Abstract
The disc diffusion test is used for antimicrobial susceptibility testing worldwide. In this study, the performance of both Bio-Rad® antibiotic discs (as compared with Oxoid® discs) and the ADAGIO™ automated system for the reading of disc diffusion test results was evaluated with American Type Culture Collection (ATCC) quality control (QC) and wild strains of bacteria. Inhibition zones of both disc brands were read manually and through use of the ADAGIO™ system. Categorized interpretation of the results for each strain and antibiotic combination was summarized according to the Clinical Laboratory Standards Institute MS-100 (2017 update) manual and ADAGIO™ readings. Eight ATCC QC strains and 120 different wild strains were evaluated, to give a total of 1226 antibiotic/bacteria combinations and 2486 manual readings. One major error and four minor errors (0.08% and 0.34%, respectively) were detected via manual readings of the Bio-Rad® discs as compared with the Oxoid® discs. For the same number of antibiotic/bacteria combinations, five minor errors and one major error (0.42% and 0.08%, respectively) were detected with the Bio-Rad® discs read by the ADAGIO™ system. In addition, the number of times the automatic reading needed manual edition with Bio-Rad® discs was statistically lower than it did with Oxoid® discs (3.7% vs. 5.7%, p < 0.05). These findings support the hypothesis that Bio-Rad discs are not inferior to Oxoid® discs, and the performance of the ADAGIO™ system is comparable to that of manual readings with both disc brands.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The disc diffusion test is currently used for antimicrobial susceptibility testing worldwide [1,2,3]. This method is simple, flexible, and inexpensive. In most laboratories, the results are read manually and require a significant amount of hands-on time. The main disadvantages of this manual, non-automated method are the lack of standardization and documentation of the readings, human and transcription errors, and lack of reagent traceability, which is mandatory in an era of certification and accreditation of clinical laboratories. In the last few years, full automation of the process has been suggested as a fair solution to the above-mentioned pitfalls [4]. Moreover, for those laboratories where full automation is still not an option, a camera-based system that could automatically read and analyze disc diffusion tests may be a useful tool for standardizing and documenting the results and batch numbers of the plates and antibiotics.
ADAGIO™ is an automated system built around data management software and an imaging device that measures the size of the inhibition zone around antibiotic discs. It is characterized by speed, accuracy, and reproducibility and, thereby, less hands-on time, no transcription errors, and standardized antimicrobial susceptibility test readings. Antibiotic discs and media batch numbers can be recorded for full traceability in every single test. The system automatically recognizes Bio-Rad® antibiotic discs (Bio-Rad, Marnes-la-Coquette, France) regardless of the position on the agar plate, leading to a significant improvement in the reading process and increased confidence in the interpretation of results. The other systems on the market do not automatically recognize the contents of the discs; therefore, the disc position template on the agar plate should be fixed to avoid a mismatch of the results [5,6,7,8,9,10,11].
ADAGIO™ offers a built-in expert system, based on readings that can detect potential errors and suggest edits to the results, which are based on the phenotypes of known antibiotic resistance mechanisms. The ADAGIO™ software also includes a powerful tool to monitor resistance trends and nosocomial infections.
As compared with manual reading, the use of ADAGIO™ with Bio-Rad® discs improves the reading process and increases confidence in the interpretation of results. Thus, in the first phase of this study, the performance of Bio-Rad® discs was compared to that of Oxoid® discs (Oxoid Ltd, Basingstoke, Hans, UK), which have been recognized as the superior brand in earlier studies [12].
In a previous study, the performance of discs from nine manufacturers (including Bio-Rad® and Oxoid®) was compared in 2014 and 2017 using the European Committee on Antimicrobial Susceptibility Testing (EUCAST) criteria [13]. That study showed that although a significant improvement was observed in the performance of Bio-Rad® discs between 2014 and 2017, it was still slightly inferior to that of Oxoid® discs in 2017 [13].
The aim of the first phase of the present study was to evaluate Bio-Rad® discs as compared with Oxoid® discs, by testing a larger number of strains using the Clinical Laboratory Standards Institute (CLSI) criteria. The Oxoid® discs and CLSI criteria are both routinely used in Israel and were therefore chosen as the gold standard for the present study.
In the second phase of the study, the performance of the ADAGIO™ system was evaluated with Bio-Rad® and Oxoid® discs against American Type Culture Collection (ATCC) quality control (QC) and wild strains and compared to manual reading. To the best of our knowledge, only one other study has compared the performance of the ADAGIO™ system with that of manual reading for fastidious bacteria alone. In this study, we considerably increased the number of bacterial strains evaluated [14].
Material and methods
Bio-Rad® and Oxoid® discs were evaluated against the following ATCC QC strains: Escherichia coli 25922, Klebsiella pneumoniae 700603, Pseudomonas aeruginosa 27853, Staphylococcus aureus 25923, Streptococcus pneumoniae 49619, Haemophilus influenzae 49247, and Haemophilus influenzae 49766.
All tests were performed according to the CLSI disc diffusion methodology [15]. Discs were tested on Mueller–Hinton agar (MHA), or MHA supplemented with 5% defibrinated horse blood (both from Hy Laboratories, Rehovot, Israel), or HTM (MHA supplemented with hemin and β-NAD from Novamed Ltd, Jerusalem, Israel), depending on the microorganism tested. Each combination of agent and QC strain was tested in triplicate. Discs used for the triplicate tests were consistently from the same lot and the same vial. All triplicate tests were performed on the same day using three individually prepared inoculum suspensions. For each agent, one Oxoid® disc and one Bio-Rad® disc were placed on the same 90 mm circular agar plate to minimize variations due to differences in inoculum size, media, and incubation conditions. Zone diameters were measured to the nearest millimeter with a caliper. The mean readings were calculated and reported for each strain and antibiotic combination, whether it fell within the range or not.
As compared with Oxoid® discs, the performance of Bio-Rad® discs was evaluated against wild strains of Enterobacteriaceae, P. aeruginosa, Acinetobacter spp., Shigella spp./Salmonella enterica, Haemophilus spp., Moraxella catarrhalis, Staphylococcus spp., Streptococcus spp., and Enterococcus spp. Zone diameters were measured twice by two technicians and the mean readings were calculated for each strain and antibiotic combination. Categorized interpretation of the results (as susceptible, intermediate, or resistant) was summarized for each strain/antibiotic combination according to the CLSI MS-100, 2017 update. Based on the ISO 20776–2 guideline, category agreement was established according to the following: very major error for false susceptible interpretation, major error for false resistant interpretation, and minor error for false categorization involving intermediate results.
In the second phase of the study, the ADAGIO™ system, used for the automatic reading of disc diffusion results, was evaluated and compared with manual reading. Both Bio-Rad® and Oxoid® discs were used against four ATCC strains (E. coli 25922, S. aureus 25923, S. pneumoniae 49619, and H. influenzae 49247) and wild strains of Enterobacteriaceae, P. aeruginosa, Acinetobacter spp., Shigella spp., S. enterica, Haemophilus spp., M. catarrhalis, Staphylococcus spp., Streptococcus spp., and Enterococcus spp.). For each strain, two 90 mm circular agar plates were inoculated with the same bacterial inoculum at the same time, and Bio-Rad® or Oxoid® discs with relevant antibiotics were placed on each plate. Plates were incubated together under the same conditions. Following automatic reading, diameters were manually edited by changing the radius of the inhibition zone on the ADAGIO™ screen, only in those cases the laboratory technician who reviewed the results considered it necessary. The number of manual corrections was recorded. The ADAGIO™ readings were categorized as susceptible, intermediate, or resistant and compared against manual readings of the Oxoid® discs. Error categories were defined for all readings as described above. The statistical significance of differences was calculated using the Fisher’s exact test.
Results
In the first part of the study, eight ATCC QC strains were tested in triplicate, giving a total of 34 strain/antibiotic combinations and 102 individual readings. As shown in Table 1, only the same two out of 34 combinations (5.9%) were not within the expected range with both Oxoid® and Bio-Rad® discs. Table 2 shows the performance of Bio-Rad® discs as compared with Oxoid® discs against wild strains. From a total of 1192 antibiotic/bacteria combinations (2384 duplicate readings) with 120 different wild strains, only one major error and four minor errors (0.08% and 0.34%, respectively) were detected with the Bio-Rad® discs as compared with the Oxoid® discs.
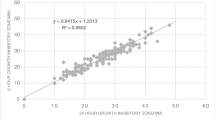
In the second part of the study, among 32 strain/antibiotic combinations (64 readings) of four ATCC strains read by ADAGIO™, only the one strain (3.1%) was out of the expected range (by 1 mm) with the Bio-Rad® discs. The same diameter was read manually, suggesting that the ADAGIO™ reading was correct (Table 3).
Table 4 shows the performance of ADAGIO™ readings with Bio-Rad® and Oxoid® discs, as compared with manual readings with Oxoid® discs against wild strains. From a total of 1192 strain/antibiotic combinations with 120 different wild strains read by ADAGIO™, five minor errors and one major error (0.42% and 0.08%, respectively) were detected with Bio-Rad® discs, and one minor error and no major errors (0.08% and 0%, respectively) were detected with Oxoid® discs (Table 5).
Table 5 also shows the number of times the automatic reading was manually edited for both Bio-Rad® and Oxoid® discs (3.7% vs. 5.7%, respectively). The number of times that the automatic reading was manually edited with Bio-Rad® discs was significantly lower than was necessary with Oxoid® discs (p < 0.05).
Discussion
The first part of this study presents an evaluation of the Bio-Rad® discs for antimicrobial susceptibility testing by disc diffusion. As seen in Table 1, no significant differences were observed between the performance of Bio-Rad® and Oxoid® discs against ATCC QC strains. The same two strain/antibiotic combinations (S. pneumoniae 49619 with meropenem and H. influenzae 49766 with ertapenem) were out of range with both disc brands. The fact that all triplicate readings of both brands in the two cases showed similar slightly out-of-range results may suggest that the problem was related to other causes (e.g., isolate, culture media, or incubation conditions) and not to the quality of the discs. Nevertheless, the performance of both brands was similar. The previous study [13], which used the EUCAST criteria, checked the performance of antibiotic discs from different brands and showed that the average readings of Bio-Rad® and Oxoid® discs against ATCC strains were all within the expected range. The present study confirms these findings.
Bio-Rad® discs performed very well against wild strains; the percentage of minor and major errors was very low, 0.34% and 0.08%, respectively, with no very major errors. The number of minor and major errors detected with Bio-Rad® discs was not statistically higher than that detected with Oxoid® discs (p = 0.20 and p = 0.99, respectively) (Table 5). In contrast to the aforementioned EUCAST study [13], in which Oxoid® discs were found to be significantly superior to Bio-Rad® discs, the present study evaluated a larger number of QC and wild strains, and the results suggest that Bio-Rad® discs are not inferior and can be used without compromising patient safety.
In the second part of the study, the performance of the ADAGIO™ system was evaluated, first with ATCC QC strains and then with a large number of wild strains. Only one of the QC strain/antibiotic combinations showed out-of-range results (H. influenzae against trimethoprim/sulfamethoxazole) by only 1 mm, and the results were again similar for both disc brands.
The performance of the ADAGIO™ system with wild strains also showed very good results, with only five minor errors and one major error with the Bio-Rad® discs (0.42% and 0.08%, respectively) out of 1192 strain/antibiotic combinations. No very major errors were observed with either brand. With the Oxoid® discs, only one minor error (0.08%) was observed, and the difference between both brands was not statistically significant (Tables 4 and 5). Based on these results, we can assume that both disc brands are equivalent and can be used with the ADAGIO™ system in a similar manner.
As previously stated, the advantage of the Bio-Rad® discs with the ADAGIO™ system is the automatic recognition of the disc contents, regardless of its position on the agar plate, without the need to maintain a fixed pattern. This leads to a significant improvement in the reading process, greater confidence in the interpretation of results, and reduction in the overall duration of hands-on time required. In addition, as shown in Table 5, manual editing of automatic readings was required in a significantly lower number of cases with the Bio-Rad® discs than with the Oxoid® discs (3.7% and 5.7%, respectively, p = 0.02).
To summarize, the findings of the present study support the hypothesis that Bio-Rad® discs are not inferior to Oxoid® discs for antimicrobial susceptibility testing by disc diffusion. To our knowledge, this is the first study to include a wide range of bacteria and shows the excellent performance of the ADAGIO™ system with Bio-Rad® discs. This method would potentially lead to a significant improvement in hands-on time, accuracy, reagent traceability, and patient safety. Additional conclusions based on the evaluation include the fact that the system is user-friendly, requires minimal maintenance, and a very short duration for personnel training.
Although the time needed for reading the results was not thoroughly evaluated in this study, it was evidently shorter with the ADAGIO™ system than with manual measurement. This study also demonstrated the ability of the ADAGIO™ system to read different types of agar plates, even those supplemented with blood. The only disadvantage we found with the system is its ability to read only open plates without lids. This is a potential safety issue that requires further consideration.
In conclusion, based on the results of the present study, the ADAGIO™ system in combination with Bio-Rad® antibiotic discs yielded results that were not inferior to manual reading of Oxoid® discs. This combination may be an excellent alternative to current manual techniques and could thereby improve standardization, traceability, and patient safety in clinical microbiology laboratories.
References
Jorgensen JH, Ferraro MJ (2009) Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin Infect Dis 49(11):1749–1755
Périllaud C, Pilmis B, Diep J, de Ponfilly GP, Vidal B, Couzigou C, Mizrahi A, Lourtet-Hascoët J, Le Monnier A, Nguyen Van JC (2019) Prospective evaluation of rapid antimicrobial susceptibility testing by disk diffusion on Mueller-Hinton rapid-SIR directly on blood cultures. Diagn Microbiol Infect Dis 93(1):14–21
Chandrasekaran S, Abbott AN, Campeau S, Zimmer BL, Weinstein MP, Thrupp L, Hejna J, Walker L, Ammann T, Kirn TJ, Patel R, Humphries RM (2018) Direct-from-blood-culture disk diffusion to determine antimicrobial susceptibility of gram-negative bacteria: preliminary report from the Clinical and Laboratory Standards Institute methods development and standardization working group. J Clin Microbiol 56(3):e01678–e01617
Hombach M, Jetter M, Blöchliger N, Kolesnik-Glodmann N, Böttger EC (2017) Fully automated disc diffusion for rapid antibiotic susceptibility test results: a proof-of-principle study. J Antimicrob Chemother 72(6):1659–1668
Hombach M, Zbinden R, Böttger EC (2013) Standardisation of disk diffusion results for antibiotic susceptibility testing using the sirscan automated zone reader. BMC Microbiol 13(1):225
Kolbert M, Chegrani F, Shah PM (2004) Evaluation of the OSIRIS video reader as an automated measurement system for the agar disk diffusion technique. Clin Microbiol Infect 10(5):416–420
Lestari ES, Severin JA, Filius PMG, Kuntaman K, Offra Duerink D, Hadi U, Wahjono H, Verbrugh HA (2008) Comparison of the accuracy of disk diffusion zone diameters obtained by manual zone measurements to that by automated zone measurements to determine antimicrobial susceptibility. J Microbiol Methods 75(2):177–181
Sánchez M, Sánchez del Saz B, Loza E, Baquero F, Cantón R (2001) Evaluation of the OSIRIS video reader system for disk diffusion susceptibility test reading. Clin Microbiol Infect 7(7):352–357
Medeiros AA, Crellin J (2000) Evaluation of the Sirscan automated zone reader in a clinical microbiology laboratory. J Clin Microbiol 38(4):1688–1693
Andrews JM, Boswell FJ, Wise R (2000) Evaluation of the Oxoid Aura image system for measuring zones of inhibition with the disc diffusion technique. J Antimicrob Chemother 46(4):535–540
Korgenski EK, Daly JA (1998) Evaluation of the BIOMIC video reader system for determining interpretive categories of isolates on the basis of disk diffusion susceptibility results. J Clin Microbiol 36(1):302–304
Joshi A, Iyer V, Balasubramaniam U, Kagal A, Bharadwaj R (2008) Comparison of efficacy of three commercially available antibiotic discs. Indian J Med Microbiol 26(2):160–162
Åhman J, Matuschek E, Kahlmeter G (2019) The quality of antimicrobial discs from nine manufacturers—EUCAST evaluations in 2014 and 2017. Clin Microbiol Infect 25(3):346–352
Idelevich EA, Becker K, Schmitz J, Knaack D, Peters G, Köck R (2016) Evaluation of an automated system for reading and interpreting disk diffusion antimicrobial susceptibility testing of fastidious bacteria. PLoS One 11(7):e0159183
Weinstein MP, Patel JB, Campeau S, et al. (2018) Performance Standards for Antimicrobial Susceptibility Testing M100 (28th edn) A Clinical and Laboratory Standards Institute publication. http://homenew.clalit.org.il/sites/Communities/logi/rechesh/Reagent/CLSI/M100Ed28E.pdf Wayne USA. www.clsi.org. Accessed 1 Apr 2018.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Strauss, M., Zoabi, K., Sagas, D. et al. Evaluation of Bio-Rad® discs for antimicrobial susceptibility testing by disc diffusion and the ADAGIO™ system for the automatic reading and interpretation of results. Eur J Clin Microbiol Infect Dis 39, 375–384 (2020). https://doi.org/10.1007/s10096-019-03735-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-019-03735-4