Abstract
The incidence of infections caused by antimicrobial-resistant Enterobacteriaceae in Thailand is increasing and human intestinal flora is an important reservoir for these organisms. This study was carried out to determine the intestinal carriage of bla CTX-M extended spectrum ß-lactamase-positive Enterobacteriaceae (ESBL + E) and AmpC-positive Enterobacteriaceae in a community setting in Northern Thailand, and to identify potential risk factors for carriage. A total of 307 fecal samples were collected from healthy volunteers in Phitsanulok province, and cefotaxime-resistant Enterobacteriaceae (CtxRE) were isolated using selective media. Polymerase chain reaction (PCR) was used to detect ESBL and AmpC genes. Risk factors were analyzed using multiple logistic regression. Genotyping was performed by multilocus sequence typing (MLST) analysis. Two hundred ninety-one CtxRE isolates were obtained and Escherichia coli was the predominant organism (66.3%). The intestinal carriage rates of bla CTX-M ESBL + E and AmpC-positive Enterobacteriaceae were 52.1% and 6.2%, respectively. Comparative levels of bla CTX-M group 1 and bla CTX-M group 9 were found while bla CMY-2 was the predominant genotype among AmpC genes. Co-existence of two ß-lactamase genes in a single isolate was found in 6.5% of isolates. Consumption of undercooked meat was strongly associated with intestinal carriage of bla CTX-M ESBL + E (p = 0.003, OR = 2.133, 95% CI = 1.289–3.530). Phylogenetic grouping and MLST analysis of E. coli isolates revealed the presence of E. coli B2-ST131 (n = 8). Of these, seven carried bla CTX-M-group 9 and 1 carried bla CMY-2. Our results suggest that residents in Thailand are at high risk for developing endogenous infections caused by antibiotic-resistant Enterobacteriaceae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The incidence of multidrug-resistant Enterobacteriaceae (MDRE) is increasing rapidly and has now become a global threat [1]. Treatment of diseases caused by MDRE is far more challenging where treatment options are becoming limited. In particular, resistance to expanded-spectrum cephalosporins, monobactam and carbapenems among Enterobacteriaceae is increasingly reported and is usually mediated by the production of extended-spectrum ß-lactamase (ESBL), AmpC and carbapenemase [1].
Due to the fact that most ESBL-, AmpC- and carbapenemase-encoding genes reside on mobile genetic elements (MGEs), i.e. plasmids and transposons, they are readily transferrable among Enterobacteriaceae. Furthermore, MGEs invariably carry genes encoding for resistance to other classes of antibiotics, such as aminoglycosides and fluoroquinolones, hence facilitating the emergence and spread of MDRE [1]. MDRE are now widespread in hospital settings, communities and the environment [2]. The human intestine is increasingly recognized as an important reservoir for MDRE and thus the human-to-human transmission and dissemination of MDRE in communities can occur where living conditions have poor sanitation [3]. Furthermore, the acquisition of MDRE in healthy individuals traveling to endemic regions have been documented, further enhancing the international spread of MDR organisms [3].
Previous studies have shown that intestinal carriage by ESBL-positive Enterobacteriaceae (ESBL + E) has been associated with developing infection due to ESBL producers [4, 5]. Many studies have reported the fecal carriage of ESBL + E among healthy populations [6,7,8,9]. Moreover, the high carriage rates of ESBL + E (53.9–75.5%) have been reported among healthy residents in central Thailand [10,11,12,13]. However, these rates were slightly lower (32%) in Northern and Southern parts of Thailand [11].
Since the incidence of infections due to ESBL + E and AmpC-positive Enterobacteriaceae in Thailand has continuously increased [14], this study was carried out to gain a better understanding of the intestinal carriage of ESBL + E and identify risk factors for ESBL + E carriage in a community setting in Northern Thailand. Furthermore, this study was extended to include the carriage rate of AmpC-positive Enterobacteriaceae. In addition, characterization of resistant genes and molecular analysis of resistant isolates were investigated.
Materials and methods
Study setting and ethical approval
This prospective cohort study was conducted on healthy volunteers in Wang Thong District which is located in the rural area of Phitsanulok province, Northern Thailand. All subjects were at least 15 years of age. Participants who had diarrheal diseases within the previous 1 month were excluded from the study. Participant recruitment and sample collection were assisted by local public health personnel. This study was approved by the Naresuan University Institutional Review Board (COA no. 49/2013). Written informed consent was obtained from each subject participating in the study. For the subjects who were under 18 years old, consent was provided by their parent or guardian.
Sample collection and questionnaires
Fresh fecal sampling was performed between October and December 2013. A total of 307 healthy volunteers participated in this study. Fecal samples were collected by subjects using sterile stool containers with a sterile plastic spoon. One sample was obtained from each participant. Microbiological analysis was carried out within 24 h.
Volunteers were asked to fill in a comprehensive questionnaire to collect information regarding demographic data (age, gender, education, occupation, etc.), consumption of undercooked meat and fresh vegetables, type of drinking water, previous antibiotic use, and history of hospitalization.
Isolation and identification of cefotaxime-resistant Enterobacteriaceae
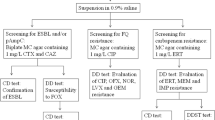
The fecal samples were directly cultured on Chrome UTI agar (Oxoid Ltd., Basingstoke, Hamshire, UK) supplemented with 25 mg/L vancomycin and 1 mg/L cefotaxime (Sigma Aldrich, St. Louis, MO, USA) and incubated under aerobic condition for 24 h at 37 °C. Colonies of different morphology were selected and subcultured onto EMB agar for further studies. Species identification was performed by using RapID™ ONE System (REMEL Inc., Lenexa, KS, USA) according to the manufacturer’s instruction. Isolates showing ambiguous results were further identified by sequencing of the 16S rRNA gene.
Antimicrobial susceptibility testing and ESBL production
All isolates were subjected to susceptibility testing against 12 antimicrobial agents (amoxicillin/clavulanic acid, cefoxitin, ceftazidime, cefepime, aztreonam, imipenem, meropenem, gentamicin, amikacin, ciprofloxacin, doxycycline and trimethoprim-sulfamethoxazole) by disk diffusion method according to Clinical Laboratory Standards Institute (CLSI) protocols and the results were interpreted according to CLSI criteria [15]. An isolate was defined as MDR if it was resistant to at least three or more antimicrobial classes [16].
Screening for ESBL production was performed by combination disk method according to CLSI guidelines using cefotaxime and ceftazidime alone or in combination with clavulanic acid [15].
Determination of genes encoding for CTX-M, AmpC and carbapenemase by PCR and sequencing
The presence of genes encoding for CTX-M, AmpC and carbapenemase was performed by multiplex PCR using previously published primers and conditions [17,18,19]. An individual colony was suspended in 1 mL of water and used as template in PCR. Amplification was performed in a total volume of 20 μL containing 1 μL of template DNA, 1X PCR buffer, 1.5 mM MgCl2, 0.2 mM dNTPs and 0.5 μM of each primer and 1 U of Taq polymerase (Vivantis Technologies, Selangor, Malaysia). DNA amplification was performed in a Veriti®Thermal Cycler (Applied Biosystems, CA, USA). PCR products were analyzed by agarose gel electrophoresis.
Selected PCR products were purified using a DNA purification kit (GF-1 Nucleic Acid Extraction Kit; Vivantis Inc., Chino, CA, USA) and were sent to a commercial facility for sequencing (First BASE Laboratories Sdn Bhd, Selangor, Malaysia). Sequences were compared with those available in the GenBank database using the BLAST algorithm available on the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov).
Phylogenetic grouping and multilocus sequence typing (MLST) analysis
Phylogenetic grouping (A, B1, B2 and D) of Escherichia coli was performed by a multiplex PCR assay for chuA, yjaA and DNA fragment TspE4C2 as previously described [20]. MLST was performed by amplification and sequencing of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA and recA) according to the protocols from the E. coli MLST website (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli).
Repetitive palindromic polymerase chain reaction (rep-PCR)
Selected E. coli isolates were typed by rep-PCR as described previously [21]. The PCR reaction mixture consisted of 1 μL template DNA, 1X PCR buffer, 0.2 mM dNTPs, 0.5 μM of each primer and 1 U of Taq polymerase, in a total volume of 25 μL. DNA banding patterns were assessed by visual inspection.
Statistical analysis
Data were collected and analyzed using SPSS version 17.0 (SPSS, Chicago, IL, USA). Univariate analysis was performed using chi-square for categorical variables. Factors that were statistically significant in univariate analysis were included in multivariate logistic regression analysis to determine risk factors associated with intestinal carriage of bla CTX-M ESBL + E. The results were presented as odds ratios (OR) with 95% confidence intervals. A p value <0.05 was considered a significant difference.
Results
Characteristics of participants
A total of 307 healthy volunteers were enrolled in this study. Volunteer demographics are shown in Table 1. The median age of participants was 51 years (range, 15–88). The majority of them were female, farmers and educated from primary school.
Isolation of cefotaxime-resistant Enterobacteriaceae from volunteers, antimicrobial susceptibility testing and ESBL production
From 307 fecal samples, cefotaxime-resistant Enterobacteriaceae (CtxRE) isolates were detected in 227 samples. One hundred sixty-three subjects carried one species while 64 subjects carried two species of CtxRE resulting in a total of 291 CtxRE isolates. Species identification showed that the majority of isolates were Escherichia coli (66.3%, 193/291) followed by Enterobacter cloacae (14.8%, 43/291) and Klebsiella pneumoniae (10.3%, 30/291). We also recovered Citrobacter spp. (3.4%), Enterobacter spp. (2.1%), Klebsiella spp. (1.4%), Pantoea spp. (0.7%), Kluyvera spp. (0.7%) and Shigella spp. (0.3%).
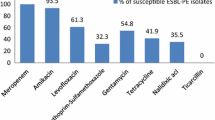
The 291 CtxRE isolates showed additional resistance to cefepime (61.9%), aztreonam (51.2%), trimethoprim/sulfamethoxazole (48.5%), ceftazidime (43.3%) and gentamicin (42.6%) (Fig. 1). A total of 10.7% of isolates were resistant to imipenem. In addition, 138 isolates (47.4%, 138/291) were considered as MDR [16]. Phenotypic analysis showed that 58.6% (180/307, 95% CI = 52.9%–64.2%) of volunteers carried ESBL + E in their intestinal tract.
Resistance rates of cefotaxime-resistant Enterobacteriaceae isolated from healthy volunteers (n = 291). AMC amoxicillin/clavulanate, FOX cefoxitin, CAZ ceftazidime, FEP cefepime, ATM aztreonam, IPM imipenem, MEM meropenem, CN gentamicin, AK amikacin, CIP ciprofloxacin, DO doxycycline, SXT trimethoprim/sulfamethoxazole
Detection of genes encoding for CTX-M, AmpC and carbapenemase
Of the 291 CtxRE, 169 isolates from 160 persons were bla CTX-M ESBL + E by phenotypic (combination disk method) and genotypic characterization (PCR and sequencing). Hence the prevalence of bla CTX-M ESBL + E in healthy volunteers was 52.1% (160/307, 95% CI = 46.7–58.1%).
Genotyping of the bla CTX-M-positive isolates revealed that bla CTX-M group 1 and bla CTX-M group 9 were found in 51.5% (87/169) and 57.4% (97/169) of isolates, respectively. Among these, 10.1% (17/169) of isolates carried both bla CTX-M group 1 and bla CTX-M group 9. In addition, two Klebsiella spp. isolates carried bla CTX-M group 8 (Table 2).
The presence of AmpC- and carbapenemase-encoding genes was investigated. Twenty-one isolates (mostly E. coli) from 19 subjects carried AmpC genes: 17, 3 and 1 carried bla CMY-2, bla DHA-1 and bla MIR, respectively (Table 2). Therefore, the prevalence of AmpC-positive Enterobacteriaceae in healthy volunteers was 6.2% (19/307). The genes encoding for AmpC were found either alone or in combination with bla CTX-M. No carbapenemase genes (bla IMP, bla VIM, bla NDM, bla KPC and bla OXA-48) were detected.
Risk factors for intestinal carriage of bla CTX-M ESBL + E
Using univariate analysis, no significant differences in terms of age, gender, education, occupation, drinking tap water, consumption of fresh vegetables and history of hospitalization were associated with the carriage of bla CTX-M ESBL + E (Table 1). In contrast, consumption of undercooked meat and previous antibiotic use were significantly associated with bla CTX-M ESBL + E carriage (p < 0.05). Accordingly, these two variables were included in multivariate logistic regression analysis. The result showed that consumption of undercooked meat was the only independent risk factor associated with bla CTX-M ESBL + E colonization (p = 0.003, OR = 2.133, 95% CI = 1.289–3.530) (Table 3).
Phylogenetic grouping, MLST analysis and rep-PCR
Phylogenetic grouping was investigated for all 193 E. coli isolates. The results showed that the predominant groups were A (106 isolates, 54.9%), followed by groups B1 and D (each group comprised 39 isolates, 20.2%) and B2 (9 isolates, 4.7%). Nine B2 isolates were identified as ST131 (n = 8) and ST95 (n = 1). The carriage rate of E. coli B2-ST131 among healthy volunteers was 2.6% (8/307). We also observed that five isolates of E. coli ST131 showed identical rep-PCR pattern (Supplemental Fig. 2).
Discussion
In this study, the intestinal carriage of antibiotic-resistant Enterobacteriaceae in a community setting in Phitsanulok province, Northern Thailand was investigated. Our results showed a large number of CtxRE isolates in asymptomatic carriers. Several species were identified; however, the most common species was E. coli followed by E. cloacae and K. pneumoniae. In addition, some of these isolates showed resistance to other classes of broad-spectrum antibiotics, including cefepime, ceftazidime, aztreonam, gentamicin, and trimethoprim/sulfamethoxazole. Furthermore, nearly half (47.4%) of isolates were classified as MDR [16].
Several reports on the intestinal carriage of ESBL + E in community settings have been documented. Studies in the European countries such as the Netherlands and Norway revealed a small number (<10%) of ESBL + E in healthy subjects [7, 9]. The relatively low carriage rates of ESBL + E among healthy populations were reported from Asia, including Japan [22] and Korea [23]. Our results showed the high prevalence of intestinal carriage of bla CTX-M ESBL + E (52.1%) in the community. These results were consistent with those reported from China [8], Southeast Asian countries, e.g. Laos and Vietnam [6], as well as central Thailand [10,11,12], although there are some differences in methodologies or number of subjects who participated in each study. Our results confirmed previous observations that Thailand is a highly endemic area for bla CTX-M ESBL + E carriage within the community [3]. Previous reports from Thailand showed that bla CTX-M group 9 was the predominant ESBL-gene found in ESBL + E among healthy volunteers [10,11,12]. In this study we found the comparative level of bla CTX-M group 1 (51.5%) and bla CTX-M group 9 (57.4%) in ESBL + E, suggesting a shift in the prevalance of bla CTX-M genotypes. The bla CTX-M group 8, which is uncommon in Thailand, was also found at a low frequency.
Several factors have been shown to be associated with intestinal carriage of bla CTX-M ESBL + E in healthy volunteers, including education status, previous antibiotic usage, history of hospitalization and foreign travel [3, 8, 12]. In our study, consumption of undercooked meat was strongly identified as a risk factor associated with bla CTX-M ESBL + E colonization (p = 0.003) suggesting that carriage of bla CTX-M ESBL + E is driven from meat or farming sectors. A possible explanation could be due to fact that bla CTX-M ESBL + E is frequently found in food-producing animals and their role as major reservoirs for ESBL + E has been well documented [24]. In addition, several studies have shown the presence of ESBL + E, including those containing bla CTX-M, in several types of meat and food-producing animals in Thailand [13, 24, 25]. Therefore the possibility exists that transfer of ESBL + E to human intestinal flora could occur by consumption of undercooked meat.
In contrast to ESBL, data on the prevalence of AmpC-positive Enterobacteriaceae, to date, in community settings are limited although their presence in clinical isolates, environments and animals have been reported [26]. Our study is the first investigating AmpC-positive Enterobacteriaceae carriage in Thailand, the prevalence of which was 6.2%. This value is higher than that previously reported from European countries such as Spain and the Netherlands (<1%) [7, 27]. Our molecular studies of AmpC-encoding genes showed that bla CMY-2 is predominant, consistent with the study conducted in a Thai hospital [28]. Risk factors associated with the carriage of AmpC-positive Enterobacteriaceae could not be identified, probably due to the small number of carriage rate. However, our results suggested that the AmpC-producers have spread beyond hospital settings in Thailand, as seen in other studies elsewhere [7, 9]. In addition, the co-existence of two ß-lactamase genes in a single isolate were found in 6.5% of isolates (19/291) (Table 2). This may enhance the level of resistance or render an organism resistant to more ß-lactams. Furthermore, bla CTX-M and bla AmpC were not detected in some CtxRE isolates. Resistance in these isolates may be due to the presence of other enzymes such as inhibitor-resistant ß-lactamases or porin alteration [1].
No carbapenemase-encoding genes were found in our study despite the fact that 10.7% of isolates showed resistance to imipenem (Fig. 1). These results were similar to previous findings in the Netherlands and Switzerland [7, 29]. Although the mechanisms of carbapenem resistance remain unclear, the findings of carbapenem-resistant isolates among healthy volunteers are of concern since carbapenems are antibiotic of choice for treatment of infections caused by ESBL + E. Further experiments are underway to investigate the mechanism of carbapenem resistance in these isolates.
Our study showed that eight isolates of E. coli belonged to the highly pathogenic group B2-ST131. Of the eight E. coli ST131 isolates, seven were ESBL-producers, carried bla CTX-M-group 9 and showed ciprofloxacin resistance. The other isolate was a non-ESBL-producer, possessed bla CMY-2 and was ciprofloxacin susceptible. Recently, E. coli ST131 has been found in different origins (animals, environments and humans) and has been reported to cause community and hospital infections in several countries including Thailand [30, 31]. Transmission of E. coli ST131 among members living in the same household has been reported [32]; nevertheless, little is known about the intestinal carriage of E. coli ST131 among healthy subjects. Previous studies in Australia, China and France revealed that 4–7% of healthy subjects carried E. coli ST131 in their digestive tracts [33, 34]. In this study, we found the low carriage rate (2.6%) of E. coli ST131 among healthy volunteers. Moreover, we observed that five bla CTX-M-group 9-positive ST131 isolates, which were recovered from different participants from different families, showed identical rep-PCR pattern suggesting successful clonal spread of ST131 within the community.
In conclusion, our data indicate a high rate of intestinal carriage of bla CTX-M ESBL + E in a Thai community, and that carriage is strongly associated with the consumption of undercooked meat. The key concern from these findings is the E. coli B2-ST131 carrying either bla CTX-M or bla CMY-2. Our results suggest that residents in Thailand are at high risk for developing infections caused by antibiotic-resistant Enterobacteriaceae. The data from this study have profound ramifications for public health and biosecurity containment in the Thai environment. Accordingly, practices such as sanitation, domestic hygiene, and infection control policies need to be enforced to prevent the spread of antibiotic-resistant human pathogens within communities.
References
Ruppé É, Woerther PL, Barbier F (2015) Mechanisms of antimicrobial resistance in gram-negative bacilli. Ann Intensive Care 5:61
da Costa PM, Loureiro L, Matos AJ (2013) Transfer of multidrug-resistant bacteria between intermingled ecological niches: the interface between humans, animals and the environment. Int J Environ Res Public Health 10:278–294
Woerther PL, Burdet C, Chachaty E, Andremont A (2013) Trends in human fecal carriage of extended-spectrum ß-lactamases in the community: toward the globalization of CTX-M. Clin Microbiol Rev 26:744–758
Liss BJ, Vehreschild JJ, Cornely OA, Hallek M, Fätkenheuer G, Wisplinghoff H et al (2012) Intestinal colonization and blood stream infections due to vancomycin-resistant enterococci (VRE) and extended-spectrum ß-lactamase-producing Enterobacteriaceae (ESBLE) in patients with haematological and oncological malignancies. Infection 40:613–619
Vehreschild MJ, Hamprecht A, Peterson L, Schubert S, Häntschel M, Peter S et al (2014) A multicentre cohort study on colonization and infection with ESBL-producing Enterobacteriaceae in high-risk patients with haematological malignancies. J Antimicrob Chemother 69:3387–3392
Nakayama T, Ueda S, Huong BT, Tuyen le D, Komalamisra C, Kusolsuk T et al (2015) Wide dissemination of extended-spectrum ß-lactamase-producing Escherichia coli in community residents in the Indochinese peninsula. Infect Drug Resist 8:1–5
van Hoek AH, Schouls L, van Santen MG, Florijn A, de Greeff SC, van Duijkeren E (2015) Molecular characteristics of extended-spectrum cephalosporin-resistant Enterobacteriaceae from humans in the community. PLoS One 10:e0129085
Zhang H, Zhou Y, Guo S, Chang W (2015) High prevalence and risk factors of fecal carriage of CTX-M type extended-spectrum ß-lactamase-producing Enterobacteriaceae from healthy rural residents of Taian, China. Front Microbiol 6:239
Ulstad CR, Solheim M, Berg S, Lindbæk M, Dahle UR, Wester AL (2016) Carriage of ESBL/AmpC-producing or ciprofloxacin non-susceptible Escherichia coli and Klebsiella spp. in healthy people in Norway. Antimicrob Resist Infect Control 5:57
Sasaki T, Hirai I, Niki M, Nakamura T, Komalamisra C, Maipanich W et al (2010) High prevalence of CTX-M ß-lactamase-producing Enterobacteriaceae in stool specimens obtained from healthy individuals in Thailand. J Antimicrob Chemother 65:666–668
Luvsansharav UO, Hirai I, Niki M, Sasaki T, Makimoto K, Komalamisra C et al (2011) Analysis of risk factors for a high prevalence of extended-spectrum ß-lactamase-producing Enterobacteriaceae in asymptomatic individuals in rural Thailand. J Med Microbiol 60:619–624
Luvsansharav UO, Hirai I, Nakata A, Imura K, Yamauchi K, Niki M et al (2012) Prevalence of and risk factors associated with faecal carriage of CTX-M ß-lactamase-producing Enterobacteriaceae in rural Thai communities. J Antimicrob Chemother 67:1769–1774
Boonyasiri A, Tangkoskul T, Seenama C, Saiyarin J, Tiengrim S, Thamlikitkul V (2014) Prevalence of antibiotic-resistant bacteria in healthy adults, foods, food animals, and the environment in selected areas in Thailand. Pathog Glob Health 108:235–245
Jean SS, Hsueh PR, SMART Asia-Pacific Group (2017) Distribution of ESBLs, AmpC ß-lactamases and carbapenemases among Enterobacteriaceae isolates causing intra-abdominal and urinary tract infections in the Asia-Pacific region during 2008-14: results from the study for monitoring antimicrobial resistance trends (SMART). J Antimicrob Chemother 72:166–171
Clinical and Laboratory Standards Institute (CLSI) (2013) Performance standards for antimicrobial susceptibility testing; Twenty-third informational supplement. CLSI Document M100-S23. CLSI, Wayne, PA
Falagas ME, Karageorgopoulos DE (2008) Pandrug resistance (PDR), extensive drug resistance (XDR), and multidrug resistance (MDR) among gram-negative bacilli: need for international harmonization in terminology. Clin Infect Dis 46:1121–1122
Pérez-Pérez FJ, Hanson ND (2002) Detection of plasmid-mediated AmpC ß-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol 40:2153–2162
Woodford N, Fagan EJ, Ellington MJ (2006) Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum ß-lactamases. J Antimicrob Chemother 57:154–155
Poirel L, Walsh TR, Cuvillier V, Nordmann P (2011) Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 70:119–123
Clermont O, Bonacorsi S, Bingen E (2000) Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66:4555–4558
Versalovic J, Koeuth T, Lupski JR (1991) Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res 19:6823–6831
Luvsansharav UO, Hirai I, Niki M, Nakata A, Yoshinaga A, Moriyama T et al (2011) Prevalence of fecal carriage of extended-spectrum ß-lactamase-producing Enterobacteriaceae among healthy adult people in Japan. J Infect Chemother 17:722–725
Ko YJ, Moon HW, Hur M, Park CM, Cho SE, Yun YM (2013) Fecal carriage of extended-spectrum ß-lactamase-producing Enterobacteriaceae in Korean community and hospital settings. Infection 41:9–13
Changkaew K, Intarapuk A, Utrarachkij F, Nakajima C, Suthienkul O, Suzuki Y (2015) Antimicrobial resistance, extended-spectrum ß-lactamase productivity, and class 1 integrons in Escherichia coli from healthy swine. J Food Prot 78:1442–1450
Trongjit S, Angkittitrakul S, Chuanchuen R (2016) Occurrence and molecular characteristics of antimicrobial resistance of Escherichia coli from broilers, pigs and meat products in Thailand and Cambodia provinces. Microbiol Immunol 60:575–585
Liebana E, Carattoli A, Coque TM, Hasman H, Magiorakos AP, Mevius D et al (2013) Public health risks of enterobacterial isolates producing extended-spectrum ß- lactamases or AmpC ß-lactamases in food and food-producing animals: an EU perspective of epidemiology, analytical methods, risk factors, and control options. Clin Infect Dis 56:1030–1037
Garrido A, Seral C, Gude MJ, Casado C, González-Domínguez M, Sáenz Y et al (2014) Characterization of plasmid-mediated ß-lactamases in fecal colonizing patients in the hospital and community setting in Spain. Microb Drug Resist 20:301–304
Singtohin S, Chanawong A, Lulitanond A, Sribenjalux P, Auncharoen A, Kaewkes W et al (2010) CMY-2, CMY-8b, and DHA-1 plasmid-mediated AmpC ß-lactamases among clinical isolates of Escherichia coli and Klebsiella pneumoniae from a university hospital, Thailand. Diagn Microbiol Infect Dis 68:271–277
Nüesch-Inderbinen M, Zurfluh K, Hächler H, Stephan R (2013) No evidence so far for the dissemination of carbapenemase-producing Enterobactericeae in the community in Switzerland. Antimicrob Resist Infect Control 2:23
Nicolas-Chanoine MH, Bertrand X, Madec JY (2014) Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 27:543–574
Netikul T, Sidjabat HE, Paterson DL, Kamolvit W, Tantisiriwat W, Steen JA et al (2014) Characterization of an IncN2-type bla NDM-1−carrying plasmid in Escherichia coli ST131 and Klebsiella pneumoniae ST11 and ST15 isolates in Thailand. J Antimicrob Chemother 69:3161–3163
Ender PT, Gajanana D, Johnston B, Clabots C, Tamarkin FJ, Johnson JR (2009) Transmission of an extended-spectrum ß-lactamase-producing Escherichia coli (sequence type ST131) strain between a father and daughter resulting in septic shock and emphysematous pyelonephritis. J Clin Microbiol 47:3780–3782
Leflon-Guibout V, Blanco J, Amaqdouf K, Mora A, Guize L, Nicolas-Chanoine MH (2008) Absence of CTX-M enzymes but high prevalence of clones, including clone ST131, among fecal Escherichia coli isolates from healthy subjects living in the area of Paris, France. J Clin Microbiol 46:3900–3905
Li B, Sun JY, Liu QZ, Han LZ, Huang XH, Ni YX (2011) High prevalence of CTX-M ß-lactamases in faecal Escherichia coli strains from healthy humans in Fuzhou, China. Scand J Infect Dis 43:170–174
Acknowledgements
This work was funded by Naresuan University (R2558B035), Royal Golden Jubilee-PhD program from Thailand Research Fund, Rajamangala University of Technology Lanna and Naresuan University (PHD/0054/2555, UT; PHD/0181/2557, AK). The authors would like to thank public health personnel for their help in sample collection and all participants who provided samples for this study. We would also like to thank Panuka Mate for her technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Written informed consent was obtained from all participants prior to their participation.
Conflict of interest
The authors declare that they have no conflict of interest.
Data availability
All data analyzed during the present study are included in this article or available from the corresponding author on reasonable request.
Electronic supplementary material
ESM 1
(DOCX 303 kb)
Rights and permissions
About this article
Cite this article
Niumsup, P.R., Tansawai, U., Na-udom, A. et al. Prevalence and risk factors for intestinal carriage of CTX-M-type ESBLs in Enterobacteriaceae from a Thai community. Eur J Clin Microbiol Infect Dis 37, 69–75 (2018). https://doi.org/10.1007/s10096-017-3102-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-017-3102-9