Abstract
Extreme environments are the main source of industrially suitable biocatalysts. The non-cultivable approach of searching enzymes is known to provide ample scope to accomplish novelty for their industrial applications. Lip479 clone out of seven lipase-producing clones obtained from Taptapani hot spring was found to be optimally active at pH 8.0 and temperature 65 °C. The recombinant Lip479 was highly stable in organic solvents, methanol, DMF, DMSO, acetone, and dichloromethane. Lip479 lipase activity was enhanced in the presence of K+, Mn2+, Na+, Zn2+, and Ca2+ except for Fe3+. The ability of Lip479 lipase to act on long carbon chain of 4-nitrophenyl myristate suggests it might be a true lipase. Lip479 clone was found to have ORF of 1251 bp encoding 416 amino acid residues of 42.57 KDa size (theoretically calculated). The presence of conserved motif Ala-His-Ser-Gln-Gly and Zn2+-binding consensus sequence (GAAHAAKH) of the clone assigns the protein to lipase family 1.5. Phylogenetic lineage of the protein sequence of Lip479 was traced to family 1.5 as it was clubbed up with those of reported lipases of the same family. The above biochemical features indicated that Lip479 lipase can be a potential biocatalyst for its use in various industries.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Lipases (triacylglycerol acyl hydrolases, EC 3.1.1.3) are the second largest produced industrial enzymes which catalyze the hydrolysis of a wide range of lipid substrates. Their remarkable ability to catalyze hydrolysis reactions in aqueous as well as non-aqueous conditions make them very suitable for industry (Sharma and Kanwar 2014). Unique biochemical properties of thermostable lipases, such as broad substrate specificity, high stability under a wide range of pH, temperature, salt, detergents, and organic solvents, are advantageous in their potential application in biotechnology industries (Glogauer et al. 2011; Jeon et al. 2011; Biver and Vandenbol 2013).
Conventional pure culture methods have limited scope for exploration of novel biomolecules from their respective ecosystems because almost 99% of the microorganisms present in any ecological niche are not cultivable. To overcome the limitations of declining diversity during in vitro cultivation, metagenomics has become the choice of approach for exploring microbial community over the last few decades (Duan and Feng 2010). It is a technique in which direct extraction and cloning of DNA of the accumulated microorganisms from any habitat are performed bypassing in vitro culture in an artificial medium (Glogauer et al.2011; Khan et al. 2013). Metagenomic approach, lipases have been successfully discovered from metagenomic DNA libraries of different sources viz, hot spring (Tirawongsaroj et al. 2008; López-López et al. 2015; Yan et al. 2017), marine sediment (Lee et al. 2006; Hårdeman and Sjöling 2007), soil sample (Lee et al. 2004; Lee et al. 2010), and animal gut (Liu et al. 2009; Bayer et al. 2010). However, industrially desirable biochemical properties of lipases obtained from thermophilic environments are mostly preferred due to their stability in high temperature, alkaline, acidic, saline, and other harsh environments.
Taptapani is one among several hot springs of Odisha, India, which is situated on the eastern slope of eastern ghat at the crest of the hill within the green forest having a temperature (48 ± 5 °C), pH (8.63 ± 0.21) and location (Latitude 19° 30′ 16.8″ N, Longitude 84° 24′ 4.6″ E). This tropical sulfur hot spring so far has not been given much attention to explore and understand the bacterial diversity, screening, and functional characterization of isolates/clones/genes responsible for the production of enzymes with special reference to lipase. In this study, seven clones were screened from the metagenomic library prepared from the Taptapani sediment, a tropical hot spring of Odisha, India, following non-cultivable approach. Lip479 showed maximum activity out of the seven isolates. Further, enzymatic characterization of this recombinant lipase was performed which includes substrate specificity, stability at high temperature, and pH, as well as effect of salts, solvents, chelating agents, and detergents so as to find its suitability to harsh environmental conditions found in industrial fermentation processes.
Materials and methods
Materials and strains
The substrates 4-nitrophenyl esters were purchased from Sigma, USA. Microbiological media and chemicals (analytical grade) were purchased from Himedia, India. Molecular biology chemicals, vector, and GeneJet extraction kit were purchased from Thermo Scientific, India.
Isolation of metagenomic DNA
The sampling was done from hotspring sediment in triplicate from Taptapani hot spring. The temperature and pH were measured to be 50 °C and 8.5 respectively. Metagenomic DNA was extracted as described by Sahoo (2016). Sediment sample (5 g) was mixed with 10-ml lysis buffer [100-mM Tris–HCl (pH 8.0), 10-mM sodium EDTA (pH 8.0), and 100 μl of lysozyme (100 mg/ml)] by horizontal shaking at 200 rpm for overnight at 37 °C. 100 μl of proteinase K (10 mg/ml) was added and incubated at 37 °C for 1 h. After incubation, 1 ml of 10% (w/v) SDS was added and incubated at 37 °C for 1 h with gentle endeavor-end inversions every 15–20 min. Then, the sample was again incubated at 90 °C for 1 h with gentle endeavor-end inversion every 15 min, and extraction was done using 25:24:1, v/v/v ratio of phenol: chloroform: isoamyl alcohol. The aqueous layer was collected after centrifugation at 10,000 g for 10 min at 25 °C. The aqueous layer was precipitated with 0.3 M sodium acetate and two volumes of chilled ethanol at − 20 °C for 1 h. After centrifugation at 10,000 g for 10 min at 25 °C, the pellet was washed with cold 70% ethanol and resuspended in sterile deionized water. DNA was visualized by 0.7% agarose gel electrophoresis (Sahoo 2016) .
Construction of metagenomic library and sequence analysis
High molecular weight metagenomic DNA (5 μg) was partially restricted with Sau 3A1 restriction enzyme. After partial digestion, GeneJET gel extraction kit was used to recover 3–8 Kb DNA fragments from the agarose gel (Thermo Scientific, India). The purified digested DNA fragments were incubated with pUC19/BamH1 vector and T4 DNA ligase at 16 °C for overnight. The ligated products were transformed into electro-competent E.coli DH5α by using Micropulser (BIO-RAD, USA). Libraries were screened for lipolytic activity, by spreading the transformants on Luria-Bertani agar (LB) in plate with ampicillin (50 μg/ml) as well as Rhodamine B (0.01% (w/v) and olive oil (1% (v/v). After 48-h incubation at 37 °C, the lipase positive clones were identified by halo orange zone on exposure with ultraviolet light. The positive lipolytic clones were sequenced (Xcelris Pvt. Ltd. Ahmadabad, India) and sequence homology was carried out with BLASTP program. Open reading frame and signal peptides were predicted employing ORF Finder tool (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and SignalP 4.1 programs respectively. The nucleotide sequence was deposited in the NCBI database, and the accession number was KR232658. Phylogenetic tree of the lipase protein was constructed using by neighbour-joining method with the 1000 bootstrap replicates using Molecular Evolutionary Genetics Analysis (MEGA 6.0) (Sahoo et al. 2014). Multiple sequence alignment was carried out using Geneious prime version 2019.0.4 software (https://www.geneious.com) by aligning of lip479 with other lipase of family 1.5.
Lipolytic activity assay
The lipase producing metagenomic clone was incubated overnight at temperature 37 °C in ampicillin supplemented Luria broth. Two milliliter of overnight-incubated lipase positive clone was harvested by centrifugation and pellets were suspended in nuclease-free water. The cell suspension was lysed by sonication at 20 KHz (VCX500, Sonics, USA) on ice in a 1.5-ml centrifuge tube for 5 to 10 min in 10 s interval. After sonication, the lysate solution was centrifuged for 10 min at 10,000 g at 25 °C. After centrifugation, the cell extract was used as a crude lipase. The activity of crude lipase was measured by quantifying the amount of 4-nitrophenol at 405 nm released at 15 min of incubation (Sahoo et al.2014). Quantity of lipase which has generated 1 μM of 4-nitrophenol in 1 min represent one international unit (IU) for its activity.
Biochemical characterization of lipase clone
The optimum pH of crude lipase was determined using 4-nitrophenyl laurate as substrate at 50 °C for incubation temperature. The different buffers (0.1 M) used for determination of optimum pH were acetate buffer (for pH 4.0 and 5.0), sodium phosphate buffer (for pH 6.0 and 7.0), Tris Buffer (for pH 8.0 and 9.0), and glycine-NaOH buffer (for pH 10.0). pH stability of the crude lipase was measured after incubating the lipase for 0–6 h at different pH from 6.0 to 10.0. In order to determine the optimum condition for temperature, the lipase activity was quantified at temperature from 30 to 90 °C keeping the pH at optimum. Incubation was also carried out for several time interval varying from 0 to 6 h to find out the stability at different temperatures ranging between 55 and 95 °C.
Influence of metal ions such as, CaCl2, CoCl2, HgCl2, CuSO4, NaCl, ZnCl2, MgCl2, FeCl2, MnCl2, KCl, AgNO3, and Al2(SO4)3 on lipase activity was determined by pre-incubating the enzyme in presence of metal (each at 10 mM) for 1 h. Influence of organic solvents; dimethyl sulfoxide, dimethylformamide, n-hexane, dichloromethane, chloroform, ethyl ether, ethanol xylene, acetone, glycerol, isoamyl alcohol, isopropanol, methanol, butanol, ethyl acetate, and benzene at concentration of 25% (v/v) were tested after pre-incubating enzyme solution for 1 h. Similarly, the effect of detergents CTAB, Triton X-100, Tween-20, and SDS (each at 1 % w/v) and other chemical reagents phenyl methyl sulfonyl fluoride (PMSF), and ethylene diamine tetra acetic acid (EDTA) at 10-mM concentration on lipase activity was also determined.
Determination of substrate specificity
The substrate specificity of crude lipase was found out against 4-nitrophenyl esters with variable chain length: C2 (acetate), C4 (butyrate), C8 (caprylate), C12 (laurate), C14 (myristate), C16 (palmitate), and C18 (stearate) (each at 1 mM) as substrate. All the measurements were carried out in triplicate at optimum condition of pH and temperature.
Results and discussion
Functional screening of metagenomic library for lipase
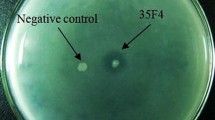
The extracted metagenomic DNA from Taptapani hot spring sediment was utilized for metagenomic library construction employing E.coli DH5α host and pUC19 vector. The library consisting of ~ 13,000 clones were screened for lipase by exhibiting fluorescence illumination and zone formation in rhodamine B and tributyrin supplemented plates respectively (Fig. S1). Out of ~ 13,000 clones, only seven clones showed clear zone on tributyrin as well as fluorescence on rhodamine plates. These lipase producing clones were further analysed in BHB medium supplemented with olive oil. pUC-lip-479 clone exhibited highest lipase activity out of the 7 analysed clones which were further processed for characterization. Previously, Zheng et al. (2013) reported only one functional lipase clone out of 20,000 transformants, and Khan et al. (2013) obtained two lipase positive clones out of 2000 transformants and established their novelty. In comparison to conventional methods used previously, this method is more advantageous for discovering novel functional genes from natural resources although their number is small (Ferrer et al. 2005).
Sequence analysis of the pUC-lip-479 clone insert DNA of 1251 bps length ORF encoding a polypeptide of 416 amino acid residues with a conserved motif of Ala-His-Ser-Gln-Gly sequence (Arpigny and Jaeger 1999) and a Zn2+-binding consensus sequence (GAAHAAKH) (Fig.S2). The size of the protein was found to be 42.57 kDa (theoretically calculated) and the number of amino acids was 387 (Fig.S2). Based on the conserved motif and Zn-binding consensus sequence, it could be assigned to family 1.5 of microbial lipase/esterase classification system (Arpigny and Jaeger 1999; Chakravorty et al. 2011). Blast analysis showed that Lip479 shares 95% amino acid similarity with lipase from uncultured bacterium (AEL99899). Phylogenetic analysis and multiple sequence alignment of amino acid sequence of Lip479 and those of previously reported lipase families showed that Lip479 could be a member of the family 1.5, as it was found to be clubbed up with protein sequence of other lipases belonging to family 1.5 (Fig. 1a, b). These lipases belong to alkalo- and thermophilic Geobacillus species, and they usually exhibit maximum activity at 65 °C and pH 9.0. They also have a molecular mass of about 45 kDa. (Arpigny and Jaeger 1999).
a Phylogenetic analysis of Lip479, its homologues and other lipases. Phylogenetic relationship of Lip479 and lipase proteins of six different sub family of family 1 was performed using the program MEGA 5.1. Except for Lip479, the other protein sequences for previously identified families of bacterial lipolytic and esterolytic enzymes were retrieved from gene bank. The numbers at node indicate the bootstrap percentages of 1000 resampling(Source: Sahoo 2016). b Multiple sequence alignment of Lip479 with other lipases. Zinc-binding site and conserved motif were marked in blue and red colour outline respectively
Biochemical characterization of metagenomic-derived lipase
Effect of pH and stability study
The enzyme assay using different parameters like pH, temperature, organic solvent, and metal ions was performed to understand their effect on lipase activity. Highest lipase activity by Lip479 clone was observed at pH 8.0 in 0.1 M Tris-Cl buffer with 4-nitrophenol laurate as substrate (Fig. 2a) which was found to be matching with lipase activity reported earlier from other metagenomes (Couto et al. 2010; Fan et al. 2011; Zheng et al. 2013). Lipase from metagenomic library of Galicia hot spring showed optimum activity at pH 7.5 (López-López et al. 2015). The clone retained 100% relative activity at incubation period of 30 min, pH 8.0, and 50 °C temperature. At pH 7.0, 9.0, 10.0, and 11.0, activity was found to be 99, 99, 95, and 84% respectively which got declined with the incubation period up to 6 h (Fig. S3). With the increase in pH from pH 7.0–11.0, lipase activity was found to be reduced from 87 to 1.4%. A similar trend of stability at a wide range of alkaline pH was also observed with lipase from soil metagenome (Khan et al. 2013).
Effect of temperature
Lip479 showed highest activity at temperature 65 °C which continuously decreased at 75 °C and 85 °C (Fig. 2b). In contrast to Lip479 lipase, lipase of SMlipD and SMlipB were reported to be active only at 50 °C and 30 °C (Khan et al. 2013). Similarly, Biver and Vandenbol (2013) isolated lipase clones SBlip5.1 and SBLip2 from forest metagenome having optimum activity at 50 °C and 45 °C respectively. As compared with lipase from other hot spring metagenome, lip479 showed optimum activity at higher temperature (López-López et al. 2015; Yan et al. 2017). Thermal stability analysis showed that Lip479 lipase was stable at 65 °C, and it retained 101.46%, 95.12%, and 92.49% relative enzyme activity up to 30 min, 3 h, and 6 h respectively at this temperature (Fig. S4). On the other hand, at 75, 85, and 95 °C, it showed 98, 97, and 91% activity only up to 30 min of incubation which sharply decreased to 60, 30, and 1.3% after 6 h incubation. The stability exhibited by present lipase clone Lip479 was found to be higher than the previous ones published from different metagenomic libraries (Fan et al. 2011; Khan et al. 2013). This feature of exhibiting stability at higher temperature (65 °C) for longer incubation period proves its prospective industrial utility.
Substrate specificity and activity
To find the substrate utilization potential of lipase, fatty acids of different chain lengths (C2-C18) were used as the substrate. Lip479 lipase hydrolyzed a broad range of substrates preferentially between chain lengths varying from C2 to C18 attached with 4-nitrophenyl ester. The maximum activity of lipase was achieved with 4-nitrophenyl myristate (C14) followed by 4-nitrophenyl palmitate (C16), 4-nitrophenyl laurate (C12), and 4-nitrophenyl stearate (C18) (Fig. 3). Least lipase activity was displayed with 4-nitrophenyl butyrate (C4). Ability to hydrolyse long chain carbon 4-nitrophenyl esters indicated that Lip479 clone was producing lipase which is closely similar to true lipases (Jaeger et al. 1999; Glogauer et al. 2011).
Effect of metal ions, chelating agents, and detergents
Metal ions are known for enhancing the lipases activity as well as its stability (Ahmed et al. 2010). To find out the effect of metal ions on lipase produced by Lip479, the influence of following metal ions Na+, Al3,+ Co2+, Cu2+, Mg2+, Mn2+, K+, Zn2+, Fe3+, and Ca2+ (each at 10 mM) were tested and summarized in Table 1. Lip479 lipase activity was enhanced in the presence of K+ (127.13%) and Mn2+ (115.85%), followed by Na+ (105.39%), Zn2+ (102.27%), and Ca2+ (101.82%). The activity of crude lipase in presence of all metal ions were found to be statistically significant (p < 0.05) with respect to activity in absence of metal ions (control). Many of the lipases were active when Ca2+ and Mg 2+ were supplemented (Jinwal et al. 2003; Khan and Jithesh 2012; Zheng et al. 2013; Bose and Keharia 2013). Rahman et al. (2005) advocated the binding of Ca2+ to the active site of the lipases which results in conformational change in the protein. Bose and Keharia (2013) also reported that the stability of the enzyme could be due to formation of cross-linked bridge amongst Ca2+/Mg2+ and carboxylate side chain group of enzyme polypeptides. More than 65.11% of lipase activity was retained when Mg2+ was present but the enzyme activity was decreased to 45.34, 21.24, 18.95, and 16.9% in presence of Co2+, Cu2+, Fe3+, and Al3+ respectively. The inhibitory effect of metal ions may be due to conformational changes in the protein (Rahman et al. 2005). Similarly, stable lipase activity in the presence Zn2+ may be accredited to the presence of metal-binding site responsible for protein thermostability (Chakravorty et al. 2011). In a response similar to lipase derived from Pseudomonas aeruginosa AAU2 (Bose and Keharia 2013), EDTA, metal chelating agent reduce the relative activity of the enzyme 60.09% (at 10 mM) (Table 1). Only 36.75% lipase activity could be retained (Table 1) at 10-mM concentration of PMSF. This is probably due to the inhibitory effect of PMSF which blocks the accession of the serine residue of lipase active site. In presence of non-ionic surfactants such as Triton X-100 and Tween 20, Lip479 lipase showed 2.48 and 1.63% relative activity, but it was reduced to 0.17%, in presence of ionic surfactants CTAB (cationic) and SDS (anionic) (each at 1% w/v) (Table 1). The expansion of the lipid-water interfacial area is boosted by both non-ionic and ionic surfactants which in turn enhances the rate of reaction of lipase (Chen et al. 2007). Non-ionic surfactant having low hydrophilic/lipophilic balance (HLB) values have a magnifying effect on the activity of lipase in comparison to ionic surfactant (SDS) with high HLB (Bose and Keharia 2013).
Effect of organic solvents
The stable nature of lipase enzyme in presence of organic solvents is considered to be a desirable trait for industrial production of cosmetics, chemical, pharmaceuticals, and biodiesel. (Salihu and Alam 2015). Hence, the stability of RK-Lip-479 in the presence of various organic solvents was studied. Synthesis of pharmaceuticals, chemicals, cosmetics, and biodiesel production requires lipases which are tolerant to methanol, ethanol, acetone, DMF, DMSO, and dichloromethane (Salihu and Alam 2015). Moreover stability of lipase in the presence methanol is critical to biodiesel production (Korman et al. 2013). Varying degrees of stability of the enzyme in different organic solvents were studied. For acetone, the activity was relatively highest (126.3 ± 5.5%) followed by DMF (122.34 ± 2.39%), DMSO (114.73 ± 3.08%), dichloromethane, and methanol (100.59 ± 3.74%) (each at 25% v/v) as shown in Table 2. Selvin et al. (2012) reported that lipase from marine sponge metagenome was stable in presence of acetone, DMSO, and methanol. Lipase from Pseudomonas aeruginosa LX1(Ji et al. 2010) showed stability in the presence of glycerol, DMF, and DMSO which was similar to our finding. Lip479 lipase was more than 60% stable in the presence of ethanol, glycerol, and isopropanol (each at 25% v/v) (Table 2). However, chloroform at 25% (v/v) was found to suppress the activity of the clone Lip479. The ability of lipase to catalyze reactions in non-aqueous environments has gained a lot of attention because of its numerous advantages like easy enzyme recovery and reusability without immobilization, elimination of the risk of microbial contamination, inhibition of side reactions dependent on water (Castilho et al. 2000). Logarithm of partition co-efficient of an organic solvent (log p) is usually considered an ideal parameter to co-relate enzyme activity with the organic solvent used (Aldercreutz and Mattiasson 1987). Higher log p values have been associated with better enzyme activity in non-aqueous environments (Aldercreutz and Mattiasson 1987). However, as our study shows (Table 2), we could find a clear linear relationship between log p values of organic solvents and enzyme activity only for the first three organic solvents glycerol, DMSO, and DMF which had log p values of − 1.76, − 1.35, and − 1.04 and enzyme activity 62.88 ± 3.28, 114.73 ± 3.08, and 122.34 ± 2.39 respectively. We did not find any clear relationship between the log p values and the enzyme activity for the remaining organic solvents. It has been suggested that other major determinants like hydrophobicity (Torres and Otero 1996), polarity index (Gupta et al. 2004), polarity of the solvent, and denaturation capacity (Khmelnitsky et al. 1988) determine the catalytic potential and stability of a biocatalyst in an organic medium. Many findings also state the inability to find a genuine correlation between lipase catalysis in organic solvents even after taking the abovementioned determinants into account (Castro and Knubovets 2003). Understanding how lipase catalyzes reactions in a non-aqueous environment a fascinating phenomenon which shall be a part of our future study.
Conclusion
The Lip479 lipase isolated from hot spring metagenome showed a remarkable difference in biochemical characteristics from other reported lipases isolated from various metagenomic libraries. This lipase was observed to be closely similar to a true lipase belonging to family 1.5. The unique features of Lip479 are its thermostability, alkali stability, and stability in the broad range of organic solvents and metal ions. Therefore, it can be successfully used in most of the industries after further characterization preferably where high-temperature industrial operations are performed.
References
Ahmed EH, Raghavendra T, Madamwar D (2010) A thermostable alkaline lipase from a local isolate Bacillus subtilis EH 37: characterization, partial purification, and application in organic synthesis. Appl Biochem Biotechnol 160:2102–2113. https://doi.org/10.1007/s12010-009-8751-4
Aldercreutz P, Mattiasson B (1987) Aspects of biocatalyst stability in organic solvents. Biocatal Biotransform 1:99–108
Arpigny JL, Jaeger KE (1999) Bacterial lipolytic enzymes: classification and properties. Biochem J 343 Pt 1:177–183. https://doi.org/10.1042/0264-6021:3430177
Bayer S, Kunert A, Ballschmiter M, Greiner-Stoeffele T (2010) Indication for a new lipolytic enzyme family: isolation and characterization of two esterases from a metagenomic library. J Mol Microbiol Biotechnol 18:181–187. https://doi.org/10.1159/000315459
Biver S, Vandenbol M (2013) Characterization of three new carboxylic ester hydrolases isolated by functional screening of a forest soil metagenomic library. J Ind Microbiol Biotechnol 40:191–200. https://doi.org/10.1007/s10295-012-1217-7
Bose A, Keharia H (2013) Production, characterization and applications of organic solvent tolerant lipase by Pseudomonas aeruginosa AAU2. Biocatal Agric Biotechnol 2:255–266. https://doi.org/10.1016/j.bcab.2013.03.009
Castilho LR, Polato CMS, Baruque EA, Sant’Anna GL Jr, Freire DMG (2000) Economic analysis of lipase production by Penicillium restrictum in solidstate and submerged fermentations. Biochem Eng J 4(3):239–247
Castro GR, Knubovets T (2003) Homogeneous biocatalysis in organic solvents and water-organic mixtures. Crit Rev Biotechnol 23(3):195–231
Chakravorty D, Parameswaran S, Dubey VK, Patra S (2011) In silico characterization of thermostable lipases. Extremophiles 15:89–103. https://doi.org/10.1007/s00792-010-0337-0
Chen S, Qian L, Shi B (2007) Purification and properties of enantioselective lipase from a newly isolated Bacillus cereus C71. Process Biochem 42:988–994. https://doi.org/10.1016/j.procbio.2007.03.010
Couto GH, Glogauer A, Faoro H, Chubatsu LS, Souza EM, Pedrosa FO (2010) Isolation of a novel lipase from a metagenomic library derived from mangrove sediment from the south Brazilian coast. Genet Mol Res 9:514–523. https://doi.org/10.4238/vol9-1gmr738
Duan C-J, Feng J-X (2010) Mining metagenomes for novel enzymes. Biotechnol Lett 32:1765–1775
Fan X, Liu X, Wang K, Wang S, Huang R, Liu Y (2011) Highly soluble expression and molecular characterization of an organic solvent-stable and thermotolerant lipase originating from the metagenome. J Mol Catal B Enzym 72:319–325. https://doi.org/10.1016/j.molcatb.2011.07.009
Ferrer M, Martínez-Abarca F, Golyshin PN (2005) Mining genomes and “metagenomes” for novel catalysts. Curr Opin Biotechnol 16:588–593. https://doi.org/10.1016/j.copbio.2005.09.001
Glogauer A, Martini VP, Faoro H, Couto GH, Müller-Santos M, Monteiro RA, Mitchell DA, de Souza EM, Pedrosa FO, Krieger N (2011) Identification and characterization of a new true lipase isolated through metagenomic approach. Microb Cell Factories 10:54. https://doi.org/10.1186/1475-2859-10-54
Gupta R, Gupta N, Rathi P (2004) Bacterial lipases: an overview of production, purification, and biotechnological properties. Appl Microbiol Biotechnol 64:763–781
Hårdeman F, Sjöling S (2007) Metagenomic approach for the isolation of a novel low-temperature active lipase from uncultured bacteria of marine sediment. FEMS Microbiol Ecol 59:524–534. https://doi.org/10.1111/j.1574-6941.2006.00206.x
Jaeger KE, Dijkstra BW, Reetz MT (1999) Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu Rev Microbiol 53:315–351. https://doi.org/10.1146/annurev.micro.53.1.315
Jeon JH, Kim SJ, Lee HS, Cha SS, Lee JH, Yoon SH, Koo BS, Lee CM, Choi SH, Lee SH, Kang SG, Lee JH (2011) Novel metagenome-derived carboxylesterase that hydrolyzes β-lactam antibiotics. Appl Environ Microbiol 77:7830–7836
Ji Q, Xiao S, He B, Liu X (2010) Purification and characterization of an organic solvent-tolerant lipase from Pseudomonas aeruginosa LX1 and its application for biodiesel production. J Mol Catal B Enzym 66:264–269. https://doi.org/10.1016/j.molcatb.2010.06.001
Jinwal UK, Roy U, Chowdhury AR, Bhaduri AP, Roy PK (2003) Purification and characterization of an alkaline lipase from a newly isolated Pseudomonas mendocina PK-12CS and chemoselective hydrolysis of the fatty acid ester. Bioorg Med Chem 11:1041–1046. https://doi.org/10.1016/S0968-0896(02)00516-3
Khan M, Jithesh K (2012) Expression and purification of organic solvent stable lipase from soil metagenomic library. World J Microbiol Biotechnol 28:2417–2424. https://doi.org/10.1007/s11274-012-1051-0
Khan M, Jithesh K, Mookambikay R (2013) Cloning and characterization of two functionally diverse lipases from soil metagenome. J Gen Appl Microbiol 31:21–31
Khmelnitsky YL, Levashov AV, Klyachko NL, Martinek K (1988) Engineering biocatalytic systems in organic media with low water content. Enzym Microb Technol 10(12):710–724
Korman TP, Sahachartsiri B, Charbonneau DM, Huang GL, Beauregard M, Bowie JU (2013) Dieselzymes: development of a stable and methanol tolerant lipase for biodiesel production by directed evolution. Biotechnol Biofuels 6:70. https://doi.org/10.1186/1754-6834-6-70
Lee SW, Won K, Lim HK, Kim JC, Choi GJ, Cho KY (2004) Screening for novel lipolytic enzymes from uncultured soil microorganisms. Appl Microbiol Biotechnol 65:720–726. https://doi.org/10.1007/s00253-004-1722-3
Lee MH, Lee CH, Oh TK, Song JK, Yoon JH (2006) Isolation and characterization of a novel lipase from a metagenomic library of tidal flat sediments: evidence for a new family of bacterial lipases. Appl Environ Microbiol 72:7406–7409. https://doi.org/10.1128/AEM.01157-06
Lee MH, Hong KS, Malhotra S, Park JH, Hwang EC, Choi HK, Kim YS, Tao W, Lee SW (2010) A new esterase EstD2 isolated from plant rhizosphere soil metagenome. Appl Microbiol Biotechnol 88:1125–1134. https://doi.org/10.1007/s00253-010-2729-6
Liu K, Wang J, Bu D, Zhao S, McSweeney C, Yu P, Li D (2009) Isolation and biochemical characterization of two lipases from a metagenomic library of China Holstein cow rumen. Biochem Biophys Res Commun 385:605–611. https://doi.org/10.1016/j.bbrc.2009.05.110
López-López O, Knapik K, Cerdán M-E, González-Siso M-I (2015) Metagenomics of an alkaline hot spring in Galicia (Spain): microbial diversity analysis and screening for novel lipolytic enzymes. Front Microbiol 6:1291
Rahman RNZRA, Baharum SN, Basri M, Salleh AB (2005) High-yield purification of an organic solvent-tolerant lipase from Pseudomonas sp. strain S5. Anal Biochem 341:267–274. https://doi.org/10.1016/j.ab.2005.03.006
Sahoo RK (2016) Pure culture and metagenomic approaches to investigate Taptapani hot spring for lipase gene, Dissertation, Siksha O Anusnadhan University, India
Sahoo RK, Subudhi E, Kumar M (2014) Quantitative approach to track lipase producing Pseudomonas sp. S1 in nonsterilized solid state fermentation. Lett Appl Microbiol 58:610–616. https://doi.org/10.1111/lam.12235
Salihu A, Alam MZ (2015) Solvent tolerant lipases: a review. Process Biochem 50:86–96. https://doi.org/10.1016/j.procbio.2014.10.019
Selvin J, Kennedy J, Lejon DPH, Kiran G, Dobson ADW (2012) Isolation identification and biochemical characterization of a novel halo-tolerant lipase from the metagenome of the marine sponge Haliclona simulans. Microb Cell Factories 11:72. https://doi.org/10.1186/1475-2859-11-72
Sharma S, Kanwar SS (2014) Organic solvent tolerant lipases and applications. Sci World J 2014:1–15. https://doi.org/10.1155/2014/625258
Tirawongsaroj P, Sriprang R, Harnpicharnchai P, Thongaram T, Champreda V, Tanapongpipat S, Pootanakit K, Eurwilaichitr L (2008) Novel thermophilic and thermostable lipolytic enzymes from a Thailand hot spring metagenomic library. J Biotechnol 133:42–49
Torres C, Otero C (1996) Influence of the organic solvents on the activity in water and the conformation of Candida rugosa lipase: description of a lipase activating pretreatment. Enzym Microb Technol 19(8):594–600
Yan W, Li F, Wang L, Zhu Y, Dong Z, Bai Z (2017) Discovery and characterizaton of a novel lipase with transesterification activity from hot spring metagenomic library. Biotechnol Rep 14:27–33
Zheng J, Liu C, Liu L, Jin Q (2013) Characterisation of a thermo-alkali-stable lipase from oil-contaminated soil using a metagenomic approach. Syst Appl Microbiol 36:197–204. https://doi.org/10.1016/j.syapm.2012.12.008
Funding
Present research work received the financial support from the DBT, New Delhi through project grants no: BT/PR7944/BCE/8/1036/2013).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they do not have any conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Fig. S1
Plate showing lipase positive clones. a LB agar containing olive oil and Rhodamine B, b LB agar containing tributyrin (PNG 1002 kb)
Fig. S2
Lip479 clone (NCBI Accession No. KR232658) of 1251 bp length which codes 416 amino acid residues. Signal peptide was marked in red colour outline. Zinc-binding site and conserved motif sequence were marked in brown and blue colour outline respectively. The gene sequence has ATG as start codon (green outline) and TAA as stop codon (black outline). Small alphabets represent nucleotides and block alphabets represent the amino acids (PNG 1772 kb)
Fig. S3
pH stability of Lip479. pH stability was investigated by incubating the crude enzyme at pH 7, 8, 9, 10, and 11 for a period ranging from 0 to 6 h (PNG 131 kb)
Fig. S4
Thermostability of Lip479. Thermostability was measured by incubating the crude enzyme at different temperatures 55, 65, 75, 85, and 95 °C for a period ranging from 0 to 6 h (PNG 137 kb)
Rights and permissions
About this article
Cite this article
Sahoo, R.K., Das, A., Sahoo, K. et al. Characterization of novel metagenomic–derived lipase from Indian hot spring. Int Microbiol 23, 233–240 (2020). https://doi.org/10.1007/s10123-019-00095-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10123-019-00095-z