Abstract
Centipedes contain pharmacologically active compounds used as important medicinal material. However, the poisons produced by centipedes can cause human diseases; therefore, its use as a food ingredient is prohibited. This is the first report to develop a real-time PCR method for detection of centipedes. The primer and probe targeting the mitochondrial cytochrome c oxidase subunit 1 (COI) gene were newly designed. The specificity was verified using ten species and was confirmed to amplify only the centipede species. The real-time PCR method exhibited good linearity with a high-determination coefficient (R2 = 0.999) and a detection limit was 0.001 ng. The performance of our method was also verified using five real-time PCR platforms under Universal and Fast PCR conditions. Finally, its applicability to processed food was evaluated using binary insect mixtures, and at least 0.1% of centipedes was detected. Therefore, our method can specifically and sensitively detect centipedes in food, contributing to food safety.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Arthropods of the Scolopendra genus are extensively distributed globally, especially in Southeast Asia, North or Central America, and North Africa (Kang et al., 2017; 2019). Scolopendra species have been used for medicinal purposes at long time with Bombyx mori, Cordyceps militaris, and Protaetia brevitarsis seulensis in Asia (Kang et al., 2017). The centipedes are arthropods in the Scolopendra genus used as an important source of herbal medicine due to their high-medical value such as anti-inflammatory, analgesic, and antitumor activities (Han et al., 2018; Kang et al., 2019; Liu et al., 2020). However, they produce venomous and toxic substances that contain numerous active ingredients, including enzymatic proteins (acid/alkaline phosphatases, esterases, and hyaluronidases), non-enzymatic proteins (cardiotoxins and mycotoxins), and non-peptide active ingredients (histamine and serotonin), which pose a high risk to humans (Guo et al., 2013; He et al., 2019). Furthermore, infection with centipede venom can cause symptoms such as myocardial ischemia, infarction, hemoglobinuria, fever, chills, rash, eosinophilic cellulitis, and anaphylaxis (Kong et al., 2013).
Arthropods have been used for medicinal purposes long time in Asia, with the most commonly consumed arthropods worldwide include silkworm, white-spotted flower chafer, and centipedes (Kang et al., 2017). However, the regulatory agency in Korea strictly prohibits the manufacture of centipedes as food except to use herbal medicine because centipedes are poisonous (Ministry of Food and Drug Safety, 2010). Although centipedes were mostly imported from China as herbal medicines, domestic breeding became possible with the development of breeding technology (Rural Development Administration, 2011; Yoon, 2014). In addition, it was included as the insect industry in the category of livestock, it is bred in the form of a farm (Ministry of Agriculture, Food and Rural Affair, 2019). For these reason, centipedes are easily accessible to the public at herbal markets without a special permit, resulting in a high likelihood of misuse as a folk remedy or functional food. In reality, there has been a case of the illegal manufacture and selling of functional foods mixed with steroids and centipede and athlete who was actually taking it was caught for doping (Ministry of Food and Drug Safety, 2011). Thus, considering consumer protection and food safety, it is crucial to rapidly and accurately detect unintentional contamination and intentional product fraud from non-approved centipedes as food ingredients (Hong et al., 2017).
Many techniques have been applied to detect adulteration and fraud in food, such as thin layer chromatography (TLC), matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS), polymerase chain reaction (PCR), real-time PCR, PCR-restriction fragment length polymorphism (RFLP), loop-mediated isothermal amplification (LAMP), and near-infrared (NIR) spectroscopy (Hong et al., 2017; Kim et al., 2019a; 2019b; Kim et al., 2022; Li et al. 2019a; 2019b; Lopez-Calleja et al., 2005; Ochiai and Komiya, 2021; Suh et al., 2020; Ulrich et al., 2017). However, the protein-based methods have weaknesses, such as less sensitivity when applied to thermally treated foods due to protein denaturation and alteration in specific epitopes (Li et al. 2019a; 2019b; Melgar-Lalanne et al., 2019). Conversely, DNA-based PCR methods have been mostly applied in food adulteration and fraud detection due to their higher thermal stability than protein-based methods (Kim et al., 2019a; 2019b). Furthermore, real-time PCR has significant specificity for fresh, digested, or processed samples than other techniques enabling it to rapidly and accurately detect target DNA in food (Kim et al., 2019a; 2019b). In particular, centipedes were identified by the morphological classification, near-infrared spectroscopy, and sequencing methods in previous studies (Kang et al., 2017, 2019), but PCR-based methods, including real-time PCR, that can detect centipedes rapidly and sensitively has not been reported.
In order to improve the specificity and sensitivity of PCR-based methods, the selection of target genes or target sequences is a crucial factor (Kim et al., 2021). Mitochondrial genomes have been targeted to eukaryote by real-time PCR methods (Li et al. 2019a; 2019b; Sul et al., 2020). Among the mitochondrial genes, the cytochrome c oxidase subunit 1 (COI) gene was mainly used as a genetic marker to distinguish eukaryotic species (Kim et al., 2019a; 2019b). Although the COI gene is highly conserved in the same species, it has a high-DNA variation between different species (Hebert et al., 2003). Toxic substances produced by centipedes can cause the illness to humans, so toxin-producing genes could be a candidate for primer targets. However, this gene has a low copy number, and missing expression of a toxic gene can lead to false-negative results (Kang et al., 2017). On the other hand, the mitochondrial genome can sensitively detect target species even when the DNA in the samples is degraded minutely with processed foods due to the high-copy number of DNA in the cell than nuclear DNA like toxic-producing gene (Girish et al, 2004). Therefore, in many studies, mitochondrial genomes have been targeted to eukaryotes by real-time PCR methods (Li et al. 2019a; 2019b; Sul et al., 2020).
In this study, we designed a new primer and probe set targeting the COI gene in centipedes. Additionally, we developed the real-time PCR method to rapidly and accurately detect centipedes in foods, preserving the consumers from their presence.
Materials and methods
Sample preparations
The four types of animals ingredient used at herbal medicines (centipede, Scolopendra mutilans; earthworm, Lumbricina sp.; gecko, Gekko gecko; blister beetle, Hycleus cichorii) and six types of edible insects (two-spotted cricket, Gryllus bimaculatus; migratory locust, Locusta migratoria; rhinoceros beetle larvae, Allomyrina dichotoma; honey pupa, Apis mellifera; rice grasshopper, Oxya chinensis; silkworm, Bombyx mori) were obtained from the National Institute of Biological Resources (NIBR, Incheon, Korea), the Rural Development Administration (RDA, Jeonju, Korea), and the herbal medicine local market in Korea. All specimens were homogenized in liquid nitrogen and stored at − 20 °C until use.
DNA extraction
DNA of each sample was extracted using a DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions with minor modifications. Briefly, 25 mg of ground sample was lysed with 400 μl of ATL buffer and 20 μl of proteinase K (20 mg/ml) at 56 °C for 2 h. After adding 4 μl of RNase A, the reaction mixture was incubated at room temperature for 2 min. Then, 400 μl of chloroform was added and centrifuged at 11,600×g for 15 min. Finally, after washing with washing buffers, the column-bound DNA was eluted with distilled water. The DNA concentration and purity were measured using a UV/VIS Spectrophotometer (Mecasys, Daejeon, Korea).
Design of primer and probe
The mitochondrial COI genes of centipede (Scolopendra mutilans, accession no. MG991738.1), Blister beetle (Hycleus cichorii, accession no. NC039657.1), Earthworm (Lumbricina sp., accession no. HQ405776), Gecko (Gekko gecko, accession no. AY282753), Rhinoceros beetle (Allomyrina dichotoma, accession no. LC074686.1), Honeybee pupa (Apis mellifera, accession no. L06178.1), Silkworm (Bombyx mori, accession no. NC002355.1), Migratory locust (Locusta migratoria, accession no. NC001712.1), Rice grasshopper (Oxya chinensis, accession no. EF437157.1), Rice grasshopper (Oxya japonica, accession no. KC261378.1), Two-spotted cricket (Gryllus bimaculatus, accession no. KR071876.1) were obtained from the GenBank database. The sequences of centipedes and the non-target species were aligned using the online ClustalW2 software. The primer and probe set for identification of centipede was designed using Primer Designer version 3.0 (Scientific and Educational Software, Durham, NC, USA). The primer and probe were synthesized by Bionics (Seoul, Korea). The nucleotide sequences of the primer and probe used in this study are shown in Table 1.
Specificity and sensitivity of real-time PCR method
The specificity of designed primer was confirmed using target species and nine non-target species. The standard curve was generated using serially diluted DNA extracted from centipedes (from 10 to 0.001 ng).
A real-time PCR method was performed with a 7500 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). The reaction mixture contained 12.5 μl of TaqMan™ Universal PCR Master Mix (Applied Biosystems), 400 nM of primer, 200 nM of probe, 10 ng of template DNA, and distilled water to a final volume of 25 μl. Amplification was performed with a denaturation step at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 60 s. Finally, the PCR results were analyzed using the 7500 Software version 2.3 (Applied Biosystems). Notably, all reactions were performed in triplicates.
Verification of the developed real-time PCR method
Four real-time PCR platforms (ViiA7 Real-Time PCR System, 7500 Fast Real-Time PCR System, StopOnePlus Real-Time PCR System, and QuantStudio 3 Real-Time PCR System) were used to verify the performance of the developed method and compared under Universal and Fast real-time PCR conditions.
The conditions of the ViiA7 Real-Time PCR System (Applied Biosystems) are shown in ‘Specificity and sensitivity of real-time PCR method’ section. PCR amplification conditions using the 7500 Fast Real-Time PCR System (Applied Biosystems), StepOnePlus Real-Time PCR System (Applied Biosystems), and QuantStudio 3 Real-time PCR System (Applied Biosystems) are as follows: 500 nM of primers, 250 nM of probe, 10 μl of TaqMan™ Universal Fast PCR Master Mix (Applied Biosystems), 10 ng of the template DNA, and distilled water to a final volume of 20 μl. Real-time PCR condition involved a holding stage at 95 °C for 20 s, followed by 40 cycles at 95 °C for 3 s and 60 °C for 30 s. The Universal and Fast real-time PCR conditions are shown in Table 2.
Applicability of the real-time PCR method
The binary mixture sample was prepared to measure the detection limit of centipedes in processed foods. First, binary mixture samples containing 0.1%, 0.5%, 1%, 5% and 10% of centipede powder in silkworm powder were prepared, and then 100 mg of the binary mixture sample was immediately extracted for analysis. The detection limit of the binary mixture sample was confirmed using five real-time PCR platforms (7500 Real-Time PCR system, ViiA7 Real-Time PCR System, 7500 Fast Real-Time PCR system, StepOnePlus Real-Time PCR system, and QuantStudio 3 Real-Time PCR system) under the Universal and Fast conditions.
Results and discussion
Specificity test
The accuracy of real-time PCR depends on the specificity of the target gene or sequence used in the experiment (Kim et al., 2021). Mitochondrial genes are highly conserved in different animal species (Kang et al., 2017; van der Kuyl et al., 1995). Mitochondrial DNA has been used to develop detection methods since its high-copy number, and small circular shape permit greater stability in numerous processing conditions (Kim et al., 2019a; 2019b; Sentandreu and Sentandreu, 2014). Furthermore, mitochondrial genes, such as the D-LOOP gene, 12S rRNA and 16S rRNA genes, COI gene, and the cytochrome b gene, have been used for animal species identification in foods (Li et al., 2019a; 2019b; Ghovvati et al., 2009; Wang et al., 2021). The COI gene plays an important role in eukaryote metabolism and has been mostly used as a target gene for distinguishing closely related species.
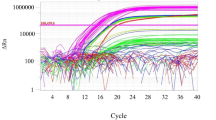
In this study, new primer and TaqMan probe set was designed to target the COI gene of Scolopendra mutilans (accession number MG991738.1) after aligning with the mitochondrial COI region of nine non-target species (Fig. 1). The amplicon size was 154 bp, increasing the target DNA detectability in processed foods. The specificity was evaluated using a target centipede (Scolopendra mutilans) and nine non-target species. The target DNA of the centipede was successfully amplified (Ct value = 18.83 ± 0.08), and no amplification was observed in the nine non-target DNA tests, revealing that the primer and probe were specific for centipede detection (Table 3). These results indicate that our method is specific and accurate assay for detecting centipede DNA.
Sensitivity test
A standard curve for the centipede (Scolopendra mutilans) was constructed using serial dilution of centipede DNA. A standard curve had a slope of − 3.57, and the coefficient of correlation (R2) was 0.999 (Fig. 2). The amplification efficiency, calculated from the equation, E = [10 (− 1/slope) − 1], was 90.64%, further highlighting this method's suitability in detecting centipede DNA. To generate a valid primer and probe set, the R2 and slope values for the standard curve should be ≥ 0.98 and − 3.1 to − 3.6, respectively (Broeders et al., 2014). Hence, the real-time PCR method is a highly efficient technique for centipede (Scolopendra mutilans) detection.
The detection limit in centipede DNA was 0.001 ng, with a Ct value of 32.99 ± 0.08 (mean ± standard deviation). Previous studies have identified the detection limit of real-time PCR methods to detect eukaryotes used for the edible or medicinal purposes (Kim et al., 2019a; 2019b; Quek et al., 2018). Kim et al. (2019a; 2019b) obtained a detection limit value of 0.001 ng for edible rice grasshopper (Oxya chinensis) DNA by real-time PCR method. Additionally, Quek et al. (2018) developed a real-time PCR method to quantify an edible bird (Aerodramus fuciphagus) with a detection limit of 0.004 ng. Therefore, the detection limit value obtained from our study is similar to or slightly lower than in previous studies.
Verification of the real-time PCR method
In general, the detection values of real-time PCR platforms from the same company do not show significant differences, but there may be differences in detection values between different platforms (Lu et al., 2010). The performance of the real-time PCR method was verified using four different real-time PCR platforms (ViiA7 Real-Time PCR System, 7500 Fast Real-Time PCR System, StepOnePlus Real-Time PCR System, and QuantStudio3 Real-Time PCR System). The ViiA7 Real-Time PCR System was performed under the same Universal conditions as the 7500 Real-Time PCR System. Conversely, the 7500 Fast Real-Time PCR System, StepOnePlus Real-Time PCR System, and QuantStudio3 Real-Time PCR System were amplified under Fast real-time PCR conditions.
The slopes for the ViiA7 Real-Time PCR System, 7500 Fast Real-Time PCR System, StepOnePlus Real-Time PCR System, and QuantStudio3 Real-Time PCR System were − 3.30, − 3.54, − 3.56, and − 3.22, respectively, and the R2 value was 0.998, 0.998, 0.997, and 0.993, showing high linearity. The amplification efficiencies of the four real-time PCR platforms ranged from 90.91 to 104.824%, revealing a high efficiency (Broeders et al., 2014). Moreover, the detection limit in centipede DNA was 0.001 ng in both Universal and Fast real-time PCR conditions. Therefore, the primer and probe desgined in this study were able to sensitively and accurately detect centipede DNA under Universal conditions (within 100 min) and Fast real-time PCR conditions (within 40 min).
Application of real-time PCR method in binary mixture samples
Furthermore, binary mixture samples were applied to evaluate the applicability to food samples contaminated with centipedes. The analytical detection limit for food samples contaminated with centipedes was tested three times on different days using binary mixture samples containing 0.1%, 0.5%, 1%, 5% and 10% centipedes mixed with silkworm powder. The detection limit of five real-time PCR platforms were 0.1% centipede in silkworm powder (Table 4). Both the Universal and Fast conditions shoewed the same detection limit. To verify the applicability of processed food, in the previous studies, various types of binary mixture samples were prepared, and limit of detection was confirmed. Amaral et al. (2014) allowed 0.1% sensitivity for a binary mixture sample containing red deer in pork meat. Also, Kim et al. (2019a; 2019b) obtained a 0.1% detection limit value for a binary mixture sample containing rice grasshopper in the Bombay locust by rice grasshopper-specific real-time PCR method, consistent with our study. These results propose that the sensitivity of this method is sufficient for detecting centipedes in food samples.
This assay is the first to report a real-time PCR method to detect centipede (Scolopendra mutilans) DNA in food. Target DNA was successfully detected under both Universal (within 100 min) and Fast real-time PCR conditions (within 40 min). The analysis showed a sensitivity of 0.1% for binary mixture samples containing centipedes in silkworm, suggesting its usefulness in identifying unintentional contaminants and intentional product fraud using centipedes. Although binary mixture samples were used to evaluate the applicability of food samples contaminated with centipedes, it will be necessary to detect centipedes contained in various food matrices in the future. The real-time PCR method might be a rapid, sensitive, and efficient method to manage illegal health functional foods or unintentional adulteration of centipedes in food.
References
Amaral J, Santos C, Melo V, Oliveira M, Mafra I. Authentication of a traditional game meat sausage (Alheira) by species-specific PCR assays to detect hare, rabbit, red deer, pork and cow meats. Food Research International. 60: 140-145 (2014)
Broeders S, Huber I, Grohmann L, Berben G, Taverniers I, Mazzara M, Roosens N, Morisset D. Guidelines for validation of qualitative real-time PCR methods. Trends in Food Science & Technology. 37(2): 115-126 (2014)
Ghovvati S, Nassiri M, Mirhoseini S, Moussavi A, Javadmanesh A. Fraud identification in industrial meat products by multiplex PCR assay. Food Control. 20(8): 696-699 (2009)
Girish P, Anjaneyulu A, Viswas K, Anand M, Rajkumar N, Shivakumar B, Bhaskar S. Sequence analysis of mitochondrial 12S rRNA gene can identify meat species. Meat Science. 66(3): 551-556 (2004)
Guo J, Zhang R, Wang J, Miao L. Molecular cloning and characterization of a new cDNA encoding a trypsin-like serine protease from the venom gland of Scolopendra subspinipes mutilans. African Journal of Pharmacy and Pharmacology. 7(17): 1054-1060 (2013)
Han T, Lee Y, Kim S, Yoon H, Park I, Park H. Genetic variation of COI gene of the Korean medicinal centipede Scolopendra mutilans Koch, 1878 (Scolopendromorpha: Scolopendridae). Entomological Research. 48(6): 559-566 (2018)
He X, Lee K, Kim B, Lee K, Ko h, Jia J, Yoon H, Jin B. Centipede (Scolopendra subspinipes mutilans) venom toxin peptide exhibits cytotoxic and cell growth effects in a concentration-dependent manner. Journal of Asia-Pacific Entomology. 22(1): 19-24 (2019)
Hebert P, Ratnasingham S, DeWaard J. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely reated species. Proceedings of the Royal Society of London. Series B: Biological Scienses. 270 (2003)
Hong E, Lee S, Jeong J, Park J, Kim B, Kwon K, Chun H. Modern analytical methods for the detection of food fraud and adulteration by food category. Journal of the Science of Food and Agriculture. 97(12): 3877-3896 (2017)
Kang S, Deng H, Chen L, Zeng X, Liu Y, Chen K. Rapid identification and quality evaluation of medicinal centipedes in China using near-infrared spectroscopy integrated with support vector machine algorithm. Journal of Spectroscopy. 2019 (2019)
Kang S, Liu Y, Zeng X, Deng H, Luo Y, Chen K, Chen S. Genus Scolopendra in China using integrated methods of external morphology and molecular phylohenetics. Scientific Reports. 7(1): 1-14 (2017)
Kim S, Kim M, Jung S, Kim H. Development od a fast real-time PCR assay based on TaqMan probe for identification of edible rice grasshopper (Oxya chinensis) in processed food products. Food Research International. 116: 441-446 (2019a)
Kim M, Kim S, Jung S, Kim M, Kim H. Development and validation of ultrafast PCR assay to detect six species of edible insects. Food Control. 103: 21-26 (2019b)
Kim E, Yang S, Kim D, Kim H. Real-time PCR method for qualitative and quantitative detection of Lactobacillus sakei group species targeting novel markers based on bioinformatics analysis. International Journal of Food Microbiology. 355: 109335 (2021)
Kim H, Suh S, Kang H, Shin S, Kim H. Duplex loop-mediated isothermal amplification assay for peanut (Arachis hypogaea) and almond (Prunus dulcis) detection of allergen coding genes. Food Control. 138: 109003 (2022)
Kong Y, Hui J, Shao Y, Huang S, Chen H, Wei J. Cytotoxic and anticoagulant peptide form Scolopendra subspinipes mutilans venom. African Journal of Pharmacy and Pharmacology. 7(31): 2238-2245 (2013)
Li T, Jalbani Y, Zhang G, Zhao Z, Wang Z, Zhao X. Detectio of goat meat adulteration by real-time PCR based on a reference primer. Food Chemistry. 277: 554-557 (2019a)
Li J, Li J, Xu S, Xiong S, Yang J, Chen X, Wang S, Qiao X, Zhou T. A rapid and reliable multiplex PCR assay for simultaneous detection of fourteen animal species in two tubes. Food Chemistry. 295: 395-402 (2019b)
Liu Z, Liang J, Lan X, li T, Zhang J, Zhao F, Li G, Chen P, Zhang Y, Lee W, Zhao F. Comparative analysis of diverse toxins from a new pharmaceutical centipede, Solopendra mojiangica. Zoological Research. 41(2): 138 (2020)
Lopez-Calleja I, Gonzaiez Alonso I, Fajardo V, Rodriguez M, Hernandez P, Garcia T, Martin R. PCR detection of cow’s milk in water buffalo milk and mozzarella cheese. International Dairy Journal. 15(11): 1122-1129 (2005)
Lu S, Smith A, Moore D, Lee N. Different real-time PCR systems yield different gene expression values. Molecular and Cellular Probes. 24(5): 315-320 (2010)
Melgar-Lalanne G, Alvarez A, Salinas-Castro A. Edible insects processing: Traditional and innovative technologies. Comprehensive Reviews in Food Science and Food Safety. 18(4): 1166-1191 (2019)
Ministry of Food and Drug Safety. Regulations on raw materials that cannot be used in health functional foods. Available from: https://www.law.go.kr/LSW/admRulLsInfoP.do?admRulSeq=2000000056454#AJAX. Accessed Apr. 13, 2010.
Ministry of Food and Drug Safety. Illegal food manufacturers and serllers caught by mixing steroids and centipedes. Available from: https://www.korea.kr/news/pressReleaseView.do?newsId=155800260. Accessed Dec. 1, 2011.
Ministry of Agriculture, Food and Rural Affair. Act on the promotion and support of the insect industry. Available from: https://www.law.go.kr/%EB%B2%95%EB%A0%B9/%EA%B3%A4%EC%B6%A9%EC%82%B0%EC%97%85%EC%9D%98%EC%9C%A1%EC%84%B1%EB%B0%8F%EC%A7%80%EC%9B%90%EC%97%90%EA%B4%80%ED%95%9C%EB%B2%95%EB%A5%A0. Accessed July 1, 2019.
Ochiai M, Komiya Y. Detection of edible insect derived phospholipids with polyunsaturated fatty acids by thin-layer chromatography, gas chromatography, and enzymatic methods. Journal of Food Composition and Analysis. 99: 103869 (2021)
Quek MC, Chin NL, Tan SW, Yusof YA, Law CL. Molecular identification of species and production origins of edible bird’s nest using FINS and SYBR green I based real-time PCR. Food Control. 84: 118-127 (2018)
Rural Development Administration. Mass breeding and industrialization of medicinal insects. Available from: https://www.rda.go.kr/board/board.do?mode=view&prgId=day_farmlcltinfoEntry&dataNo=100000456559. Accessed Sept. 19, 2011.
Sentandreu M, Sentandreu E. Authenticity of meat products: Tools against fraud. Food Research International. 60: 19-29 (2014)
Suh S, Kim M, Kim H, Kim H, Kim H. A multiplex PCR assay combined with capillary electrophoresis for the simultaneous detection of tropomyosin allergens from oyster, mussel, abalone, and clam mollusk species. Food Chemistry. 317: 126451 (2020)
Sul S, Kim M, Lee J, Kim S, Kim H. Development of a rapid on-site method for the detection of chicken meat in processed ground meat products by using a direct ultrafast PCR system. Journal of Food Protection. 83(6): 984-990 (2020)
Ulrich S, Kuhn U, Biermaier B, Piacenza N, Schwaiher K, Gottschalk C, Gareis M. Direct identification of edible insects by MALDI-TOF mass spectrometry. Food Control. 76: 96-101 (2017)
van der Kuyl A, Kuiken C, Dekker J, Goudsmit J. Phylogeny of African monkeys based upon mitochondrial 12S rRNA sequences. Journal of Molecular Evolution. 40(2): 173-180 (1995)
Wang L, Cheng H, Zhou Y, Ma M. Clinical significance of the D-loop gene mutation in mitochondrial DNA in laryngeal cancer. Onco Targets and Therapy. 14: 3461 (2021)
Yoon CH. Method and apparatus for growing centipede. KR20140064568A (2014)
Acknowledgements
This research was supported by Grant 17162MFDS065 from the Ministry of Food and Drug Safety in Korea.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Suh, SM., Kim, E., Kim, MJ. et al. Development of real-time PCR method for rapid and accurate detection of Centipedes (Scolopendra mutilans) in food. Food Sci Biotechnol 32, 979–985 (2023). https://doi.org/10.1007/s10068-022-01231-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10068-022-01231-8