Abstract
Different reports of Pneumocystis jirovecii pneumonia (PcP) outbreaks on oncology and transplant units suggest the possibility of a person-to-person transmission. Based on these reports, we searched retrospectively for possible PcP clusters in UZ Leuven in 2013. A movement and transmission map was established for all patients (n = 21) with a positive PcP PCR on BAL fluid. BAL fluid samples from all patients with a positive PCR on the mitochondrial large subunit mRNA of P. jirovecii and possible cross exposure were typed with multilocus sequence typing (MLST). Five patients with a positive PcP PCR could have contact with another PcP patient. Another five patients with a weak positive PcP PCR on BAL fluid during the same period were also included. Based on the MLST typing of the BAL samples of these ten patients, there was no evidence of a PcP outbreak in UZ Leuven in 2013. MLST has proven to be a useful tool in genotyping and outbreak detection. From this case series, it could be concluded that current infection control precautions for P. jirovecii are appropriate in UZ Leuven. However, there is need for an international Pneumocystis database and more clarity in the geographic distribution of different P. jirovecii genotypes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pneumocystis jirovecii (P. jirovecii) is an opportunistic fungal pathogen and an important causative agent of pulmonary infections in immunocompromised hosts characterized by fever, cough, dyspnea and ground-glass opacity on CT-scan. Since the introduction of highly active anti-retroviral therapy (HAART) the incidence of PcP is significantly decreased in the HIV population. Unfortunately, the incidence of PcP is increasing in non-HIV immunocompromised patients [1].
Formerly it was thought that Pneumocystis infection was caused by reactivation of latent infection. This theory is supported by the high prevalence of anti-pneumocystis antibodies in the population. Different studies report a prevalence of antibodies ranging from 70 to 100 % in healthy children [1]. In healthy adults, colonization can range from 0 to 20 % [2].
In the last decade, human and animal studies revealed the possibility of interhuman transmission of P. jirovecii. The isolation of trimethoprim-sulfamethoxazole (TMP-SMX) resistant P. jirovecii strains caused by mutations in the dihydropteroate synthase locus from patients who have never been exposed to TMP-SMX, PcP outbreak reports and the appearance of genotype variation in recurrent infections are all arguments supporting this theory [1].
Choukri et al. were the first to demonstrate the presence of P. jirovecii in air samples from rooms of infected patients. In 15 of 19 patients with Pneumocystis pneumonia, P. jirovecii was also detected in 79 % of air samples collected at 1 m of the patient’s head. Moreover, the fungal burden in the air decreased with distance from the patients. In four rooms, no P. jirovecii was detected, probably because of a low fungal burden in these patients’ lungs. One year later, Choukri et al. examined the air shedding of Pneumocystis in a rat model. Interestingly P. jirovecii was detected in the air starting from 1 week after infection, indicating the presence of a short latency period between infection and fungal shedding. An increase of fungal load in the air was determined until day 28 before stabilizing [3, 4].
The first report of interhuman transmission dates from World War II and was observed in malnourished children. Many other cluster reports (small epidemics) followed, especially on oncology and transplant units [5–11]. Although, it wasn't until the early 1990s that molecular typing methods were introduced [12, 13].
A variety of typing methods have been examined in previous years. Multilocus sequence typing (MLST) is considered the gold standard typing method because it offers the advantage of reproducibility and has a Simpson index of diversity between 0.987 and 0.996 [14]. Maitté et al. used a MLST method relying on eight loci: large subunit mitochondrial rRNA gene (mt26S), large subunit of the rRNA gene (26S), internal transcribed spacer 1 (ITS1), beta-tubulin (β-TUB), superoxide dismutase (SOD), cytochrome b (CYB), dihydropholate reductase (DHFR) and dihydropteroate synthase (DHPS). They applied this approach for genotyping respiratory samples of 33 patients. Locus ITS and 26S were the least efficient. In a more clinical setting, they proposed a simplified scheme with SOD, mt26S and CYB (Simpson-index 0.987) or ITS1, 26S, mt26S and β-Tub as MLST method. Remarkably, in 10 of 33 patients infections with several genotypes were observed [14].
UZ Leuven is a large (1900 bed) tertiary care hospital conducting both solid and hematopoietic stem cell transplantation and also an AIDS Reference Center. As such there is a considerable patient population at risk for PCP. With this retrospective study we aimed to investigate the presence of a P. jirovecii outbreak in UZ Leuven from January 2013 until January 2014 and in case of confirmation, the possible implications on our current isolation policy. At present, infected patients are placed in contact isolation on wards where immunocompromised patients are hospitalized (e.g. hematology-oncology). Isolation is continued until discharge and patients wear a mask when leaving their room.
Based on a thorough database search using the Laboratory and Medical Information system, a transmission map was made of patients positive for P. jirovecii on BAL samples, who have had a possible cross-exposure to P. jirovecii. The BAL samples of the selected patients were subjected to MLST typing. These results were compared to MLST patterns of control samples and reference sequences to make a final conclusion whether there was evidence for an outbreak in 2013.
Materials and methods
Patient inclusion criteria
In UZ Leuven, an in-house, semi quantitative ‘real time’ PCR was used with mitochondrial rRNA as target. Based on clinical validation, a Ct value ≤28; >28 and ≤35; >35 and <45 is compatible with a positive, weak positive and very weak positive result, respectively. During the specified time period, 1023 BAL samples were analyzed for P. jirovecii. A total of 898 samples were negative, 23 samples (from 21 patients) were positive, 44 weak positive, 52 very weak positive and for six samples inhibition of the PCR reaction occurred. All 21 patients of the positive group were included. Based on the patients’ medical files a movement and transmission map was established focusing on possible contacts between these 21 patients. As mentioned above the incubation period of P. jirovecii ranges from 3 to 12 weeks with a latency period of 7 days post infection. Therefore, a period of maximum 11 weeks prior to onset of symptoms is taken into account in our search for possible cross-exposure. Further exclusion will be done based on the results of the transmission map. As a control group ten patients with a positive P. jirovecii result, hospitalized in 2014, were taken. Four of them were hospitalized in UZ Leuven, six in other Flemish hospitals but whose BAL samples were analyzed in UZ Leuven. Four had a weak positive result, the other six had a positive result. From these latter ten control patients, no patient characterization and no transmission map was established because we did not have access to their medical history. In case of the four patients hospitalized in UZ Leuven, there was evidence of a non-nosocomial infection. In case of identical genotypes, the results were interpreted according to the patients’ postal code of residence.
MLST typing
DNA extraction was performed on the Nuclisens EasyMAG (Biomerieux) platform. A 220-μL sample in lysis buffer was subjected to DNA extraction. PCR was carried out in 25-μL reactions containing 5 μL of DNA extract, 0.3 μM of each primer, 0.2 μL Platinum Taq (Life Technologies), 0.2 mM dNTP and 2.0 mM MgCl2. Primers were adopted from Maitté et al. [14].
A single round PCR on an Applied GeneAmp 9700 (Applied Biosystems) was conducted under the following conditions: 7 min at 94° followed by 40 cycles including 1 min at 94°, 1 min at 60° and a final elongation step of 1 min at 72°.
After PCR amplification, a PCR purification was performed using the QIAquick PCR purification kit (Qiagen). The purified PCR products together with the forward primers were sent to Macrogen (Amsterdam, The Netherlands) for sequencing of six fragments. Sequencing products were analyzed on an ABI 3730 XL (Applied biosystems). No sequencing analysis was performed on fragment DHPS because the wild type is most prevalent, leading to lack in differentiation. Based on a pilot study fragment 26S was not included because no good amplification product was visible on electrophoresis, probably because of a too high annealing temperature for this target. Afterwards the sequences were compared to reference sequences with accession number U07220 (ITS1), AF320344 (CYB), M58605 (mt26S), AF146753 (SOD), AF170964 (B-TUB) and AF090368 (DHFR). The following criteria were used for agreement classification: when the sequenced targets are the same for all six, the certainty of agreement is 100 %. When one or more targets are different, the strains are different. When not all targets are successfully sequenced, we assumed there was probable agreement when one target was not determined and the other targets were identical. When more than 1 target was not sequenced, no assumption was made.
Results
Movement and transmission map
A database search of the 21 patients with positive PCR result led to the exclusion of five patients, based on respiratory failure on admission and no former visits to our hospital or because of transfer from another hospital. By systematically looking into the dates of hospital visits of the remaining 16 patients, five patients could have had contact with other positive P. jirovecii patients. All of them were diagnosed in June, July and August 2013. Another five patients with a weak positive P. jirovecii result, diagnosed in the same period, were also included.
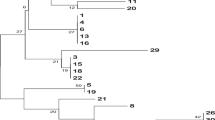
Patients 1 to 5 had a positive result for P. jirovecii, and patients 6 to 10 a weak positive result (Fig. 1). Six patients were diagnosed with hematological malignancies, one patient had received a heart transplantation, two patients underwent liver transplantation, one patient was hospitalized with status epilepticus and pulmonary embolism, but no immunosuppressive factor could be withheld. None of the patients with a weak positive result received therapy for Pneumocystis. For patients 6, 7 and 8 no exact moment of aggravating respiratory symptoms could be traced in their medical history, because they all suffered from other pulmonary infections. There was no evidence of geographic clustering according to postal code.
Figure 1 shows the transmission map of the ten patients with a positive or weak positive PCR result for P. jirovecii. When more than one sample of the same patient was analyzed, only the date of the first result was included in the transmission map. It could not be traced whether the patients were in the waiting room of the radiology department at the same time or just behind each other in the examination room. The map revealed different possibilities of transmission.
Considering that excretion of P. jirovecii is only occurring from 7 days post infection onwards and that the incubation period is at least 3 weeks, patient 5 could have received P. jirovecii from patient 4 because they had a consultation on the cardiology department on the same day. Patient 1 could have been the index patient for patient 6, because of a contact moment on the radiology department. Furthermore, patient 6 could be the index patient for patients 4, 5 and 8. There were other patient contacts but transmission was impossible because the symptoms started too early after contact.
Genotyping
Table 1 shows the results for each target based on sequencing results using only the forward primer. The loci we used were taken from Esteves et al., also used by Maitté et al. [14]. Unfortunately, not every target could be successfully sequenced, especially the ITS target, which failed for all 20 samples. Patient 2, 4, 11, 14 and 19 have multiple strains according to the mt26S target. Samples 1 and 20 have the same DHFR 312 loci and show probable agreement. However, this is between a patient of the study group and the control group, so agreement is of no significance. Samples 2, 5 and 9 also show probable agreement between strains. Samples 3 and 12 show agreement between an included patient and a control patient. Unfortunately for sample 7, three targets could not be sequenced, prohibiting a conclusion about strain agreement/concordance, and this was likewise, for samples 4 and 8, where two targets could not be sequenced successfully. For sample 6 and sample 13 the typing for CYB could not be defined with certainty because locus 279 at the beginning of the sequencing product was not clearly sequenced as only the forward primer was used. For the same reason, only locus 282 was sequenced for B-TUB. No concordance was found in the control group, suggesting a considerable geographical variability.
Discussion
MLST allows us to discriminate different genotypes. However, there is need to establish an international database of Pneumocystis genotypes independently of the used typing technique.
In total six targets were sequenced, which was successful for mt26S and CYB in most of the cases. ITS 1 amplification failed for all samples. This high failure rate was previously reported in other studies, questioning the use of this target for genotyping. One of the possible explanations could be the presence of homologous regions in yeast, present in the respiratory tract [14]. Furthermore, it is possible that the amplification of this target needs further optimization.
Interpreting both the transmission map and the sequencing results of this study, it is clear that there is no evidence for an outbreak. There were identical genotypes in the study population, but only in two patient pairs ((2-5;2–9)). The transmission map did reveal that there was no possible contact between these patients. Therefore an adjustment of the isolation policy in UZ Leuven does not seem warranted at this moment.
However, genotyping results are not always straightforward. Helweg-Larsen et al. analyzed P. jirovecii on BAL fluid and autopsied lungs of three patients pre as well as post mortem respectively. Interestingly, not all genotypes present in the lungs post mortem were found in BAL fluid ante mortem. Also the genotype is reported to change during single and recurrent episodes of PcP. The start of therapy and subsequent selection of certain genotypes could be the cause [16, 17]. Another explanation is the limitation of sampling in only one part of the lung with bronchoscopy. Hauser et al. examined BAL samples of 212 patients in ten European hospitals. In 75 % of the cases two or more genotypes were found, illustrating the diversity of P. jirovecii genotypes and consequently the limitation of genotyping [18].
The importance of colonized immunocompetent subjects in transmission of P. jirovecii is not well described. Different studies report the possible role of immunocompetent carriers in transmission to immunocompromised patients [19]. If this is the case, the search for a possible source is difficult and every healthcare worker colonized with P. jirovecii could be the source of transmission and eventually lead to an outbreak. Also, possible contacts with visitors or patients in cafeteria and other places in the hospital are difficult to trace and to include in a transmission map.
Conclusion
Both a transmission map and MLST allowed us to exclude the presence of a P. jirovecii outbreak in a large tertiary care hospital. This indicates that the current isolation policy for this pathogen can be maintained. This study shows that MLST is a useful tool for epidemiologic studies and outbreak detection. Nevertheless, a better knowledge of the geographic distribution of different P. jirovecii genotypes and an international database would be helpful for the interpretation of MLST data about this pathogen.
References
Morris A, Norris KA (2012) Colonization by Pneumocystis jirovecii and its role in disease. Clin Microbiol Rev 25(2):297–317
Ponce CA, Gallo M, Bustamante R, Vargas SL (2010) Pneumocystis colonization is highly prevalent in the autopsied lungs of the general population. Clin Infect Dis 50(3):347–53
Choukri F, Menotti J, Sarfati C, Lucet JC, Nevez G, Garin YJ, Derouin F, Totet A (2010) Quantification and spread of Pneumocystis jirovecii in the surrounding air of patients with Pneumocystis pneumonia. Clin Infect Dis 51(3):259–65
Choukri F, el Aliouat M, Menotti J, Totet A, Gantois N, Garin YJ, Bergeron V, Dei-Cas E, Derouin F (2011) Dynamics of Pneumocystis carinii air shedding during experimental pneumocystosis. J Infect Dis 203(9):1333–6
Hopkin JM (1991) Pneumocystis carinii. Oxford University Press, Oxford
Le Gal S, Damiani C, Rouillé A, Grall A, Tréguer L, Virmaux M, Moalic E, Quinio D, Moal MC, Berthou C, Saliou P, Le Meur Y, Totet A, Nevez G (2012) A cluster of Pneumocystis infections among renal transplant recipients: molecular evidence of colonized patients as potential infectious sources of Pneumocystis jirovecii. Clin Infect Dis 54(7):e62–71
Schmoldt S, Schuhegger R, Wendler T, Huber I, Söllner H, Hogardt M, Arbogast H, Heesemann J, Bader L, Sing A (2008) Molecular evidence of nosocomial Pneumocystis jirovecii transmission among 16 patients after kidney transplantation. J Clin Microbiol 46(3):966–71
de Boer MG, van Coppenraet LE B, Gaasbeek A, Berger SP, Gelinck LB, van Houwelingen HC, van den Broek P, Kuijper EJ, Kroon FP, Vandenbroucke JP (2007) An outbreak of Pneumocystis jiroveci pneumonia with 1 predominant genotype among renal transplant recipients: interhuman transmission or a common environmental source? Clin Infect Dis 44(9):1143–9
Rostved AA, Sassi M, Kurtzhals JA, Sørensen SS, Rasmussen A, Ross C, Gogineni E, Huber C, Kutty G, Kovacs JA, Helweg-Larsen J (2013) Outbreak of pneumocystis pneumonia in renal and liver transplant patients caused by genotypically distinct strains of Pneumocystis jirovecii. Transplantation 96(9):834–42
Gianella S, Haeberli L, Joos B, Ledergerber B, Wüthrich RP, Weber R, Kuster H, Hauser PM, Fehr T, Mueller NJ (2010) Molecular evidence of interhuman transmission in an outbreak of Pneumocystis jirovecii pneumonia among renal transplant recipients. Transpl Infect Dis 12(1):1–10
Hauser P, Rabodonirina M, Nevez G (2013) Pneumocystis jirovecii genotypes involved in pneumocystis pneumonia outbreaks among renal transplant recipients. Clin Infect Dis 56(1):165–6
Wakefield AE, Pixley FJ, Banerji S, Sinclair K, Miller RF, Moxon ER, Hopkin JM (1990) Detection of Pneumocystis carinii with DNA amplification. Lancet 336(8713):451–3
Helweg-Larsen J, Tsolaki AG, Miller RF, Lundgren B, Wakefield AE (1998) Clusters of Pneumocystis carinii pneumonia: analysis of person-to-person transmission by genotyping. QJM 91(12):813–20
Maitte C, Leterrier M, Le Pape P, Miegeville M, Morio F (2013) Multilocus sequence typing of Pneumocystis jirovecii from clinical samples: how many and which loci should be used? J Clin Microbiol 51(9):2843–9
Esteves F, Gaspar J, Marques T, Leite R, Antunes F, Mansinho K, Matos O (2010) Identification of relevant single-nucleotide polymorphisms in Pneumocystis jirovecii: relationship with clinical data. Clin Microbiol Infect 16(7):878–84
Helweg-Larsen J, Lee CH, Jin S, Hsueh JY, Benfield TL, Hansen J, Lundgren JD, Lundgren B (2001) Clinical correlation of variations in the internal transcribed spacer regions of rRNA genes in Pneumocystis carinii f.sp. hominis. AIDS 15(4):451–9
Hughes WT (2007) Transmission of Pneumocystis species among renal transplant recipients. Clin Infect Dis 44(9):1150–1
Hauser PM, Blanc DS, Sudre P, Senggen Manoloff E, Nahimana A, Bille J, Weber R, Francioli P (2001) Genetic diversity of Pneumocystis carinii in HIV-positive and -negative patients as revealed by PCR-SSCP typing. AIDS 15(4):461–6
Medrano FJ, Montes-Cano M, Conde M et al (2005) Pneumocystis jirovecii in general population. Emerg Infect Dis 11(2):245–50
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
No funding sources
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
For this type of study formal consent is not required.
Informed consent
Informed consent was not obtained, because of the retrospective character of the study and no mention of personal patient information. Leftovers of stored broncho-alveolar lavage fluids were used for analysis. Ethical approval was given by the ethical board hospital hygiene of the Catholic University Leuven, approved on March 20, 2015, without need for informed consent of the patients.
Rights and permissions
About this article
Cite this article
Depypere, M., Saegeman, V. & Lagrou, K. Typing of Pneumocystis jirovecii by multilocus sequencing: evidence of outbreak?. Eur J Clin Microbiol Infect Dis 35, 911–916 (2016). https://doi.org/10.1007/s10096-016-2615-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-016-2615-y