Abstract
Oplopanax elatus and Aralia cordata, commonly referred to as “Dureub” in Korea, are generally used as medicinal or food raw materials. Although O. elatus, a rare and endangered plant, is typically sold at high prices, the more abundant A. cordata is comparatively inexpensive. Given their common names and morphological root similarities, both plants can easily be confused, thereby providing potential opportunities for fraudulent use in food products. Species-specific molecular markers that can be used for quantitative real-time PCR (qPCR) analysis were developed. Verification of the six primer pairs revealed a correlation coefficient greater than 0.99, with a slope between -3.33 and -3.56. The assay confirmed specificity based on an analysis of 14 non-target plant species and verified its practicality using 10 commercial products with reliability based on a blind test. Thus, qPCR assays can contribute to food safety and protect consumer rights and interests.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
According to a report by the World Health Organization (WHO), a majority of the populations in developing countries are dependent on herbs for medicinal purposes (WHO, 2007). In addition to developing countries, as phytonutrients and dietary supplements, herbal medicines are also used in many developed countries to treat different health problems, and demand is rapidly increasing (WHO, 2004). However, with the increasing demand for these herbal products, there has been a concomitant increase in public health and safety concerns, owing to the lack of quality control or inadequate labeling of these products (Raynor et al., 2011).
Plants in the family Araliaceae, which comprises 43 genera and approximately 1,500 species, have for long been widely used for medicinal purposes or as raw food materials. The bark and leaves of these plants contain saponins, alkaloids, calcium, phosphorus, and vitamin C, which have been demonstrated to have medicinal efficacy in the treatment of a number of adult diseases and disorders, including diabetes, high blood pressure, and rheumatism (Shin, 2006). Among these plants, Oplopanax elatus and Aralia cordata, which in Korea are commonly referred to “Dureub,” are generally used medicinally or as food raw materials.
O. elatus, which is widely, although sparsely, distributed in Korea and China, is known to have physiological activities similar to those of ginseng, and is mainly used to treat fevers, coughs, and inflammation (Kim et al., 2012). In addition, the roots of O. elatus contain large amounts of falcarindiol and oplopandiol, both of which exhibit strong anticancer activity (Huang et al., 2010). However, although O. elatus, which has diverse pharmacological effects, has been used medicinally as a whole plant, owing to its requirement for specific growth conditions, it is difficult to propagate. Consequently, it has been designated as a rare and endangered plant in Korea (Seong et al., 2019). A. cordata is widely distributed in mountainous regions of Asia, including Korea and China. Its roots, which are used for medicinal purposes, have been established to have efficacy in the treatment of inflammation, cough, and diabetes (Lee et al., 2015). Unlike the O. elatus, the growth of the whole plant is vigorous. In addition, the yield tends to be high due to its strong regenerative ability (Kim et al., 1999).
Given their common name “Dureub” and the morphological similarity of their roots, which are used as medicinal materials, O. elatus and A. cordata can readily be confused. However, whereas O. elatus, a rare and endangered plant, is sold at a high price, A. cordata is comparatively inexpensive. Accordingly, to prevent the misleading substitution of O. elatus with A. cordata, it would be desirable to have a reliable discriminatory assay. To date, however, there have been no such assays developed that could be used to differentiate between these two species.
A diverse range of techniques, including DNA-based, spectroscopic, sensory, and chromatographic analyses, have been developed to detect counterfeit constituents in complex food products (Hong et al., 2017), among which, DNA-based quantitative real-time PCR (qPCR) analysis has high sensitivity and specificity, thereby facilitating the detection of very small amounts of target components (Kane and Hellberg, 2016; Kim et al., 2022). qPCR analysis is generally divided into probe-based TaqMan and fluorescent dye-based SYBR Green assays, which have high accuracy and sensitivity (Arya et al., 2005). Of these two approaches, it is relatively difficult to design probes and primers for genotyping when using probe-based TaqMan assays, as this necessitates the presence of numerous SNPs and indels between the target and non-target species (Navarro et al., 2015, Safdar and Junejo, 2015; Şakalar and Kaynak, 2016). In contrast, the SYBR Green assay-based detection of target species tends to be less expensive and more convenient than probe-based qPCR, as independent fluorescent dyes bind to DNA duplexes and can be used to detect target species with measured fluorescence values. However, false-positive results may occur (Safdar and Junejo, 2015).
In this study, we developed a molecular marker system to differentiate between O. elatus and A. cordata using qPCR analysis. The reliability of the developed assay was validated based on blind assessments and specificity tests, and its practical applicability was examined using commercial products.
Materials and methods
Sample preparation and genomic DNA extraction
The leaves and stems of O. elatus were provided by the Bioactive Natural Product Chemistry Laboratory of Kangwon National University (Chuncheon, Gangwon-do, Korea). The leaves and stems of A. cordata were kindly provided by the Gangwon-do Agricultural Promotion Agency (Chuncheon, Gangwon-do, Korea). Both samples were stored in a -70 °C deep freezer until use. All commercial plant-based products used in this study were purchased from a local market and stored at room temperature (23–24 °C).
Genomic DNA was extracted from samples using a DNeasy Plant Pro Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The concentration and purity of the extracted DNA were measured using a SPECTROstar Nano Leader microplate reader (BMG Labtech, Otenberg, Germany), and if the measured purity had a value of less than 1.7, a Wizard DNA Clean-up System (Promega, Madison, WI, USA) was used to obtain higher quality DNA. The purity of all gDNA used in this study was between 1.7 and 2.0.
Preparation of binary mixtures of O. elatus and A. cordata
Reference binary mixtures containing different amounts (0.1, 1, 10, and 100%, total 2 g) of O. elatus were mixed with A. cordata powder (99.9, 99, 90, and 0%). In addition, to evaluate the applicability of the developed markers in detecting differently processed medicinal herbs, binary mixture samples were treated as follows: (1) drying in an oven at 55 °C for 48 h, (2) boiling at 100 °C for 15 min, and (3) autoclaving at 120 °C for 15 min at 150 kPa. The gDNA of the reference binary mixtures was then extracted according to a modified large-scale CTAB-based method (Minas et al., 2011).
Sequence analysis and species-specific primer design
The sequence of the chloroplast genome of A. cordata (accession number NC_040964) was downloaded from the National Center for Biotechnology Information (NCBI) database, and the chloroplast gene sequence of O. elatus was obtained from the National Instrumentation Center for Environmental Management database (NICEM, Seoul, Korea; http://nicem.snu.ac.kr). To develop species-specific primer pairs based on sequences of the chloroplast genes of the two species, the nucleotide sequences were aligned using ClusteralW2 (EMBL-EBI, Hinston, Cambridgeshire, UK) and primers were designed on the basis of SNPs between the two species using BioEdit v.7.2 (Ibis Biosciences, Carlsbad, CA, USA). The designed primers were synthesized commercially by Macrogen (Seoul, Korea).
Optimization of the qPCR assay
Real-time PCR analysis was performed using an AccuPower® 2 × GreenStar qPCR Master Mix (Bioneer, Daejeon, Korea). The reaction mixture for this assay contained 1 μL of 10 ng μL−1 genomic DNA, 0.5 μL of each primer (10 μM), 0.25 μL of ROX Dye, 10 μL of 2 × GreenStar Master Mix, and distilled water to a final volume of 20 μL. The resulting mixture was subjected to qPCR using a Quantstudio 3 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). The qPCR conditions were as follows: an initial denaturation at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 30 s, annealing at 56–59 °C (melting temperature of each primer) for 30 s, and extension at 72 °C for 30 s. All qPCR experiments were performed in triplicate.
Construction of a qPCR standard curve and data analysis
The efficiency of the developed primer pairs was confirmed using five concentrations (10 ng/µL to 1 pg/µL) obtained by tenfold dilutions of the target DNA. The threshold level of logarithmic fluorescence was defined as the cycle threshold (Ct) in the qPCR analysis. Default parameters were used to determine the correlation between the Ct standard curve and diluted DNA. The standard curve was determined as y = − ax + b, where a is the standard curve slope and b is the y-intercept. The reaction efficiency was calculated using the following formula: E = (10–1/a − 1) × 100. The permissible range of amplification efficiency should be between 110 and 90%, which corresponds to a slope of between − 3.1 and − 3.6 (ENGL, 2015).
Inter-laboratory validation
To confirm the applicability of the developed qPCR assay, inter-laboratory validation was performed by two independent laboratories. The instruments used in this assessment experiment were a CFX Connect Real-time PCR detection system (Bio-Rad, Hercules, CA, USA) and an Applied Biosystems 7500 Fast Real-Time PCR System (Applied Biosystems), respectively. qPCR was performed in triplicate under the same conditions as those used for the developed assay.
Specificity test
To verify the cross-reactivity of the six developed primer pairs, specificity assays were conducted for 14 non-target plant species (Table 2). Plant samples of the 14 species were purchased from a local market and used in the experiments. Samples were homogeneously ground using liquid nitrogen and a mortar for subsequent DNA extraction. The extracted DNA was used to assess and verify the amplification efficiency using 18S rRNA as a positive control.
Cloning and sequencing of PCR products
Conventional PCR for sequencing the target species was performed in a VeritiPro Thermal Cycler (Thermo Fisher Scientific, Waltham, MA, USA) using TaKaRa Ex Taq™ DNA polymerase (TaKaRa Bio Company, Kusatsu, Shiga, Japan). The PCR conditions were pre-denaturation at 95 °C for 5 min; followed by 35 cycles of 95 °C for 10 s, 53–58 °C for 30 s (depending on the primer), and 72 °C for 1 min; and a final extension at 72 °C for 5 min. The PCR products thus obtained were processed as previously described (Uh and Jang, 2023).
Results and discussion
Design of DNA markers based on sequence variation
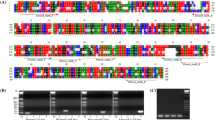
Primers for distinguishing A. cordata from O. elatus were designed based on the chloroplast genes, acetyl-CoA carboxylase subunit D (accD), β” subunit of RNA polymerase (rpoC2), ATP synthase beta subunit (atpB), and Cytochrome b6 subunit (petB), which have frequently been used for species identification in other studies. Following alignment of the sequences of A. cordata and O. elatus, primers were designed based on SNPs detected between the respective genes in the two species (Supplementary Fig. 1). Processing of different medicinal materials, such as drying, heating, and autoclaving, may reduce the quality of the DNAs contained in complex food products (Lo and Shaw, 2018). Primer pairs were designed to amplify short sequences of 92–188 bp (Table 1), given that low-quality DNA (e.g., that attributable to fragmentation) reduces the efficiency of qPCR.
Amplification efficiency and reproducibility of the developed primers
To assess the sensitivity of the six developed primer pairs (AC_rpoC2, AC_atpB, AC_accD, OE_accD, OE_rpoC2, and OE_petB), we initially performed regression analysis using a tenfold serial dilution of DNA samples (10–0.001 ng/µL). The correlation coefficient (R2) of the six primer pairs was higher than 0.99, with slopes ranging from -3.3 to -3.56. In addition, on the basis of the slope, we also assessed the amplification efficiency, which was estimated to be between 90.86% and 101.13% (Fig. 1). According to the guidelines issued by European Network of GMO Laboratories (ENGL), amplification efficiencies should be within the range between 90 and 110%, corresponding to a slope of between − 3.1 and − 3.6, with a correlation coefficient (R2) of 0.98 or more. Consequently, our results were within the acceptable range. Inter-laboratory validation was conducted using different qPCR systems in two different laboratories to assess the reproducibility of the standard curve of the six primer pairs, inter-laboratory validations were conducted using different real-time PCR systems in two different laboratories. The results, thus obtained indicated an R2 value higher than 0.99, a slope of between − 3.14 and − 3.59, and a PCR efficiency of between 90.05% and 108.27% (Supplementary Table 1). The amplification efficiency and reproducibility of all six primer pairs were accordingly considered acceptable for further study.
Threshold cycle (Ct) values were plotted against the logarithm of DNA concentration (ng) obtained from a series of tenfold serial dilutions of gDNA using primers targeting either Oplopanax elatus or Aralia cordata. The x-axis represents the log DNA concentration (ng), and the y-axis represents the mean of Ct values ± SD. Blue and orange dots indicate target and non-target species, respectively. Quantitative real-time PCR was performed in triplicate (n = 3)
Specificity validation of the developed primers
To examine the specificity of the six primer pairs, qPCR analyses were performed using 14 other plants that have been used as medicinal herbs, as well as O. elatus and A. cordata. Among these non-target plants, five Araliaceae plants, stalkless-flower eleuthero, ivy, Korean dendropanax, paperplant, and ginseng, were included on account of their close phylogenetic relationship with O. elatus and A. cordata. Furthermore, given that A. cordata is widely consumed as a vegetable plant, we also included a further five Apiaceae plants, namely, water dropwort, coriander, Korean angelica, wild carota, and parsley, which are commonly consumed as vegetable plants. In addition, as an outgroup, we examined four species from the family Compositae, which are generally used as medicinal herbs in Korea. The 18S rRNA primer pairs developed as a whole plant system (Allmann et al., 1993) were used as a positive control, and we confirmed that all amplifications were achieved prior to the 20th cycle. Thereafter, using the six newly developed primer pairs, we performed qPCR analyses of the 14 species, along with O. elatus and A. cordata (Table 2). Except for the primer pairs AC_accD and AC_rpoC2, the PCR products for the 14 test species were not amplified prior to the 40th cycles. In the case of these two primer pairs, samples of Oenanthe javanica were found to have been amplified within 40 cycles, albeit later than the designated cut-off Ct value, which was sufficient to enable an distinction of this species from the target species (A. cordata). In conclusion, all primers developed in this study showed specificity for detection of the two target species without appreciable cross-reactivity with any of the other assessed plant species.
Application of the developed qPCR assay using binary samples subjected to drying, heating, and autoclaving
Generally, commercial products of A. cordata and O. elatus undergo rigorous processing such as heating, drying, and high-pressure treatment. These processes can cause damage to the DNA of pharmaceutical constituents and complex food products. Heat treatment is known to cause particularly severe DNA damage and is accordingly unsuitable for use in PCR-based marker assays for complex food authentication (Hwang et al., 2015). Consequently, we used the developed real-time PCR assay to evaluate the applicability for commercially available herbal medicines by examining processed food samples. Initially, to assess the applicability of the designed method for commercial herbal medicines and foods, we examined the slope, R2, and efficiency using reference binary mixtures that had been subjected to drying, heating, and autoclaving. The leaves of both plants were mixed to prepare binary mixtures (0.1–100% w/w). DNA extracted from the binary mixtures was diluted to 10 ng/μL and used for the developed qPCR assay. All slopes evaluated were between − 3.23 and − 3.55, and the R2 values were higher than 0.99 for each of the six designed primer pairs. Using further dried, heated, and autoclaved leaf binary mixtures (0.1%–100% w/w), we examined the slope efficiencies for target species (Supplementary Fig. 2). Ct values were obtained for each sample treated with the three methods (Supplementary Table 2). Binary mixtures containing dried leaves had higher Ct values than untreated samples, whereas heated and autoclaved samples had higher Ct values than dried samples, which is consistent with the findings of previous studies indicating that drying leaves causes less DNA damage than heating or autoclaving. This accordingly indicates the necessity of applying different Ct values, depending on the food processing conditions, for complex food authentication (Uh and Jang, 2023). The designed real-time qPCR primer pairs showed slopes of − 3.13 to − 3.58, R2 values greater than 0.99, and efficiency values of between 90.28% and 108.78% for the dried, heated, and autoclaved binary mixtures (Table 3). An impurity of less than 0.1% in mixtures generally indicates that it cannot be acceptable an intentional mixture for economic benefit in commercial markers. Therefore, in the qPCR analysis, the target species Ct value corresponding to 0.1% in the dried binary mixture was designated as the cutoff cycle, and was considered to be sufficient for the detection of target species in commercial dry-processed products.
Blind test assessment of the developed real-time qPCR assay
To evaluate the reliability of the six developed primer pairs, we conducted a blind test using 25 randomly mixed samples, which had been prepared by an independent research group, who withheld information regarding the contents of A. cordata and O. elatus within these samples. The 18S rRNA primer set was used as a positive amplification control, resulting in low Ct values (13.31–14.27 cycles) (Table 4). The presence of O. elatus powder in the blind samples was determined based on the cut-off Ct value for the developed primer pairs (0.1% of O. elatus in a dried binary mixture). We accordingly failed to detect the cut-off Ct values of the O. elatus primer pairs in five of the 25 samples, thereby indicating that these sample comprised pure A. cordata powder and no O. elatus. Among the remaining 20 samples, we detected Ct values that were lower than the cutoff values of the primer pairs for both O. elatus and A. cordata, indicating that the blind samples contained a mixture of A. cordata and O. elatus. Thus, qPCR assays can be accurately used to authenticate the presence of A. cordata and O. elatus in complex food samples.
Application of the developed system for analysis of commercial products
To assess the applicability of the developed real-time PCR procedure for the analysis of commercial products, we analyzed nine commercially available herbal medicines labeled as containing A. cordata and one product indicated to contain O. elatus. Given its rare and endangered status, it is typically uncommon to find commercial products containing O. elatus in general food markets, and consequently we were limited to examining this single product. The qualities of the gDNA extracted from these 10 herbal medicines were confirmed using the 18S rRNA primer pair as a positive control (Allmann et al., 1993). The low Ct values (14.56–16.92) obtained using this primer pair indicated that all gDNAs extracted from the 10 commercial products were suitable for further examination (Table 5). On the basis of an evaluation of the cut-off Ct values of each of the six primer pairs, we confirmed that the DNA extracted from the O. elatus commercial product (Sample No. 1) was amplified with a lower Ct value than the cut-off of the O. elatus species-specific primers (OE_accD, OE_rpoC2, and OE_petB) at 21.11–22.04 cycles, whereas higher Ct values (or no amplification) were detected when using the A. cordata species-specific primers. These findings thus indicate that this product contains no traces of A. cordata. DNAs isolated from the purchased A. cordata products (sample nos. 2–10) showed Ct values lower than the cut-off values of the A. cordata species-specific primers (AC_rpoC2, AC_atpB, and AC_accD) (16.07 to 22.65 cycles) but higher Ct values than the cut-off value for O. elatus. Consequently, we established that none of the A. cordata products contained O. elatus as a constituent. These findings thus indicate that the newly developed primer pairs could be applied for the authentication of commercial herbal products.
Conclusion
The real-time PCR assay developed in this study provides a highly sensitive, reproducible, and rapid method for detecting the desired target species in processed herbal medicines and complex food products. To detect samples of the target species, we constructed standard curves based on a standard curve test and a two-way mixture test, and evaluated the specificity of the developed primer pairs using 14 different non-target plants. In addition, we verified the reliability of the developed DNA markers by conducting a blind test, and assessed the applicability of the developed assay in the authentication of commercial products. This real-time PCR assay would be a particularly effective method for discriminating A. cordata and O. elatus, and could accordingly make a valuable contribution to the protection of consumers’ rights and interests, as well as ensuring food safety.
Change history
19 October 2023
A Correction to this paper has been published: https://doi.org/10.1007/s10068-023-01454-3
References
Allmann M, Candrian U, Höfelein C, Lüthy J. Polymerase chain reaction (PCR): A possible alternative to immunochemical methods assuring safety and quality of food detection of wheat contamination in non-wheat food products. Zeitschrift für Lebensmittel-Untersuchung und Forschung. 196(3): 248-251 (1993)
Arya M, Shergill IS, Williamson M, Gommersall L, Arya N, Patel HR. Basic principles of real-time quantitative PCR. Expert Review of Molecular Diagnostics. 5(2): 209-219 (2005)
CBOL CPW, Hollingsworth PM, Forrest LL, Spouge, JL, Hajibabaei M, Ratnasingham S, Bank M, Chase MW, Cowan RS, Erickson DL, Fazekas AJ, Graham SW, James KE, Kim KJ, Kress WJ, Schneider H, AlphenStahl J, Barrett SCH, Berg C, Bogarin D, Burgess KS, Cameron KM, Carine M, Chacón J, Clark A, Clarkson JJ, Conrad F, Devey DS, Ford CS, Hedderson TAJ, Hollingsworth ML, Husband BC, Kelly LJ, Kesanakurti PR, Kim JS, Kim YD, Lahaye R, Lee HL, Long DG, Madriñán S, Maurin O, Meusnier I, Newmaster SG, Park CW, Percy DM, Petersen G, Richardson JE, Salazar GA, Savolainen V, Seberg O, Wilkinson MJ, Yi DK, Little DP. A DNA barcode for land plants. Proceedings of the National Academy of Sciences. 106(31): 12794-12797 (2009)
ENGL (European Network of GMO Laboratories). Definition of Minimum Performance Requirements for Analytical Methods of GMO Testing. Available online: http://gmocrl.irc.ec.ecrl.jrc.ec.europa.eu/doc/MPR%20Report%20Application%2020_10_2015.pdf (accessed on 20 October 2015)
Hong E, Lee SY, Jeong JY, Park JM, Kim BH, Kwon K, Chun HS. Modern analytical methods for the detection of food fraud and adulteration by food category. Journal of the Science of Food and Agriculture. 97(12): 3877-3896 (2017)
Huang W, Yang J, Zhao J, Wang CZ, Yuan CS, Li SP. Quantitative analysis of six polyynes and one polyene in Oplopanax horridus and Oplopanax elatus by pressurized liquid extraction and on-line SPE–HPLC. Journal of Pharmaceutical and Biomedical Analysis. 53(4): 906-910 (2010)
Hwang SG, Kim JH, Moon JC, Jang CS. Chloroplast markers for detecting rice grain-derived food ingredients in commercial mixed-flour products. Genes & Genomics. 37(12): 1027-1034 (2015)
Kane DE, Hellberg RS. Identification of species in ground meat products sold on the US commercial market using DNA-based methods. Food Control. 59: 158-163 (2016)
Kim Y, Lee H, Kim Y. Ground nut (Aralia cordata Thunberg) leaves and stems as a forage source, chemical composition of leaves, stems and roots, and dry matter digestibility in the rumen. Korean Journal of Agricultural Science. 26(1): 58-64 (1999)
Kim HY, Seong ES, Lee JG, et al. Establishment of suspension culture system to induce somatic embryo in Oplopanax elatus Nakai. Korean Journal of Medicinal Crop Science. 20: 461-465 (2012)
Kim YD, Uh YR, Jang CS. Development of real-time PCR based molecular markers for two medicinal herb Artemisia species A. capillaris and A. iwayomogi. Food Science and Biotechnology. 32: 59-69 (2023)
Lee S G, Jo DJ, Chang HJ, Kang H. Antioxidant and anti-inflammatory activities of ethanol extracts from Aralia continentalis Kitagawa. Journal of Naturopath. 4(1): 10-14 (2015)
Lo YT, Shaw PC. DNA-based techniques for authentication of processed food and food supplements. Food Chemistry. 240: 767-774 (2018)
Minas K, McEwan NR, Newbold CJ, Scott KP. Optimization of a high-throughput CTAB-based protocol for the extraction of qPCR-grade DNA from rumen fluid, plant and bacterial pure cultures. FEMS Microbiology Letters. 325: 162-169 (2011)
Raynor DK, Dickinson R, Knapp P, Long, AF, Nicolson DJ. Buyer beware? Does the information provided with herbal products available over the counter enable safe use? BioMed Central medicine. 9(1): 1-9 (2011)
Safdar M, Junejo Y. Development and validation of fast duplex real-time PCR assays based on SYBER Green florescence for detection of bovine and poultry origins in feedstuffs. Food chemistry. 173: 660-664 (2015)
Şakalar E, Kaynak A. Practical molecular detection method of beef and pork in meat and meat products by intercalating dye based duplex real-time polimerase chain reaction. International Journal of Food Properties. 19(1): 31-40 (2016)
Seong ES, Yoo JH, Kim HY, Choi HL, Seo JW, Hwang MH, Kim Mj, Yu CT. Establishment of acclimatization system and growth characteristics for regenerated plants of Oplopanax elatus Nakai. Korean Journal of Medicinal Crop Science. 27(6): 397-403 (2019)
Shin KH. Effects of Araliaceae hot water extract on blood sugar and blood components in diabetic rats. Journal of the Korean Society of Nutrition. 39(8): 721-727 (2006)
Uh YR, Jang CS. Establishing DNA markers to differentiate Agastache rugosa and Pogostemon cablin, which are confusedly used as medicinal herbs, using real-time PCR. Food Science and Biotechnology. 32: 239-247 (2023)
World Health Organization (WHO). Health of Indigenous Peoples. World Health Organization, Geneva, pp. 1132 (2007)
World Health Organization (WHO). WHO guidelines on safety monitoring of herbal medicines in pharmacovigilance systems. World Health Organization. (2004)
Acknowledgements
This study was supported by a grant (22193MFD471) from the Ministry of Food and Drug Safety in 2022.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Uh, Y.R., Kim, Y.M., Kim, M.J. et al. Development of real-time PCR-based markers for differentiation of Oplopanax elatus and Aralia cordata in commercial food products. Food Sci Biotechnol 32, 2153–2161 (2023). https://doi.org/10.1007/s10068-023-01313-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10068-023-01313-1