syst Appl Microbiol 33:383–397
Argaw A (2012a) Characterization of symbiotic effectiveness of rhizobia nodulating faba bean (Vicia faba L.) isolated from Central Ethiopia. Res J Microbiol 7(6):280–296
Argaw A (2012b) Evaluation of symbiotic effectiveness and size of resident Rhizobium leguminosarum var. viciae nodulating lentil (Lens culinaris medic) in some Ethiopian soils. Int J Agr & Agri R 2(4):18–31
Baye K, Kebede A, Assefa F (2015) Isolation and phenotypic characterization of field pea nodulating rhizobia from Eastern Ethiopia soils. World Appl Sci J 33(12):1815–1821
Belay Z, Assefa F (2011) Symbiotic and phenotypic diversity of Rhizobiumleguminosarum bv. viciae from Northern Gondar, Ethiopia. Afr J Biotechnol 10(21):4372–4379
Berkum V, Desta B, Vera FT, Keyser HH (1995) Variability among Rhizobium strains originating from nodules of Vicia faba. Appl Environ Microbiol 6:2649–2653
Bernal G, Graham PH (2001) Diversity in the rhizobia associated with Phaseolus vulgaris L. in Ecuadore and comparisons with Mexican bean rhizobia. Can J Microbiol 47:526–534
Berrada H, Nouioui I, Houssaini MI, Ghachtouli N, Maher G, Benbrahim KF (2012) Phenotypic and genotypic characterizations of rhizobia isolated from root nodules of multiple legume species native of Fez, Morocco. Afr J Microbiol Res 6(25):5314–5324
Beyene D, Kassa S, Assefa F, Gebremedhin H, Berkum P (2004) Ethiopian soils harbor natural populations of rhizobia that form symbioses with common bean (Phaseolus vulgaris L.). Arch Microbiol 181:129–136
Bhargava Y, Murthy JSR, Kumar TVR, Rao MN (2016) Phenotypic, stress tolerance and plant growth promoting characteristics of rhizobial isolates from selected wild legumes of semiarid region, Tirupati, India. Adv Microbiol 6:1–12
Bontemps C, Elliott GN, Simon MF (2010) Burkholderia species are ancient symbionts of legumes. Mol Ecol 19:44–52
Broughton WJ, Zhang F, Perret X, Staehelin C (2003) Signals exchanged between legumes and Rhizobium: agricultural uses and perspectives. Plant Soil 252:129–137
Buck JD (1982) Non-staining (KOH) method for determination of gram reactions of marine bacteria. Appl Environ Microbiol 44(4):992–993
Charabarti S, Lee MS, Gibson AH (1981) Diversity in the nutritional requirements of strains of various Rhizobium species. Soil Biol Biochem 13:349–354
Cloutier J, Prevost D, Nadeau P, Antoun H (1992) Heat and cold shock protein synthesis in arctic and temperate strains of rhizobia. Appl Environ Microbiol 58(9):2846–2853
CSA (Central Statistical Authority) (2017) Agricultural samples survey 2016/2017 report on area and production of crops. Volume I. Statistical Bulletin, Number 584, Addis Ababa, Ethiopia
Demelash H, Abera S, Teka TA (2015) Effects of processing on nutritional composition and anti-nutritional factors of grass pea (Lathyrus Sativus L.): a review. Food Science and Quality Management. 36:61–71
Drouin P, Prevost D, Antoun H (1996) Physiological adaptation to low temperatures of strains of Rhizobium leguminosarum bv. viciae associated with Lathyrus spp. FEMS Microbiol Ecol. 32(2000):111–120
Eardly B, Wang F, Berkum PV (1996) Corresponding 16S rRNA gene segments in Rhizobiaceae and Aeromonas yield discordant phylogenies. Plant Soil 186:69–74
Elizabeth D, David H, Ross B, Graham O, Rosalind D, Matthew D, Ron Y, Greg G, Elizabeth H, Lori P, Nikki S, John H, Neil B (2012) Inoculating legumes: a practical guide. Grains Research and Development Corporation
Fitouri DS, Ben JF, Zribi K, Rezgui S, Mhamdi R (2012) Effect of inoculation with osmotolerant strain of Rhizobium sullae on growth and protein production of sulla (Sulla coronarium L.) under water deficit. J Appl Biosci 51:642–3651
Gauri AS, Rajendra PB, Shailja PM, Kaur B, Ashok N (2011) Characterization of Rhizobium isolated from root nodules of Trifolium alexandrinum. J Agric Technol 7(6):1705–1723
Gebremariam A, Assefa F (2017) The effect of inter cross-inoculation host group rhizobia on the growth and nitrogen fixation of Faba Bean (Vicia faba L.) varieties in North Showa, Amhara Regional State, Ethiopia. J Agric Biotech Sustain. Dev 10(2):25–33
Giller KE, Witter E, McGrath SP (1998) Toxicity of heavy metals to microorganisms and microbial process in agricultural soils: a review. Soil Biol Biochem 30:1389–1414
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica 4(1):1–9
Hewedy OM, Eissa RA, Elzanaty AM, Nagaty HH, Abd-Elbary MI (2014) Phenotypic and genotypic diversity of rhizobia nodulating faba bean from various Egyptian locations. J Bioproces Biotechniq 4(5):1–8
Jennifer SM (2003) A brief history of grass pea and its use in crop improvement. Lathyrus/Lathyrism news Letter 3:18–20
Jida M, Assefa F (2011) Phenotypic and plant growth promoting characteristics of Rhizobium leguminosarum bv. viciae from lentil growing areas of Ethiopia. Afr. J Microbiol Res 5(24):4133–4142
Jordan DC (1984) Rhizobiaceae. In: Krieg N, Holt JG (eds) Bergey’s manual of systematic bacteriology. Williams, and Wilkins, Baltimore, pp 235–256
Keneni A, Assefa F, Prabu PC (2010) Characterization of acid and salt tolerant rhizobial strains isolated from faba bean fields of Wollo, Northern Ethiopia. J Agr Sci Tech 12:365–376
Laguerre G, Louvrier P, Allard MR, Amarger N (2003) Compatibility of rhizobial genotypes within natural populations of Rhizobium leguminosarum biovar viciae for nodulation of host legumes. Appl Environ Microbiol 69(4):2276–2283
Lakzian A, Murphy V, Turner A, Beynon JL, Giller KE (2002) Rhizobium leguminosarum bv. viciae populations in soils with increasing heavy metal contamination: abundance, plasmid profiles, diversity and metaltolerance. Soil Biol Biochem 34:519–529
Lebrazi S, Benbrahim KF (2014) Environmental stress conditions affecting the N2 fixing Rhizobium–legume symbiosis and adaptation mechanisms. Afr J Microbiol Res 8(53):4053–4061
Legesse S, Assefa F (2013) Symbiotic and phenotypic characteristics of rhizobia nodulating faba bean (Vicia Faba) from Tahtay Koraro, Northwestern zone of Tigray Regional State, Ethiopia. IJEERT 2(11):15–23
Lupwayi NZ, Haque I (1994) Working document: legume-rhizobium technology manual. Environmental science division international livestock center for Africa, Addis Ababa, pp 1–40
Maâtallah J, Berraho EB, Sanjuan J, Lluch C (2002) Phenotypic characterization of rhizobia isolated from chickpea (Cicer arietinum) growing in Moroccan soils. Agronomie 22:321–329
Mahdavi B, Seyed AM, Modarres S, Majid A (2007) Nodulation and root traits in four grass pea (Lathyrus sativus) ecotypes under root zone temperatures. Pak J Biol Sci 10(8):1243–1249
Michiels J, Verreth C, Vanderleyden J (1994) Effects of temperature stress on bean-nodulating Rhizobium strains. Appl Environ Microbiol 60:1206–1212
Mnalku A, Gebrekidan H, Assefa F (2009) Symbiotic effectiveness and characterization of Rhizobium strains of Faba bean (Vicia faba L.) collected from Eastern and Western Hararghe highlands of Ethiopia. EJNR 11(2):223–244
Moges G, Wodajo N, Gorton L, Yigzaw Y, Kalcher K, Belay A, Akalu G, Baboo MN, Solomon T (2004) Glutamate oxidase advances the selective bioanalytical detection of the neurotoxic amino acid ß-ODAP in grass pea: a decade of progress. Pure Appl Chem 76(4):765–775
Mohamed HA, Fatthy MM, Abdel-Wahab EET (2012) Isolation and characterization of a heavy-metal-resistant isolate of Rhizobium leguminosarum bv. viciae potentially applicable for biosorption of Cd2+ and Co2+. Int Biodeterior Biodegradation 67:48–55
Mohammad RM, Kharazian AA, Campbell WF, Rumbaugh MD (1991) Identification of salt- and drought-tolerant Rhizobium meliloti strains. Plant Soil 134:271–276
Monsoor MA, Yusuf HK (2002) In vitro protein digestibility of lathyrus pea (Lathyrus sativus), lentil (Lens culinaris), and chickpea (Cicer arietinum). Int J Food Sci Technol 37:97–99
Mulongoy K (2004) Technical paper 2. Biological nitrogen fixation. Can J Microbiol 21:1–19
Neelawan P (2012) Phenotypic and genotypic diversity of rhizobia. Silpakorn University, Thailand, pp 3–48
Perret X, Staehelin C, Broughton WJ (2000) Molecular basis of symbiotic promiscuity. Microbiol Mol Biol Rev 64:180–201
Rashid MH, Schäfer H, Gonzalez J, Wink M (2012) Genetic diversity of rhizobia nodulating lentil (Lens culinaris) in Bangladesh. Syst Appl Microbiol 35:98–109
Rivas R, García-Fraile P, Velázquez E (2009) Taxonomy of bacteria nodulating legumes. Rev Microbiol Insight 2:251–269
Santillana N, Ramirez-Bahena MH, Garcia-Fraile P, Velazquez E, Zuniga D (2008) Phylogenetic diversity based on rrs, atpD, recA genes 16S–23S intergenic sequence analyses of rhizobial strains isolated from Vicia faba and Pisum sativum in Peru. Arch Microbiol 189:239–247
Setargie A, Tilahun S, Alemayehu S, Dejenie T, Kiros S (2015) Isolation and phenotypic characterization of phosphate solubilizing bacteria from lentil (Lens culnaris.) rhizosphere soils from southern parts of Tigray, Ethiopia. Intl. J Microbiol Res 6(3):188–194
Shayne JJ, Hugenholtz P, Sangwan P, Osborne C, Jansen HP (2003) Laboratory cultivation of widespread and previously uncultured bacteria. Appl Environ Microbiol 69:7211–7214
Somasegaren P, Hoben HJ (1985) Methods in legumerhizobium technology. University of Hawaii NifTAL Project and MIRCEN Department of Agronomy and Soil Science. USAID, USA
Somasegaren P, Hoben HJ (1994) Hand book for rhizobia. Methods in legume-rhizobium technology. Springer – Verlag, New York, pp 1–441
Tena W, Wolde-Meskel E, Walley F (2016) Symbiotic efficiency of native and exotic Rhizobium strains nodulating lentil (Lens culinaris Medik.) in soils of Southern Ethiopia. Agronomy 6:1–11
Tena W, Wolde-Meskel E, Degefu T, Fran W (2017) Lentil (Lens culinaris Medik.) nodulates with genotypically and phenotypically diverse rhizobia in Ethiopian soils. Syst Appl Microbiol 40:22–33
Tsegaye D, Assefa F, Gebrekidan H, Keneni A (2015) Nutritional, ecophysiological and symbiotic characteristics of rhizobia nodulating faba bean (Vicia faba L.) collected from acidic soils of Ethiopia. Afr J Environ Sci Technol 9(7):646–654
Urga K, Fufa H, Biratu E, Husain A (2005) Evaluation of Lathyrus sativus cultivated in Ethiopia for proximate composition, minerals, ODAP and anti-nutritional components. Afr J Food Agric Nutr Dev 0.5(1):1–16
Vincent JM (1970) A manual for the practical study of root nodule bacteria. Blackwell sci. publ. oxford. PP.164.
Ward JH (1963) Hierarchical grouping to optimize an objective function. J Am Stat Assoc 58:236–244
Wei GH, Yang X, Zhang YZ, Lindstrom K (2008) Strain Mesorhizobium sp. CCNWGX035: a stress tolerant isolate from Glycyrriza glabra displaying a wide host range of nodulation. Pedosphere 18:102–112
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S RibosomalDNA amplification for phylogenetic study. J Bacteriol 173(2):697–703
Werner D, Newton WE (2005) Nitrogen fixation in agriculture, forestry, ecology, and the environment. Springer Publication.
White D (1995) The physiology and biochemistry of prokaryotes. Oxford University Press. PP. 34-46
Wielbo J, Kidaj D, Koper P, Kubik-Komar A, Skorupska A (2012) The effect of biotic and physical factors on the competitive ability of Rhizobium leguminosarum. Cent Eur J Biol 7(1):13–24
Wolde-Meskel E (2007) Genetic diversity of rhizobia in Ethiopian soils: their potential to enhance biological N fixation (BNF) and soil fertility for sustainable agriculture. Ethiop. J Biol Sci 6(1):77–95
Workalemahu A (2009) The effect of indigenous root-nodulating bacteria on nodulation and growth of faba bean (Vicia Faba) in the low-input agricultural systems of tigray highlands, Northern Ethiopia. MEJS 1(2):30–43
Zabaloy MC, Gomez MA (2005) Diversity of rhizobia isolated from an agricultural soil in Argentina based on carbon utilization and effects of herbicides on growth. Biol Fertil Soils 42:83–88
Zahran HH (1999) Rhizobium leguminosarum symbiosis and nitrogen fixation under severe conditions and in an Arid Climate. Microbiol Mol Biol Rev 63:968–989
Zahran HH, Abdel-Fattah M, Ahmad MS, Zaki AY (2003) Polyphasic taxonomy of symbiotic rhizobia from wild leguminous plants growing in Egypt. Folia Microbiologica 48:510–520
Zahran HH, Abdel-Fattah M, Yasser MM, Mahmoud AM, Bedmar EJ (2012) Diversity and environmental stress responses of rhizobial bacteria from Egyptian grain legumes. Aust J Basic & Appl Sci 6(10):571–558
Acknowledgments
We thank Dr. Shiping Deng for providing 16S rRNA gene primers and helpful suggestions. This work was supported by funds from Addis Ababa University, Wollo University and Oklahoma State University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest to this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 1595 kb)
Rights and permissions
About this article
Cite this article
Mohammed, M.A., Chernet, M.T. & Tuji, F.A. Phenotypic, stress tolerance, and plant growth promoting characteristics of rhizobial isolates of grass pea. Int Microbiol 23, 607–618 (2020). https://doi.org/10.1007/s10123-020-00131-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10123-020-00131-3