Abstract
The phosphoenolpyruvate-pyruvate-oxaloacetate node is a major branch within the central carbon metabolism and acts as a connection point between glycolysis, gluconeogenesis, and the TCA cycle. Phosphoenolpyruvate carboxylase, pyruvate carboxylase, phosphoenolpyruvate carboxykinase, malic enzymes, and pyruvate kinase, among others, are enzymes included in this node. We determined the mRNA levels and specific activity profiles of some of these genes and enzymes in Streptomyces coelicolor M-145. The results obtained in the presence of glucose demonstrated that all genes studied of the phosphoenolpyruvate-pyruvate-oxaloacetate node were expressed, although at different levels, with 10- to 100-fold differences. SCO3127 (phosphoenolpyruvate carboxylase gene) and SCO5261 (NADP+-dependent malic enzyme gene) showed the highest expression in the rapid growth phase, and the mRNA levels corresponding to SCO5896 (phosphoenolpyruvate-utilizing enzyme gene), and SCO0546 (pyruvate carboxylase gene) increased 5- to 10-fold towards the stationary phase. In casamino acids, in general mRNA levels of S. coelicolor were lower than in glucose, however, results showed greater mRNA expression of SCO4979 (PEP carboxykinase), SCO0208 (pyruvate phosphate dikinase gene), and SCO5261 (NADP+-dependent malic enzyme). These results suggest that PEP carboxylase (SCO3127) is an important enzyme during glucose catabolism and oxaloacetate replenishment. On the other hand, phosphoenolpyruvate carboxykinase, pyruvate phosphate dikinase, and NADP+-malic enzyme could have an important role in gluconeogenesis in S. coelicolor.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The bacterial life cycle depends on the synthesis of twelve precursor metabolites: glucose-6-phosphate, fructose-6-phosphate, ribose-5-phosphate, 3-phosphoglycerate, erythrose-4-phosphate, triose-phosphate, pyruvate, acetyl CoA, α-ketoglutarate, succinyl CoA, oxaloacetate, and phosphoenolpyruvate, which is one of the most important compounds in carbon metabolism because it is a precursor for many biomolecules (Valle et al. 1996). Therefore, the synthesis of these metabolites and the regulatory mechanisms that control them are crucial for maintaining the levels required for microorganism growth. In this sense, it is essential to understand the metabolic pathways that allow their synthesis in order to design strategies that improve the growth of microorganisms and the production of important industrial metabolites such as antibiotics (Van Keulen et al. 2011). It has been reported that glucose can be used by microorganisms for the synthesis of the twelve aforementioned precursors through the glycolytic and pentose phosphate pathways and through the tricarboxylic acid (TCA) cycle (Valle et al. 1996).
A large variety of enzymes involved in the phosphoenolpyruvate-pyruvate-oxaloacetate (PEP-PYR-OXA) node have been reported, such as phosphoenolpyruvate carboxylase (PEPC; EC 4.1.1.31), pyruvate carboxylase (PYC; EC 6.4.1.1), phosphoenolpyruvate carboxykinase (PEPCk; EC 4.1.1.32), malic enzymes (ME; EC 1.1.1.38), pyruvate kinase (PyK; EC 2.7.1.40), pyruvate dehydrogenase complex E1 (EC 1.2.4.1), pyruvate dehydrogenase complex E2 (EC 2.3.1.12), and pyruvate dehydrogenase complex E3 (EC 1.8.1.4). Therefore, this node is indispensable for the distribution of phosphoenolpyruvate, pyruvate, and oxaloacetate in numerous microorganisms; further, enzymes involved in this node vary among bacteria, and their activity depends on the culture conditions (Schniete et al. 2018; Sauer and Eikmanns 2005).
Actinomycetes are microorganisms present in the soil that are important commercially and medically. Actinomycetes produce 70% of the known antibiotics and unsurprisingly, the biosynthetic pathways of these secondary metabolites have been thoroughly studied; in contrast, much less information is available regarding their primary metabolism and regulation (Van Keulen et al. 2011; Spížek et al. 2016).
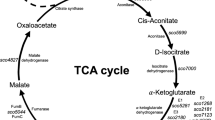
Of all actinomycetes, Streptomyces coelicolor is the most well-known and is used as a model for different metabolic studies. The complete sequence of its genome has been published, which enabled the identification of genes that encode probable enzymes of various metabolic pathways (Bentley et al. 2002). With this data, using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, it could be established that genes of the PEP-PYR-OXA node exist in the S. coelicolor genome (Fig. 1). Among these genes, the level of transcriptional and enzymatic activity in different culture conditions has been determined only for the two pyruvate kinase genes (SCO2014 and SCO5423) (Hiltner et al. 2015). On the other hand, only the specific activities have been reported for phosphoenolpyruvate carboxylase (SCO3127) and malic enzymes (SCO2951 and SCO5261) (Bramwell et al. 1993; Rodriguez et al. 2012), while phosphoenolpyruvate-utilizing enzyme (SCO5896) and pyruvate phosphate dikinase (SCO0208) have been classified as gluconeogenic enzymes based on proteomic studies (Thomas et al. 2012; Jeong et al. 2016). The functions of phosphoenolpyruvate-utilizing enzyme (SCO5896) and pyruvate phosphate dikinase (SCO0208 and SCO2494) were assigned by homology alone. However, no further details of their expression or activity have been provided.
The phosphoenolpyruvate-oxaloacetate node in Streptomyces coelicolor. Abbreviations: PYK, pyruvate kinase; PEPC, phosphoenolpyruvate carboxylase; PEPCk, phosphoenolpyruvate carboxykinase; PYC, pyruvate carboxylase; PEP-UE, phosphoenolpyruvate-utilizing enzyme, PYPDK, pyruvate phosphate dikinase; NAD+-ME, NAD+-dependent malic enzyme; NADP+-ME, NADP+-dependent malic enzyme; PDH, pyruvate: quinona oxidoreductase; PDHC, pyruvate dehydrogenase complex; MDH, malate dehydrogenase; TCA, tricarboxylic acid cycle
In our laboratory, a malate dehydrogenase deletion mutant (DSCO4827) was isolated, and we observed that it possessed the ability to grow using glucose as a carbon source, although it did not do so as effectively as the wild-type strain (Takahashi-Íñiguez et al. 2018). The growth of this mutant may only have been possible in the presence of other enzymes capable of synthesizing oxaloacetate, an intermediate of the TCA cycle that is required for several functions. This suggested the presence of alternative pathways that allowed the organism to function without the activity of malate dehydrogenase, and the genes and enzymes that participate in the PEP-PYR-OXA node were the best candidates. The enzymes involved in the oxaloacetate synthesis of this node enabled the mutant to avoid malate accumulation and to obtain oxaloacetate for the TCA cycle and other metabolic pathways necessary for growth.
Due to the lack of detailed data regarding the expression of PEP-PYR-OXA node genes and enzymes in actinomycetes, in this article, we present the expression profiles of the genes and the enzymatic activity involved in oxaloacetate synthesis in glycolytic and gluconeogenic growth conditions to determine their role in carbon metabolism by the wild-type S. coelicolor strain.
Materials and methods
Reagents
Phosphoenolpyruvate, pyruvate, NAD+, NADP+, acetyl CoA, NADH, ATP, Tris-base, EDTA, malate dehydrogenase, lactate dehydrogenase, malic acid, and ADP were obtained from Sigma-Aldrich (St. Louis, MO, USA).
Bacterial strains and culture conditions
S. coelicolor M145 was grown at 30 °C on soy flour mannitol agar for spore production or at 30 °C and 200 rpm (New Brunswick G-25) in liquid minimal medium (MM) containing 5 g NaCl, 0.3 g K2HPO4, 0.5 g MgSO4.7H2O, 0.02 g FeSO4, 0.05 g ZnSO4, 0.02 g CaCl2, 0.001 g MnCl2, 0.001 g CoCl2, 50 g PEG 8000, 20 g MOPS buffer, and 1% glucose with 2 g (NH4)2SO4, or 1% casamino acids (as the carbon and nitrogen source) per liter. The pH was adjusted to 7.0 with NaOH 1 N. Two hundred milliliters of Luria Bertani (LB) medium were inoculated with 108 fresh spores and grown at 30 °C, 200 rpm for 24 h. Fifty milliliters of LB medium were centrifuged at 5000×g for 5 min and washed twice with sterile water and used to inoculate 500 mL MM contained in siliconized Fernbach flasks (Sigma-Aldrich, St. Louis, MO, USA) and incubated at 30 °C and 200 rpm. Samples of 10–30 mL at different times were collected for RNA purification, and 50 mL samples were used to obtain a cell-free extract for determining specific activities. Two 3 mL samples were collected at different fermentation times to quantify protein in regard to growth by the Lowry method, as previously described (Flores and Sánchez 1985). Glucose and ammonium concentration in the supernatant were determined by the Miller’s (1959) and Weatherburn (1967) methods, respectively.
Preparation of cell-free extract
Mycelia were recovered at different fermentation times by centrifugation and washed twice with extraction buffer (Tris-HCl 100 mM pH 7.3, 5 mM EDTA, 1.2 mM PMSF, and 0.4 mM DTT) and the pellet was resuspended in a minimal volume of the same buffer and sonicated at 4 °C for 1 min in intervals of 10 s, with 10 s of rest while chilled, using a Soniprep 150 MSE sonicator (MSE, London, UK). Cell debris was removed by centrifugation at 22,000×g for 15 min. The supernatant was used for enzyme activity assays. Protein concentration was determined by Bradford method (Bradford 1976).
Reverse transcription quantitative PCR
The expression levels of the following genes were examined using a two-step Reverse transcription quantitative PCR (RT-qPCR) system: SCO3127 (phosphoenolpyruvate carboxylase), SCO0546 (pyruvate carboxylase), SCO4979 (PEP carboxykinase), SCO2014 (pyruvate kinase), SCO0208 (pyruvate phosphate dikinase), SCO5896 (phosphoenolpyruvate-utilizing enzyme), SCO2951 (NAD+-dependent malic enzyme), and SCO5261 (NADP+-dependent malic enzyme). The specific primers were designed by Primer-Blast (NCBI) based on the genome sequence GenBank NC_003888 and are listed in Table 1; the final PCR product lengths ranged from 150 to 200 bp. Total RNA was isolated from S. coelicolor M145 grown in MM with 1% glucose or 1% casamino acids for 12, 24, 36, 48, 60, and 72 h using a MagJET RNA kit (Thermo Scientific, Waltham, MA, USA). A 10 μg sample of total RNA was subjected to DNase digestion with RNase-free DNase I (TURBO DNA-free, Ambion, Austin, TX, USA). The RNA quality and concentration were determined by the A260 nm/A280 nm ratio (within the range of 1.9–2.1) on a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). RNA integrity was confirmed with formaldehyde agarose gel electrophoresis, and the purified RNA samples were stored at − 80 °C until use. The RT reactions were performed using M-MLV reverse transcriptase (Promega, Madison, WI, USA) and the corresponding reverse primer according to manufacturer’s instructions using 100 ng of total RNA as template. The qPCR reactions were carried out in a total volume of 15 μL containing 7.5 μL all-in-one qPCR mix (GeneCopoeia, Rockville, MD, USA), 1.25 μL cDNA, and two gene-specific primers on a Rotor-Gene 6000 following the manufacturer’s instructions (Corbett Life Science, Sydney, Australia). The standards were prepared by PCR amplification with S. coelicolor M-145 DNA as a template using the same primers as those used for qPCR. The standard curve was constructed by comparing the concentration of 10-fold dilutions of the standard versus threshold cycles (Schmittgen and Livak 2008; Bustin 2004). The absence of DNA in the samples was verified by qPCR (-RT controls) using the same system. All RT-qPCR experiments satisfied the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines (Bustin et al. 2009). Two experimental and two independent biological replicates were performed for each gene. Absolute quantification analyses of the samples were processed using the Rotor-Gene 6000 Series software v1.7, and the experimental data are expressed as the mean value ± standard deviation of the mean.
Phosphoenolpyruvate carboxylase assay
The PEPC activity encoded by the gene SCO3127 was determined spectrophotometrically by monitoring the NADH concentration at 340 nm according to Bramwell et al. (1993), with minor modifications, in a 1-mL cuvette at 25 °C in a standard assay mixture containing 0.05 M Tris-HCl pH 7.8, 5 mM EDTA, 1.2 mM phenylmethylsulfonyl fluoride, 0.4 mM dithiothreitol, 2 mM phosphoenolpyruvate (PEP), 5 mM MgCl2, 10 mM NaHCO3, 0.2 mM NADH, 0.1 mM acetyl CoA, and 10 U malate dehydrogenase.
Pyruvate carboxylase assay
The PYC activity encoded by the gene SCO0546 was determined spectrophotometrically by monitoring the NADH concentration at 340 nm according to Koffas et al. (2002), with minor modifications, in a 1-mL cuvette at 25 °C in a standard assay mixture containing 0.1 M Tris-HCl pH 7.8, 4 mM ATP, 5 mM MgCl2, 25 mM NaHCO3, 0.2 mM NADH, 0.075 mM acetyl CoA, 10 mM sodium pyruvate, and 10 U malate dehydrogenase.
Pyruvate kinase assay
The pyruvate kinase activity encoded by the gene SCO2014 was determined spectrophotometrically by monitoring the NADH concentration at 340 nm according to Petersen et al. (2001), with slight modifications, in a 1-mL cuvette at 25 °C in a standard assay mixture containing 0.1 M Tris-HCl pH 7.0, 1 mM ADP, 10 mM MgCl2, 0.2 mM NADH, 10 mM PEP, and 10 U lactate dehydrogenase.
NAD+-dependent malic enzyme assay
The NAD+-ME activity encoded by the gene SCO2951 was determined spectrophotometrically by monitoring the NADH concentration at 340 nm according to Rodriguez et al. (2012), with slight modifications, in a 1-mL cuvette at 25 °C in a standard assay mixture containing 0.05 M Tris-HCl pH 8.0, 1 mM NAD+, 10 mM MgCl2, and 20 mM malate.
NADP+-dependent malic enzyme assay
The NADP+-ME activity encoded by the gene SCO5261 was determined spectrophotometrically by monitoring the NADPH concentration at 340 nm according to Rodriguez et al. (2012), with minor modifications, in a 1-mL cuvette at 25 °C in a standard assay mixture containing 0.05 M Tris-HCl pH 8.0, 1 mM NADP+, 10 mM MgCl2, and 20 mM malate.
Phosphoenolpyruvate carboxykinase assay
The PEPCK activity encoded by the gene SCO4979 was determined spectrophotometrically by monitoring the NADH concentration at 340 nm according to Lee et al. (2013), with minor modifications, in a 1-mL cuvette at 25 °C in a standard assay mixture containing 0.01 M HEPES pH 7.0, 10 mM PEP, 10 mM MnCl2, 10 mM NaHCO3, 0.2 mM NADH, 2 mM ADP, 2 mM reduced glutathione, and 10 U malate dehydrogenase.
Results and discussion
The PEP-PYR-OXA node is a distribution point of carbon flux between the different central metabolic pathways. In silico, using KEGG database, it was possible to establish which genes participating in the PEP node are encoded in the genome of S. coelicolor M-145 (Fig. 1); 10 enzymes were found to be involved in the node but the following genes and enzymes, phosphoenolpyruvate carboxylase (SCO3127; PEPC), pyruvate carboxylase (SCO0546; PYC), phosphoenolpyruvate carboxykinase (SCO4979; PEPCK), NAD+-dependent malic enzyme (SCO2951; NAD+-ME), NADP+-dependent malic enzyme (SCO5261; NADP+-ME), pyruvate kinase (SCO2014; PYK 1), pyruvate phosphate dikinase (SCO0208; PYPDK), and phosphoenolpyruvate-utilizing enzyme (SCO5896; PEP-UE) were studied, since they are involved in the synthesis of oxaloacetate. As mentioned previously, very little information has been found regarding the conditions in which these genes are expressed or their enzymatic activity profiles in Streptomyces.
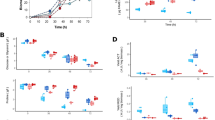
To determine the gene expression level, absolute quantification was used because the expression of the hrdB gene was not constant during the growth of S. coelicolor WT or the Δmdh in our conditions (Takahashi-Íñiguez et al. 2018). Two types of carbon sources were used: a glycolytic source (glucose) and a gluconeogenic source (casamino acids) to compare the expression of these genes considered anaplerotic or gluconeogenic. Figure 2 depicts the growth curves with glucose (panel A) and casamino acids (panel B). It was observed that glucose enabled greater growth compared with casamino acids, and this was associated with sugar and ammonium consumption. The microorganism grew over the first 12 h, after which growth was stably maintained; at 36 h, the biomass further increased with casamino acids until 60 h. The stationary phase began at 48 h with glucose and 60 h with casamino acids. In the latter, ammonium accumulated in the medium after 36 h of growth, probably due to amino acids degradation.
Biomass (white square), supernatant ammonium concentration (white triangle), and residual glucose (white circle) of Streptomyces coelicolor M-145 grown in MM with 1% glucose (a) or 1% casamino acids (b). Samples were taken at the indicated times, and growth was quantified as described in the “Materials and methods” section. Bars represent the standard deviation, and all measurements were performed at least in triplicate
Pyruvate kinase (SCO2014)
Pyruvate kinase encoded by the gene SCO2014 catalyzes the last step of glycolysis, in which a phosphoryl group is transferred from phosphoenolpyruvate to ADP to form pyruvate (Van Keulen et al. 2011; Schniete et al. 2018); therefore, it would be expected that mRNA levels of the pyk1 gene would be higher during the rapid growth of S. coelicolor when glucose is the carbon source. This corresponds to the obtained results, since the mRNA levels increased from 12 to 36 h, which coincided with growth, and declined in the last 12 h. The maximum level was 0.007 ng μg−1 total RNA, which is lower than that obtained with other genes that are considered anaplerotic like SCO0546 (Fig. 3a, c). With casamino acids, pyk1 transcriptional levels diminished during the first 12 h of growth and then increased to a maximum at 48 h. These results could be due to the fact that in the LB medium, there were high levels of mRNA expression that were not maintained in the medium with casamino acids, so this decrease was observed. The specific activity of pyruvate kinase showed the same pattern as the RNA; notably, the enzymatic activity of a glycolytic enzyme was greater under gluconeogenic conditions (Fig. 4a). Contrasting with Schniete et al. (2018) results, these authors report a constitutive expression for pyk1 gene both in minimal medio with glucose and tween. The differences could be because, in that studies, the relative quantification method was used, comparing expression in stationary phase versus log phase and normalizing to hrdB gene. It has been reported that hrdB transcript levels not always remain constant, observing changes between growth phases; its expression is considered also culture conditions dependent (Li et al. 2015).
The mRNA expression profile of PEP-pyruvate-oxaloacetate node genes (black triangle) and specific activity of the enzymes (black square) at different growth times in S. coelicolor. RNAs obtained from cells grown in MM supplemented with 1% glucose were used for RT-qPCR analysis. Gene transcript levels and specific activities were determined as described in “Materials and methods”. a SCO2014 (pyruvate kinase 1); b SCO3127 (PEP carboxylase); c SCO0546 (pyruvate carboxylase); d SCO 4979 (PEP carboxykinase); e SCO5261 (NADP+-dependent malic enzyme); f, SCO2951 (NAD+-dependent malic enzyme)
The mRNA expression profile of PEP-pyruvate-oxaloacetate node genes (black triangle) and specific activity of the enzymes (black square) at different growth times in S. coelicolor. RNAs obtained from cells grown in MM supplemented with 1% casamino acids were used for RT-qPCR analysis. Gene transcript levels and specific activities were determined as described in the “Materials and methods” section. a SCO2014 (pyruvate kinase 1); b SCO3127 (PEP carboxylase); c SCO0546 (pyruvate carboxylase); d SCO4979 (PEP carboxykinase); e SCO5261 (NADP+-dependent malic enzyme); f SCO2951 (NAD+-dependent malic enzyme)
S. coelicolor possesses two pyruvate kinase isoenzymes, PYK1 (SCO2014) and PYK2 (SCO5423), so enzyme activity obtained corresponds to both; however, it is important to mention that the Kcat of PYK1 (the enzyme analyzed in the present study) is at least 20 times greater than that of PYK2 (Schniete et al. 2018). This could be the reason why the specific activity of pyruvate kinase showed the same profile as the RNA. Notably, the enzymatic activity of a glycolytic enzyme was greater under gluconeogenic conditions, suggesting that the protein may be under posttranslational regulation when glucose was the carbon source (Figs. 3 and 4a), which is consistent with the previous proposal that transcript levels of pyk genes could be regulated at transcriptional or posttranscriptional level (Schniete et al. 2018).
Phosphoenolpyruvate carboxylase (SCO3127)
PEPC catalyzes the carboxylation of phosphoenolpyruvate to form oxaloacetate, an important metabolic compound that is the precursor of several other amino acids. The mRNA levels of SCO3127 increased from 12 to 36 h and then declined. The specific activity showed the same pattern, as levels increased during the first 36 h of growth with glucose as the carbon source (Fig. 3b). With casamino acids, the transcription level decreased from 12 to 24 h, increased at 36 h, and subsequently decreased again. Under this condition, the specific activity decreased from 12 to 48 h and then increased in the stationary growth phase. The specific PEPC activity was of the same order of magnitude in both carbon sources, while the mRNA levels were lower with casamino acids despite greater growth with glucose (Figs. 3 and 4b).
PEPC has long been considered as an active anaplerotic enzyme when S. coelicolor is grown in glucose (Bramwell et al. 1993; Van Keulen et al. 2011). However, the high mRNA levels coupled with the maximum specific activity in the S. coelicolor mycelium at 36 h of growth in glucose suggest that this enzyme significantly contributes to the formation of oxaloacetate during the rapid growth phase, which almost indicates a glycolytic role rather than anaplerotic. Further, it has been reported that the PEPC activity in C. glutamicum is activated by acetyl CoA and fructose 1,6-bisphosphate (an intermediate of glycolysis), indicating that PEPC is necessary for the catabolism of glucose and that, in turn, it is inhibited by aspartate (a precursor in amino acid synthesis) and α-ketoglutarate (an intermediate of the TCA cycle), which accumulate when the microorganism stops growing (Wada et al. 2016). In S. coelicolor, a similar condition may be present; thus, PEPC may be an important enzyme for glycolysis.
Pyruvate carboxylase (SCO0546)
PYC catalyzes a reaction similar to that of PEPC, but it uses pyruvate as a substrate for oxaloacetate synthesis. The mRNA levels of SCO0546 in S. coelicolor grown in 1% glucose increased linearly from 12 to 72 h, reaching a maximum level of 0.011 ng μg−1 total RNA, while the specific enzyme activity remained low from 12 to 36 h but then increased until 72 h (Fig. 3c). When S. coelicolor was grown in 1% casamino acids, the pyc mRNA level was up to 100 times lower compared with the level when grown in glucose; in contrast, the enzymatic activity corresponded to the growth phase when using casamino acids as the carbon source and was 4-fold higher in gluconeogenic conditions (Fig. 4c).
It has been observed that the expression and activity of PYC are influenced by the growth phase and carbon source in several organisms, such as Pseudomonas citronellolis, Corynebacterium glutamicum, and Saccharomyces cerevisiae. In addition, when using carbon sources other than glucose, this enzyme participates in gluconeogenesis, while in the presence of glucose, it plays an anaplerotic role (Taylor et al. 1975; Peters-Wendisch et al. 1998; Menéndez and Gancedo 1998; Brewster et al. 1994). PYC has been identified as a gluconeogenic enzyme in S. coelicolor in the KEGG database. However, as observed in other organisms, anaplerotic enzymes are active not only in gluconeogenesis but can also be active in the presence of glucose (Van Keulen et al. 2011). In this case, the high expression and low activity levels detected with glucose and the low expression and high specific activity with casamino acids suggest an anaplerotic and gluconeogenic function. Furthermore, the high expression level of the SCO0546 gene in the presence of glucose suggests that this carbon source does not have a negative effect on expression, as would be expected for an exclusively gluconeogenic enzyme. This indicates that SCO0546 is likely used for oxaloacetate synthesis, which is necessary for the formation of aspartic family amino acids.
Phosphoenolpyruvate carboxykinase (SCO4979)
Phosphoenolpyruvate carboxykinase catalyzes the decarboxylation of oxaloacetate to form phosphoenolpyruvate and is encoded by the SCO4979 gene. In S. coelicolor grown in glucose, the mRNA levels of this gene increased in the stationary growth phase and reached a maximum at 72 h, suggesting that glucose had a negative effect on its transcription. The activity was also low during the first hours of growth, showed an increase at 48 h, and then remained constant (Fig. 3d). In turn, with casamino acids, the mRNA levels decreased from 12 to 24 h, and then low levels were maintained. The activity followed the same pattern as the RNA levels, as it was approximately 3 times higher compared to that with glucose (Fig. 4d). These findings agree with those obtained in Escherichia coli, where it was demonstrated that the transcription of the pck gene coding for PEP carboxykinase increased in the stationary phase and was subjected to catabolic repression (Goldie and Medina 1990). Recently, in a study on carbon flux in E. coli, the enzymatic activity of phosphoenolpyruvate carboxykinase was calculated during growth in glucose and was found to apparently maintain the balance between the PEP and OXA reservoirs (Yang et al. 2003). In C. glutamicum, the enzymatic activity of this enzyme is present when grown in glucose (Jetten and Sinskey 1993) and is responsible for recycling approximately two thirds of the anaplerotically synthesized oxaloacetate to PEP, causing a cycling between carboxylating and decarboxylating reactions (Petersen et al. 2001). Studies in Mycobacterium tuberculosis have shown that phosphoenolpyruvate carboxykinase operates in an anaplerotic or gluconeogenic capacity depending on the growth conditions (Machová et al. 2014). Likewise, this enzyme has been identified as gluconeogenic in S. coelicolor (Van Keulen et al. 2011); however, as gene transcription and enzymatic activity occur in the presence of glucose, its function may be somewhat anaplerotic, thus generating PEP for amino acids synthesis.
It is important to mention that we have cloned the genes corresponding to PEP carboxykinase (SCO4979), pyruvate carboxylase (SCO0546), and PEP carboxylase (SCO3127) and expressed in Escherichia coli. The recombinant purified proteins had the respective activity (Manuscript in preparation).
NAD+- and NADP+-dependent malic enzymes (SCO2951 and SCO5261)
NAD+- or NADP+-dependent malic enzymes catalyze the oxidative decarboxylation of malate to form pyruvate with the concomitant formation of NADH or NADPH as reductive equivalents; additionally, their function in the synthesis of triacylglycerols has been established in S. coelicolor (Rodriguez et al. 2012).
The mRNA levels of SCO5261 (NADP+-dependent malic enzyme) in S. coelicolor grown in glucose increased from 12 h, reached a maximum at 36 h, and subsequently declined. The maximum concentration was 0.02 ng μg−1, one of the highest amounts found with this carbon source. The increase in mRNA level matched the stages of increased growth in glucose; therefore, its expression could be associated with growth (Fig. 2a; Fig. 3e). In turn, the specific activity of this enzyme remained almost constant during the first 36 h before subsequently declining (Fig. 3e). Similar to the other genes, the mRNA levels decreased from 12 to 24 h with casamino acids; transcription levels then increased, reached a maximum at 48 h, and subsequently declined. The specific activity under this condition decreased from 12 to 48 h, increased at 60 h, and then remained steady until the end of the incubation period (Fig. 4e).
When grown with glucose as a carbon source, the RNA expression of SCO2951 increased from 12 h and reached a maximum at 36 h. On the other hand, the specific activity increased from 12 to 36 h and then declined similar to RNA (Fig. 3f). The mRNA levels with casamino acids were similar to those obtained with glucose, but the profile differed, as levels decreased from 12 to 24 h, increased slightly at 36 h, and then declined near the end of fermentation (Fig. 4f). The specific activity with casamino acids followed the same profile as RNA during the first 48 h and then increased until 60 h. The levels with casamino acids were approximately 5-fold higher than that obtained with glucose while the RNA levels remained practically the same, suggesting that sugar may induce posttranslational modifications of the enzyme, therefore lessening its activity. Although NADP+-ME activity in both conditions was lower than that of NAD+-ME, both enzymes showed higher activity when grown in casamino acids than in glucose (Figs. 3 and 4f).
In C. glutamicum, it has been proposed that only the NADP+-dependent malic enzyme could be physiologically involved in supplying NADPH when the microorganism was grown with gluconeogenic substrates (Gourdon et al. 2000). However, with casamino acids, these enzymes have a gluconeogenic function, since they allow the flow of carbon from the TCA cycle to produce glucose. On the other hand, in S. coelicolor, metabolic studies have shown that malic enzymes play an important anaplerotic role to maintain the connection between the TCA cycle and central metabolic pathways under glycolytic conditions (Rodriguez et al. 2012); however, our results showed that SCO5261 was also transcribed at high levels during active growth of the microorganism, indicating that its activity was important for replenishing the pyruvate used as a precursor in the synthesis of amino acids (Figs. 3 and 4e, f).
Genes corresponding to phosphoenolpyruvate-utilizing enzyme and pyruvate phosphate dikinase (SCO5896 and SCO0208)
PEP-UE (encoded by SCO5896) and PYPDK (encoded by SCO0208) catalyze the phosphorylation of pyruvate to generate phosphoenolpyruvate (KEGG (Thomas et al. 2012). In this work, only the transcript levels of these genes were determined. As seen in Fig. 5a, the mRNA levels corresponding to SCO5896 in S. coelicolor grown in glucose were low but constant in the first 36 h and then increased approximately 5-fold until 72 h. The mRNA levels in S. coelicolor grown in casamino acids were much lower compared with glucose; therefore, phosphoenolpyruvate-utilizing enzyme could be considered as an anaplerotic enzyme.
The mRNA expression profile of PEP-pyruvate-oxaloacetate node genes at different growth times in S. coelicolor. RNAs obtained from cells grown in MM supplemented with 1% glucose (black triangle) or 1% casamino acids (white triangle) were used for RT-qPCR analysis. Transcript levels of the genes SCO5896 (a) and SCO0208 (b) were determined as described in the “Materials and methods” section. a SCO5896 (PEP-utilizing enzyme); b SCO0208 (pyruvate phosphate dikinase)
Similar mRNA levels of SCO0208 were found in glucose and casamino acids, but the levels in the former tended to increase towards the stationary phase. In turn, the mRNA levels with casamino acids reached their maximum at 36 h and then declined (Fig. 5b). These mRNA levels suggest that the encoded enzyme, PYPDK, would perform an anaplerotic and gluconeogenic function depending on the carbon source used for the growth of S. coelicolor.
According to the results, in the presence of glucose, all genes of the PEP-PYR-OXA node in S. coelicolor were expressed at various levels, with differences ranging from 10- to 100-fold. SCO3127 (PEP carboxylase) showed the highest expression (10−2) in the rapid growth phase, followed by SCO5261 (NADP+-dependent malic enzyme) and SCO2014 (pyruvate kinase 1). The mRNA levels corresponding to SCO5896, SCO4979, and SCO0546 increased 10-fold towards the stationary phase (from 10−3 to 10−2). These results suggest that PEP carboxylase is a very important enzyme during glucose catabolism and could be considered glycolytic along with pyruvate kinase and the NADP+-dependent malic enzyme. Additionally, the derepression of SCO4979, SCO5896, and SCO0546 could increase their expression when sugar levels are low or when microorganism growth ceases, and these enzymes could be considered anaplerotic or gluconeogenic.
The activities of the enzymes monitored in this study were generally higher when grown in casamino acids compared with glucose, suggesting that the carbohydrate serves as a negative control through posttranslational modifications, as described for Streptomyces griseus and Streptomyces roseosporus (Ishigaki et al. 2017; Liao et al. 2014). In C. glutamicum grown in the presence of glucose, the simultaneous enzymatic activities of PEP carboxylase and pyruvate carboxylase were also detected, where the generated oxaloacetate (10% produced by PEP carboxylase and 90% by pyruvate carboxylase) exceeded the amount necessary for biosynthetic reactions by 3-fold; therefore, PEP carboxykinase recycled the excess to phosphoenolpyruvate (Petersen et al. 2000). It has been proposed that this cycle could serve to consume energy, providing the organism with flexibility in balancing the reactions that replenish intermediates with catabolic reactions, or that the joint activity of these enzymes would allow the organism to maintain intracellular oxaloacetate homeostasis (Petersen et al. 2001). In S. coelicolor, only PEP carboxylase activity was observed in the first 36 h of growth, and the greater contribution of oxaloacetate arose from the activity of this enzyme during the faster growth stage of this organism in glucose. The activity level detected for PEPCK during the stationary phase would allow a certain amount of oxaloacetate to be recycled into pyruvate.
Simultaneous PEPC and PEPCK activity has been observed in E. coli. PEPC is the only anaplerotic enzyme observed when this bacterium is grown in glucose (Yang et al. 2003); however, because PEP carboxykinase activity is modulated by changes in the concentrations of PEP and oxaloacetate rather than the ATP/ADP ratio, PEP carboxykinase is likely involved in maintaining a balance between the reservoirs of these intermediates. In addition, PEP carboxykinase drains excess carbon from the TCA cycle (Yang et al. 2003). In S. coelicolor, the activity levels of malic enzymes in glucose suggest that these enzymes could fulfill the function to drain excess carbon from the TCA cycle.
In the stationary phase of S. coelicolor in the presence of glucose, the expression and activity levels of PEPCK and PYC increased. This generates a PEP-PYR-OXA cycle that allows the flow of carbon out of the TCA cycle as well as produces pyruvate, which is a precursor of the antibiotic synthesis that is expected in this growth phase (Novotna et al. 2003; Schniete et al. 2018).
When grown in casamino acids, S. coelicolor shows a clear increase in the enzymatic activity of the PEP-PYR-OXA node, which is associated with growth phases (12 h and 36–60 h). Under gluconeogenic conditions, carbon flux from the amino acids of the TCA cycle towards glucose formation is expected. Our results show two possible paths of the final TCA cycle intermediates for gluconeogenesis: a primary path through the malic enzymes that generates PYR and a secondary path through PEPCK that generates PEP. In addition, the activity of PYK, PYC, and PEPC would balance the pyruvate concentration during acetyl CoA and OXA formation to maintain the activity of the TCA cycle. In the stationary phase, the TCA cycle is inactive, so the increase in the activity of these enzymes could be related to an anaplerotic function, thus producing pyruvate for the synthesis of antibiotics.
The discrepancies observed in gene transcript levels and enzymatic activities in S. coelicolor grown in glucose or casamino acids suggest that all enzymes synthesized in the presence of sugar may have undergone posttranslational modifications that decreased their activities, while the enzymatic activities remained unaffected during growth in casamino acids (Schilling et al. 2015).
In this paper, the expression and specific activity levels of genes and enzymes involved in the phosphoenolpyruvate-pyruvate-oxaloacetate node are presented, demonstrating the importance of some of these enzymes in the carbon metabolism of Streptomyces coelicolor as well as suggesting potential sites for genetic improvement for antibiotic production.
References
Bentley SD, Chater KF, Cerdeño-Tárraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Bramwell H, Nimmo HG, Hunter IS, Coggins JR (1993) Phosphoenolpyruvate carboxylase from Streptomyces coelicolor A3(2): purification of the enzyme, cloning of the ppc gene and over-expression of the protein in a streptomycete. Biochem J 293(Pt 1):131–136
Brewster NK, Val DL, Walker ME, Wallace JC (1994) Regulation of pyruvate carboxylase isozyme (PYC1, PYC2) gene expression in Saccharomyces cerevisiae during fermentative and nonfermentative growth. Arch Biochem Biophys 311:62–71
Bustin SA (2004) Data analysis and interpretation. In: Tsigelny IF (ed) A-Z of quantitative PCR. International University Line, La Jolla, pp 451–457
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622
Flores ME, Sánchez S (1985) Nitrogen regulation of erythromycin formation in Streptomyces erythreus. FEMS Microbiol Lett 26:191–194
Goldie H, Medina V (1990) Physical and genetic analysis of the phosphoenolpyruvate carboxykinase (pckA) locus from Escherichia coli K12. Mol Gen Genet 220:191–196
Gourdon P, Baucher MF, Lindley ND, Guyonvarch A (2000) Cloning of the malic enzyme gene from Corynebacterium glutamicum and role of the enzyme in lactate metabolism. Appl Environ Microbiol 66:2981–2987
Hiltner JK, Hunter IS, Hoskisson PA (2015) Tailoring specialized metabolite production in streptomyces. Adv Appl Microbiol 91:237–255
Ishigaki Y, Akanuma G, Yoshida M, Horinouchi S, Kosono S, Ohnishi Y (2017) Protein acetylation involved in streptomycin biosynthesis in Streptomyces griseus. J Proteome 155:63–72
Jeong Y, Kim JN, Kim MW, Bucca G, Cho S, Yoon YJ, Kim BG, Roe JH, Kim SC, Smith CP, Cho BK (2016) The dynamic transcriptional and translational landscape of the model antibiotic producer Streptomyces coelicolor A3(2). Nat Commun 7:11605
Jetten MSM, Sinskey AJ (1993) Characterization of phosphoenolpyruvate carboxykinase from Corynebacterium glutamicum. FEMS Microbiol Lett 111:183–188
Koffas MAG, Jung GY, Aon JC, Stephanopoulos G (2002) Effect of pyruvate carboxylase overexpression on the physiology of <em>Corynebacterium glutamicum</em>. Appl Environ Microbiol 68:5422
Lee HJ, Kim H-J, Seo J, Na YA, Lee J, Lee J-Y, Kim P (2013) Estimation of phosphoenolpyruvate carboxylation mediated by phosphoenolpyruvate carboxykinase (PCK) in engineered Escherichia coli having high ATP. Enzym Microb Technol 53:13–17
Li S, Wang W, Li X, Fan K, Yang K (2015) Genome-wide identification and characterization of reference genes with different transcript abundances for Streptomyces coelicolor. Sci Rep 5:15840
Liao G, Xie L, Li X, Cheng Z, Xie J (2014) Unexpected extensive lysine acetylation in the trump-card antibiotic producer Streptomyces roseosporus revealed by proteome-wide profiling. J Proteome 106:260–269
Machová I, Snašel J, Zimmermann M, Laubitz D, Plocinski P, Oehlmann W, Singh M, Dostál J, Sauer U, Pichová I (2014) Mycobacterium tuberculosis phosphoenolpyruvate carboxykinase is regulated by redox mechanisms and interaction with thioredoxin. J Biol Chem 289:13066–13078
Menéndez J, Gancedo C (1998) Regulatory regions in the promoters of the Saccharomyces cerevisiae PYC1 and PYC2 genes encoding isoenzymes of pyruvate carboxylase. FEMS Microbiol Lett 164:345–352
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428
Novotna J, Vohradsky J, Berndt P, Gramajo H, Langen H, Li XM, Minas W, Orsaria L, Roeder D, Thompson CJ (2003) Proteomic studies of diauxic lag in the differentiating prokaryote Streptomyces coelicolor reveal a regulatory network of stress-induced proteins and central metabolic enzymes. Mol Microbiol 48:1289–1303
Petersen S, de Graaf AA, Eggeling L, Möllney M, Wiechert W, Sahm H (2000) In vivo quantification of parallel and bidirectional fluxes in the anaplerosis of Corynebacterium glutamicum. J Biol Chem 275:35932–35941
Petersen S, Mack C, De Graaf AA, Riedel C, Eikmanns BJ, Sahm H (2001) Metabolic consequences of altered phosphoenolpyruvatecarboxykinase activity in Corynebacterium glutamicum reveal anaplerotic regulation mechanisms in vivo. Metab Eng 3:344–361
Peters-Wendisch PG, Kreutzer C, Kalinowski J, Pátek M, Sahm H, Eikmanns BJ (1998) Pyruvate carboxylase from Corynebacterium glutamicum: characterization, expression and inactivation of the pyc gene. Microbiology 144:915–927
Rodriguez E, Navone L, Casati P, Gramajo H (2012) Impact of malic enzymes on antibiotic and triacylglycerol production in Streptomyces coelicolor. Appl Environ Microbiol 78:4571–4579
Sauer U, Eikmanns BJ (2005) The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol Rev 29:765–794
Schilling B, Christensen D, Davis R, Sahu AK, Hu LI, Walker-Peddakotla A, Sorensen DJ, Zemaitaitis B, Gibson BW, Wolfe AJ (2015) Protein acetylation dynamics in response to carbon overflow in Escherichia coli. Mol Microbiol 98:847–863
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108
Schniete JK, Cruz-Morales P, Selem-Mojica N, Fernández-Martínez LT, Hunter IS, Barona-Gómez F, Hoskisson PA (2018) Expanding primary metabolism helps generate the metabolic robustness to facilitate antibiotic biosynthesis in. MBio 9
Spížek J, Sigler K, Řezanka T, Demain A (2016) Biogenesis of antibiotics-viewing its history and glimpses of the future. Folia Microbiol (Praha) 61:347–358
Takahashi-Íñiguez T, Barrios-Hernández J, Rodríguez-Maldonado M, Flores ME (2018) Tricarboxylic acid cycle without malate dehydrogenase in Streptomyces coelicolor M-145. Arch Microbiol
Taylor BL, Routman S, Utter MF (1975) The control of the synthesis of pyruvate carboxylase in Pseudomonas citronellolis. Evience from double labeling studies. J Biol Chem 250:2376–2382
Thomas L, Hodgson DA, Wentzel A, Nieselt K, Ellingsen TE, Moore J, Morrissey ER, Legaie R, Wohlleben W, Rodríguez-García A, Martín JF, Burroughs NJ, Wellington EMH, Smith MCM (2012) Metabolic switches and adaptations deduced from the proteomes of Streptomyces coelicolor wild type and phoP mutant grown in batch culture. Mol Cell Proteomics 11:M111.013797
Valle F, Muñoz E, Ponce E, Flores N, Bolivar F (1996) Basic and applied aspects of metabolic diversity: the phosphoenolpyruvate node. J Ind Microbiol 17:458–462
Van Keulen GS, Dijkhuizen J, Lubbert (2011) Central carbon metabolic pathways in Streptomyces. In: P D (ed) Streptomyces: molecular biology and biotechnology. Caister Academic Press, Norfolk, pp 105–124
Wada M, Sawada K, Ogura K, Shimono Y, Hagiwara T, Sugimoto M, Onuki A, Yokota A (2016) Effects of phosphoenolpyruvate carboxylase desensitization on glutamic acid production in Corynebacterium glutamicum ATCC 13032. J Biosci Bioeng 121:172–177
Weatherburn MW (1967) Phenol-hypochlorite reaction for determination of ammonia. Anal Chem 39:971–974
Yang C, Hua Q, Baba T, Mori H, Shimizu K (2003) Analysis of Escherichia coli anaplerotic metabolism and its regulation mechanisms from the metabolic responses to altered dilution rates and phosphoenolpyruvate carboxykinase knockout. Biotechnol Bioeng 84:129–144
Acknowledgments
We thank Omar Rangel y Martha Cariño for technical support. This work was supported by Grant PAPIIT IN214116 (DGAPA-UNAM) and by CONACyT through scholarship 450646 (RLR).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Llamas-Ramírez, R., Takahashi-Iñiguez, T. & Flores, M.E. The phosphoenolpyruvate-pyruvate-oxaloacetate node genes and enzymes in Streptomyces coelicolor M-145. Int Microbiol 23, 429–439 (2020). https://doi.org/10.1007/s10123-019-00116-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10123-019-00116-x