Abstract
The disk diffusion (DD) method remains the most popular manual technique for antibiotic susceptibility testing (AST) in clinical microbiology laboratories. This is because of its simplicity, reproducibility, and limited cost compared to (automated) microdilution systems, which are usually less sensitive at detecting certain important mechanisms of resistance. Here, we evaluate the PREVI® Isola automated seeder system using a new protocol for spreading bacterial suspensions (eight deposits of calibrated inocula of bacteria, followed by two rounds of rotation) in comparison with manual DD reference testing on a large series of clinical and reference strains. The average time required for seeding one agar plate for DD with this new protocol was 51 s per plate, i.e., 70 agar plates/h. Reproducibility and repeatability was assessed on three reference and three randomly chosen clinical strains, as usually requested by the European Committee on Antimicrobial Susceptibility Testing (EUCAST), and was excellent compared to the manual method. The standard deviations of zones of growth inhibition showed no statistical discrimination. The correlation between the two methods, assessed using 294 clinical isolates and a panel of six antibiotics (n = 3,528 zones of growth inhibition measured), was excellent, with a correlation coefficient of 0.977. The new PREVI® Isola protocol adapted for DD had a sensitivity of 99 % and a specificity of 100 % compared to the manual technique for interpreting DD as recommended by the EUCAST.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Over the past ten years, the automation of biological specimen inoculation on agar plates, along with rapid bacterial identification by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry, has revolutionized clinical microbiology laboratories and has facilitated the rapid processing of large-volume specimens [1–4]. Automation in microbiology is an important step which will enable large numbers of clinical samples to be processed rapidly, reproducibly, and cost-effectively. The reproducibility and reliability of automation are its two main benefits. Several automated systems have now been marketed for this purpose, but for antibiotic susceptibility testing (AST), there is still a major need for improvement compared to microbial detection and identification [2, 5]. However, because of the stringent AST requirements from the European Committee on Antimicrobial Susceptibility Testing (EUCAST) and the Clinical and Laboratory Standards Institute (CLSI) regarding the quality of reproducibility and repeatability of results and their interpretation, standardization of the inoculation of agar plates for the disk diffusion (DD) method remains critical. AST can be performed either manually using DD, which is currently the most popular method in many laboratories, or via the use of automated liquid microdilution instruments [5]. Although AST using automated instruments (e.g., Phoenix (Becton Dickinson) [6], VITEK 2 (bioMérieux) [7], or MicroScan WalkAway (Siemens) [6]) that use turbidimetric and/or colorimetric growth detection has existed for decades [5], to the best of our knowledge, there is currently no peer-reviewed, automated instrument that is dedicated to the standardized inoculation of agar plates for DD.
Here, we report the use of the PREVI® Isola automated seeder system with a new protocol for spreading bacterial suspensions (eight deposits of calibrated inocula of bacteria, followed by two rounds of rotation) compared to the manual DD reference method recommended by the EUCAST.
Materials and methods
Bacterial isolates
The reference strains used in this study to validate and assess the repeatability and reproducibility of the technique included Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, and Staphylococcus aureus ATCC 25923, as recommended by the EUCAST [9].
We used 294 strains in the study, including routine clinical isolates from the Clinical Microbiology Laboratory of Timone Hospital in Marseille, France, as well as reference strains from the Timone culture collection, as follows: 42 E. coli, 16 Proteus mirabilis, 38 Klebsiella pneumoniae, six K. oxytoca, 11 Enterobacter aerogenes, eight E. cloacae, 45 P. aeruginosa, five Serratia marcescens, 15 Acinetobacter baumannii, five Morganella morganii, 20 Enterococcus faecalis, seven E. faecium, 25 S. aureus, eight S, epidermidis, 12 Streptococcus pneumoniae, six S. pyogenes, 19 S. agalactiae, and six Haemophilus influenzae. All the strains are presented in Table 1, along with the resistance mechanism.
Antibiotic susceptibility testing (AST)
After identification by MALDI-TOF mass spectrometry, AST was performed on solid medium according to the DD method [3]. Inocula were obtained from fresh, 18–24-h cultures on Trypticase soy agar (TSA) or chocolate agar plus PolyVitex (PVX) for fastidious organisms. The cultures were seeded on Mueller–Hinton agar (bioMérieux, Marcy l’Etoile, France) alone or with blood for Streptococcus (bioMérieux, Marcy l’Etoile, France). For the diffusion technique, this involved plating a suspension of 0.5 McFarland [∼108 colony-forming units (CFU)/ml] by flooding for 15 min after the inocula had already been prepared for the manually seeded plates at the same time as for the PREVI® Isola system, as recommended the by EUCAST. Suspensions of organisms should be plated with the PREVI® Isola system immediately after making the 0.5 McFarland suspensions.
PREVI® Isola automatic seeding
Validation step of the PREVI® Isola protocol
The PREVI® Isola (bioMérieux, Marcy l’Etoile, France) inoculation protocol was first evaluated using a suspension of crystal violet. A machine-seeded suspension of crystal violet on blood agar enabled the number of calibrated deposits and the number of circles due to the circular application to be visualized. The new protocol for automatic plating was as follows: 40 μl of each bacterial inoculum was prepared in sterile tubes for AST and pipetted by the instrument. Subsequently, 5 μl of the inoculum was deposited every 45°. In other words, eight deposits were made on the agar plate. The plates were then streaked by the applicator for 720°, i.e., two rounds of streaking. Antibiotic disks were then placed on the agar plate using the same protocol as for the manual technique. The time required for streaking on agar using this new protocol was measured with a chronometer to determine the average number of plates streaked per hour.
Reproducibility was analyzed as defined by the EUCAST using the three reference strains, as well as three clinical isolates that were randomly chosen from the collection of isolates. AST for each isolate was repeated five times for the manual technique and ten times for the PREVI® Isola streaking protocol, followed by intra- and inter-precision tests. The zone of growth inhibition of each antibiotic tested was measured and the mean diameters of the manual and the new PREVI® Isola protocols were compared. For the reference strains, the zones of growth inhibition were compared to the expected diameter documented by the EUCAST.
Correlation between the two methods
The correlation between the PREVI® Isola technique and the standard reference manual technique was defined for the 294 clinical isolates and a panel of six antibiotics (n = 3,528 zones of growth inhibition measured). A concordance curve was constructed by comparing the average zone diameters obtained for each antibiotic using the two techniques. A correlation coefficient was then calculated. The mean and standard deviation of the zone of growth inhibition in the three categories was then calculated, interpreted as susceptible, intermediate, or resistant, using the two methods of seeding AST. We then analyzed the results using Student’s t-test to obtain a p-value after verifying the equality of covariance (Fisher’s test). We considered that there was no significant difference between the two methods when the p-value was >0.05.
Analysis of discrepant results
A discrepant result was defined as a difference of interpretation (S, I, R) between PREVI® Isola and the manual technique according to the EUCAST guidelines. The manual technique was used as the gold standard. A very major error (VME) was defined as a result leading to a susceptible phenotype using the PREVI® Isola method but which was resistant when using the standard method. A major error (ME) was defined as a result leading to a resistant phenotype using the PREVI® Isola method but which was susceptible when using the standard method. A minor error (mE) was defined as a result leading to an intermediate phenotype using the PREVI® Isola method but to susceptibility or resistance using the standard method, and vice versa. All discrepant results were retested 12 times using each technique to assess the inter-experimental variability of the two techniques. The remaining discordance after this verification was tested specifically by the Etest® to determine the true minimum inhibitory concentrations (MICs).
Validation of the protocol with the Etest® technique
Twenty-six clinical isolates, including ten strains of P. aeruginosa, ten strains of S. aureus, three strains of E. coli, three strains of Candida albicans, and three strains of C. krusei, were used in this study. The MICs of vancomycin for S. aureus strains, colistin and ceftazidime for P. aeruginosa strains, and caspofungin, fluconazole, and voriconazole for yeast were determined using the Etest® method (bioMérieux, Marcy l’Etoile, France) and by DD on Mueller–Hinton agar, as recommended by the EUCAST. For yeast strains, we used RPMI agar (bioMérieux, Marcy l’Etoile, France) for antifungal susceptibility testing. The MIC results were compared to those obtained using the reference manual technique.
Results
Development and validation of the PREVI® Isola protocol
The average time to seed using the PREVI® Isola and the manual technique was measured ten times. The average time for seeding one plate with PREVI® Isola was 51 s, compared to 1 min 40 s per agar for the manual technique, when 0.5 McFarland suspensions had already been prepared.
Reproducibility
All zone diameters were repeatedly within the expected ranges for the three quality control strains, and no difference was noted between the PREVI® Isola protocol and the reference method (Table 2). The reproducibility of zone diameters by the two methods for the three E. coli, S. aureus, and P. aeruginosa clinical isolates was also excellent and reproducible, and no differences were noted between the PREVI® Isola protocol and the reference method (Table 1).
Correlation between the two methods on clinical isolates
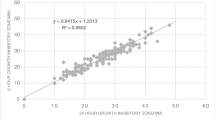
Figure 1 shows examples of the results obtained for DD using the PREVI® Isola system compared to using the manual reference technique. The raw concordance curve of measurements of the zone diameter between the two methods was excellent, with a correlation coefficient of 0.974 (Fig. 2a). The specificity of the PREVI® Isola method was 97 %, sensitivity 97 %, positive predictive value 98 %, and negative predictive value 97 %. Among the 294 strains, the percentage of discrepant results was 1.5 %, with 0.26 % VME (four diameters), 0.06 % ME (one diameter), and 1.16 % mE (18 diameters), as presented in Supplementary Table S1. All the results of the statistical analysis are presented in Table 3 and all p-values were >0.05. In conclusion, no significant differences between the zones of growth inhibition using these two methods has been demonstrated (Table 3).
The 23 discrepant results were reanalyzed 12 times using the two techniques for inter-method variability. The results showed that only six diameters continued to conflict for one antibiotic; the remainder were not discrepant and were most likely due to operating errors or real inter-method variability (Supplementary Table S2). The six remaining discrepant results were for piperacillin–tazobactam with a β-lactamase inhibitor for four isolates and imipenem for the two other strains, and were mainly due to variability in the manual technique. After correcting for discrepant results, the concordance curve showed a correlation coefficient of 0.977 (Fig. 2b), the sensitivity and negative predictive value were 99 %, and the specificity and positive predictive value were 100 %.
Validation of the protocol with the Etest® technique
The results show that the MICs, as defined by the PREVI® Isola protocol, were concordant with those obtained using the manual reference technique, with less than one dilution difference in the MICs (Table 3). Figure S1 gives an example of an MIC determination obtained after streaking with the PREVI® Isola method compared to the reference manual techniques for vancomycin for S. aureus (Fig. S1A) and for ceftriaxone for E. coli (Fig. S1B).
Discussion
We compared the use of the PREVI® Isola automated seeder system using a new protocol with the manual reference method for DD, as recommended by the EUCAST, to analyze a large series of reference and clinical strains. Although it is well known that the PREVI® Isola instrument is a robust and reproducible instrument for inoculating biological samples on a large variety of solid media [4, 8], to the best of our knowledge, this is the first study to evaluate the performance of an automated system for streaking DD plates. We show that the reproducibility of the method was excellent, with rapid plating: 51 s per agar and without the lack of reproducibility of the manual technique, which was operator-dependent. Furthermore, the accuracy of DD for a representative number of clinical bacterial isolates that were susceptible or resistant to different antibiotics meant that this method could be approved as an alternative in the clinical microbiology laboratory. This system allows six antibiotics plated on 90-mm-diameter agar plates to be tested, but it would be possible to test 12 antibiotics for AST if two agar plates were seeded at the same time. Indeed, according to current international recommendations, a technique should be considered clinically valid when the percentage of VME is lower than 1.5 %, if the MEs do not exceed 3 %, and if the new method has an overall agreement of >90 % with the reference method [5]. This was the case in our study, which analyzed a large series of bacterial species and antibacterial drugs, and had an VME of only 0.25 %, an ME of 0.06 %, and a correlation coefficient of 0.977 with the reference method. We found that streaking the plates with a standardized inoculum was highly consistent, precise, and rapid, a critical point in the accuracy and reproducibility of DD. The zone of growth inhibition with the PREVI® Isola system was observed to be 2 to 5 mm larger than the manual technique for susceptible strains, but this does not change the interpretation of resistance to the antibiotic. In order to clarify whether this phenomenon could affect the overall interpretation, we calculated the p-value for all 294 clinical isolates presented in Table 3. No statistical difference was observed. One possible explanation for such a difference could be that the volume of inoculum seeded by PREVI® Isola was lower in comparison with the manual technique. In addition, the streaking protocol was also usable for MIC determination by the Etest® but had to be placed on the sides of the agar. The center of the agar is the localization of the deposits and represents the most significant concentration in bacteria. Implementation of this protocol on any PREVI® Isola instrument is very easy because it requires a single software update, and the new protocol can be used in addition to the standard inoculation protocol, resulting in increased flexibility when using the instrument in clinical microbiology laboratories. The results obtained in this study are entirely in line with the manual reference DD method, and the EUCAST expert rules could, therefore, be applied without changes [9]. Thus, this new protocol could be considered a new element in the automation of clinical microbiology laboratories, and should accelerate inoculation, identification, and AST by DD, which are key factors in the reduction of mortality, hospital stay, and healthcare costs [10, 11].
Conclusion
Our study demonstrated that the PREVI® Isola instrument was highly concordant with the manual method for disk diffusion (DD), thus renewing interest for microbiologists wishing to continue using this method as a reference, and could be easily implemented in PREVI® Isola instruments worldwide.
References
Greub G, Prod’hom G (2011) Automation in clinical bacteriology: what system to choose? Clin Microbiol Infect 17(5):655–660
van Belkum A, Durand G, Peyret M, Chatellier S, Zambardi G, Schrenzel J, Shortridge D, Engelhardt A, Dunne WM Jr (2013) Rapid clinical bacteriology and its future impact. Ann Lab Med 33(1):14–27
Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, Raoult D (2009) Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 49(4):543–551
Mischnik A, Mieth M, Busch CJ, Hofer S, Zimmermann S (2012) First evaluation of automated specimen inoculation for wound swab samples by use of the Previ Isola system compared to manual inoculation in a routine laboratory: finding a cost-effective and accurate approach. J Clin Microbiol 50(8):2732–2736
Jorgensen JH, Ferraro MJ (2009) Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin Infect Dis 49(11):1749–1755
Winstanley T, Courvalin P (2011) Expert systems in clinical microbiology. Clin Microbiol Rev 24(3):515–556
Renaud FN, Bergeron E, Tigaud S, Fuhrmann C, Gravagna B, Freney J (2005) Evaluation of the new Vitek 2 GN card for the identification of gram-negative bacilli frequently encountered in clinical laboratories. Eur J Clin Microbiol Infect Dis 24(10):671–676
Glasson JH, Guthrie LH, Nielsen DJ, Bethell FA (2008) Evaluation of an automated instrument for inoculating and spreading samples onto agar plates. J Clin Microbiol 46(4):1281–1284
Leclercq R, Cantón R, Brown DF, Giske CG, Heisig P, MacGowan AP, Mouton JW, Nordmann P, Rodloff AC, Rossolini GM, Soussy C-J, Steinbakk M, Winstanley TG, Kahlmeter G (2013) EUCAST expert rules in antimicrobial susceptibility testing. Clin Microbiol Infect 19(2):141–160
Galar A, Yuste JR, Espinosa M, Guillén-Grima F, Hernáez-Crespo S, Leiva J (2012) Clinical and economic impact of rapid reporting of bacterial identification and antimicrobial susceptibility results of the most frequently processed specimen types. Eur J Clin Microbiol Infect Dis 31(9):2445–2452
Galar A, Leiva J, Espinosa M, Guillén-Grima F, Hernáez S, Yuste JR (2012) Clinical and economic evaluation of the impact of rapid microbiological diagnostic testing. J Infect 65(4):302–309
Acknowledgments
We are very grateful to Fanny Languasco and Linda Hadjadj for their technical assistance. This work was partly funded by IHU Méditerranée Infection.
Conflict of interest
Alex van Belkum, Corine Fulchiron, and Rachel Huguet are employees of bioMérieux, the company which manufactures and sells the PREVI® Isola instrument.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below are the links to the electronic supplementary material.
Fig. S1
(PPTX 1048 kb)
Table S1
Susceptible, intermediate, or resistant isolates, as determined by the manual technique and PREVI® Isola readings of zone diameters (DOCX 24 kb)
Table S2
Analysis of the discrepant strains (DOCX 39 kb)
Table S3
Comparison of MICs obtained using the manual technique versus the PREVI® Isola protocol (DOCX 49 kb)
Rights and permissions
About this article
Cite this article
Le Page, S., van Belkum, A., Fulchiron, C. et al. Evaluation of the PREVI® Isola automated seeder system compared to reference manual inoculation for antibiotic susceptibility testing by the disk diffusion method. Eur J Clin Microbiol Infect Dis 34, 1859–1869 (2015). https://doi.org/10.1007/s10096-015-2424-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-015-2424-8