Abstract
The nutritional requirements for antimicrobial activity of Streptomyces rimosus AG-P1441 were optimized using statistically-based experimental designs at a flask level. Based on a one-factor-at-a-time (OFAT) approach, glucose, corn starch and soybean meal were identified as the carbon and nitrogen sources having a significant effect on antimicrobial productivity. As a result of investigating the effect of glucose concentration, the highest antimicrobial activity was observed at 3% concentration. Response surface methodology (RSM) was then applied to optimize the growth medium components (corn starch, soybean meal, MgCl2 and glutamate). Antimicrobial productivity increased sharply when the medium consisted of 3% glucose, 3.5% corn starch, 2.5% soybean meal, 1.2 mM MgCl2 and 5.9 mM glutamate. The fermentation using optimized culture medium in a 5-L bioreactor allowed a significant increase in antimicrobial activity, evaluated by the paper disc assay, revealed a 29 mm inhibition zone diameter against Phytophthora capsici.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phytophthora blight caused by the oomycete Phytophthora capsici is one of the most economically destructive soil-borne diseases in chili pepper production worldwide [1]. P. capsici causes damping-off, root rot, vine blight, leaf blight or fruit rot on more than 50 host plant species including vegetables and weeds [2]. In general, it is difficult to manage diseases caused by Phytophthora spp. because of their aggressiveness and increasing resistance to chemical compounds [3]. The extensive and widespread application of chemical pesticides has not only resulted in the development of resistance of these pathogens against chemicals but become a major source of environmental pollution and ecosystem damage [4]. Thus, biological control has become an important strategy to manage soil-borne diseases and to reduce the application of chemical pesticides [2].
Numerous surveys of soil bacteria have identified strains of Streptomyces and Bacillus as potential biocontrol agents (BCAs) [5–9]. Bacterial genera, such as Streptomyces, Flavobacterium, Pseudomonas, Lysobacter and Bacillus, have been reported to suppress growth and asexual development of, and control plant diseases caused by Phytophthora sp. [4]. Actinomycetes have been recognized as producers of secondary metabolites and antibiotics, and several Streptomyces spp. have been studied as potential BCAs against fungal pathogens [5–7].
We previously studied the antimicrobial activity of S. rimosus AG–P and found strong antimicrobial activities against Fusarium oxysporum, Alternaria panax, Cylindrocarpon destructans and Alternaria porri and, in particular, Pythium ultimum and P. capsici [10]. Thus, our previous work showed that S. rimosus AG-P could be a new BCA against oomycete diseases and a potent inhibitor of Pythium spp. [5].
Specific nutritional requirements of microorganisms used in industrial fermentation processes are as complex and diverse as the microorganisms. As a method for optimizing the medium, there is a general practice of determining optimal concentration of media components by varying one-factor-at-a-time (OFAT). This method does not depict the net effect of total interactions among various media components. In this respect, media optimization is carried out by response surface methodology (RSM). Statistical optimization not only allows quick screening of a large experimental domain, but also reflects the role of each of the components [11]. The aim of the present study was to statistically optimize a production medium for improved antimicrobial compound production of S. rimosus AG-P1441 in the means of antimicrobial activity. The information obtained is expected to assist in the large-scale production of BCAs.
Materials and methods
Microorganism
AG-P1441, a mutant of S. rimosus AG-P, was used throughout this study. This strain was maintained on Bennet agar composed of (g/L): glucose, 10; yeast extract, 1; beef extract, 1; peptone, 2 and agar, 20, and stored in 20% glycerol at − 80 °C. It was a highly producing mutant isolated and identified after N-methyl-N′-nitro-N-nitrosoguanidine (NTG) and UV treatment of a parent strain AG-P in our laboratory (unpublished data).
The fungal pathogen P. capsici, a tested pathogenic strain, was obtained from the Korean Agriculture Cultural Collection, National Academy of Agricultural Sciences, Suwon, South Korea. Potato dextrose agar (Difco, Detroit, MI, USA) was used for cultivation and bioassay of the strains.
Fermentation media and culture conditions
For seed culture, a frozen stock spore suspension of the AG-P1441 mutant was inoculated into 500 mL Erlenmeyer flasks containing 50 mL of Bennet liquid medium. After shaking at 150 rpm and 28 °C for 30 h, the seed broth was used for inoculation. A 2 mL aliquot of the culture broth was inoculated into a 500 mL Erlenmeyer flask containing 50 mL GSS liquid medium (10 g soluble starch, 20 g glucose, 25 g soybean meal, 1 g beef extract, 4 g yeast extract, 2 g NaCl, 0.25 g K2HPO4 and 2 g CaCO3 per liter; pH 7.2). Fermentations for antimicrobial production were performed at 28 °C and 150 rpm for 120 h. All trials were performed in triplicate.
Bioreactor fermentation
The validation of the statistical equation obtained for antimicrobial production from S. rimosus AG-P1441 was carried out in a 5 L bioreactor (KF-5 L, KoBioTech, Korea) with a working volume of 3 L. 5 mL of the seed culture was transferred to 200 mL seed medium in 1 L Erlenmeyer flasks and 150 mL of the seed culture was inoculated. Fermentation was at 28 °C for 120 h with an aeration rate of 1 vvm and stirring at 500 rpm. The pH was not controlled. Samples of 10 ml for were taken for analysis.
Assay of antimicrobial activity
Antimicrobial activity was determined by the standard agar diffusion method using P. capsici as the test organism [12]. Antibiotic assay discs (Aldrich Whatman®, St. Louis, MO, USA) were impregnated with 50 μL of broth-culture filtrate (0.22 μm pore filter, Millipore), placed on agar plates and the inhibition zones measured (in mm) after incubation at 28 °C for 7 days. Inhibition zones were measured from the edge of the antimicrobial disk to the margin of the zone.
HPLC and LC–MS analysis
Crude antimicrobial compound was extracted from the cell-free culture broth of S. rimosus AG-P1441 using an equal volume of ethyl acetate. The antimicrobial compound was partially purified using silica column chromatography with gradient solvent system (water:methyl alcohol). Fractions were analyzed on TLC and tested for antimicrobial activity by disc diffusion method. A fraction showed antimicrobial activity was analyzed on HPLC instrument (Hitachi, Japan). The LC mobile phase is as follows: water and methanol (50:50–0:100) with linear gradient elution with the flow rate of 1 mL/min using C18 column. Filtered sample was injected into the column and the relative retention time was recorded. The partially purified antimicrobial compound was further analyzed using a LC–MS instrument (LTQ XL™ Linear Ion Trap Mass Spectrometer, Thermo Scientific, USA) with Electro Spray Ionization (ESI) source and a Cosmosil 2.5 Cholester column (Nacalai tesque, Japan) (100 × 2.0 mm) in order to determine the molecular weight.
Statistical design
The conventional OFAT method was used to select the effective factors. The production medium based on GSS medium was used to screen out the best carbon source, nitrogen source and other factors, by shaking flask culture experiments. All the experiments were done in triplicate and average values were recorded. Media components including carbon sources (arabinose, fructose, galactose, glucose, mannose, rhamnose, xylose, lactose, raffinose, cellulose, cottonseed flour, dextrin, maltodextrin, starch, glycerol, sorbitol, mannitol and inositol; 1.0% w/v), nitrogen sources (soytone, soybean meal, yeast extract, beef extract, tryptone, peptone, fish meal, urea, casamino acid, asparagine, N-Zamine, NH4NO3, NH4Cl, KNO3, (NH4)2SO4, (NH4)2CO3, Ca(NO3)2 and CH3COONH4; 1.0% w/v), mineral salts (MgCl2, CuSO4, ZnSO4, MnSO4, FeSO4, FeCl3 and NaHCO3; 0.01–1.0% w/v) and amino acids (glutamate and glutamine; 1–10 mM) were evaluated. A OFAT approach was initially implemented to determine the important process variables affecting antimicrobial production. Minitab 17.1.0 (Minitab Inc., State College, PA, USA) was used to design and analyze the data throughout the experiments. The Plackett–Burman design (PBD) was adopted for the selection of significant media components which influence production of antimicrobial compound in S. rimosus AG-P1441. In full factorial designs, the number of factors increases exponentially leading to an unmanageable number of experiments. Hence, fractional factorial design like Plackett–Burman becomes a method of choice for initial screening of medium components [13]. PBD method involves two-level fractional factorial saturated design that uses only k + 1 treatment combinations to estimate the independent effects of k factors [14]. Based on the antimicrobial activity and economic efficiency, 10 variables selected by OFAT including four inorganic components, the selected carbon and nitrogen sources, amino acids and pH were investigated. A total of 11 variables at two levels, high (+) and low (−) were involved in the 12 trials to determine their effects on antimicrobial production. After the influential variables were selected by PBD, RSM with the central composite design (CCD) was used to resolve the optimum combination and interactive effects of critical process variables that enhance the antimicrobial production [15]. Combinations of PBD and RSM have been used in a number of studies for medium formulation to give optimum amount of desired metabolites [16]. The significant variables selected by PBD such as, corn starch, soybean meal, MgCl2 and glutamate were further optimized by RSM using five-level CCD. The CCD contained a total of 30 experimental runs. Since, the theoretical relationships between the independent and dependent variables are not clear, multiple regression analysis can be applied to predict the dependent variables on the basis of a second-order equation [16].
where Y = predicted response, a 0 = intercept coefficient, a i X i = linear terms, a ij X i X j = interaction terms and a ii X 2 i = square terms.
The statistical adequacy of the model was determined through analysis of variance (ANOVA). Overall model significance was verified using Fisher’s -test and its associated probability. The quality of the polynomial model equation was judged statistically through coefficient of determination (R2) and adjusted R2. Three-dimensional (3D) response surface plots were drawn to illustrate the relationship between the responses and the experimental levels of each independent variable. An optimum level of the variables for maximum antimicrobial activity was determined by response optimizer tool of the software.
Results and discussion
HPLC and LC–MS analysis of antimicrobial compound
The active ethyl acetate extract from the cell free supernatant of fermentation broth of S. rimosus AG-P1441 was fractionated by silica gel column chromatography and the active fraction was analyzed on HPLC (Supplementary Fig. 1). The relative retention time of the fraction F42 that showed antimicrobial activity was observed to be approximately 18.5 min. The molecular weight was determined to be 293 by the ESI-mass measurements, which provided an intense quasi-molecular ion peak at m/z 294.24 [M + H]+ in positive ion mode (Supplementary Fig. 2).
Optimization of fermentation medium using a OFAT method
Effect of carbon source
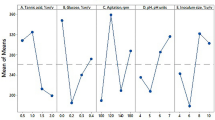
In order to identify the most suitable carbon source for antimicrobial activity of S. rimosus AG-P1441, various carbon sources were separately provided at 10 g/L, replacing glucose and starch in the GSS medium. Among the tested carbon sources, glucose, mannose, potato starch and corn starch were propitious to antimicrobial antibiotic production by the AG-P1441 strain [Fig. 1(A)].
In a medium containing mixtures of rapidly and slowly used carbon sources, the former is used up first to produce cells but little or no antibiotics are synthesized. After the rapidly assimilated compound is depleted, the ‘second-best’ carbon source is used for idiolite formation [17]. After considering the results in combination with the economic efficiency, for instance, the cost of glucose is less than mannose, glucose, as a rapidly used carbon source and corn starch, as a slowly used carbon source, were selected. Glucose, usually an excellent carbon source for growth, was selected as the basic carbon source. When glucose was added in a medium containing 1–5% the antimicrobial activity was increased as the amount of glucose increased but it did not increase when added more than 3%. Subsequent experiments were performed with 3% glucose added.
Effect of nitrogen source
In many microbial systems, antibiotic production is typically influenced by the source and concentration of nitrogen in the medium. The inhibitory effect of many N-compounds on antibiotic synthesis is observed with rapidly used nitrogen sources, such as ammonia, nitrate and certain amino acids. Therefore, proper nitrogen source selection can improve antibiotic production, particularly by using slow releasing N-compounds to avoid inhibition [18]. In the course of nitrogen source optimization, individual nitrogen sources were supplemented at 10 g/L into the medium by removing soybean meal, beef extract and yeast extract used in the GSS medium. In comparison to organic nitrogen sources, inorganic nitrogen sources gave rise to relatively lower antimicrobial activity. Among the 21 nitrogen sources examined, soytone, soybean meal, tryptone and peptone were favorable to antimicrobial production [Fig. 1(B)]. Tryptone was found to be the best nitrogen source, providing the maximum antimicrobial activity, followed by peptone, soytone and soybean meal. Although soybean meal was not the best nitrogen source, it is more affordable than other nitrogen sources. Porter and Jones reported soy flour, grits, and soybean meal as protein-rich raw materials suitable for enzyme and antibiotic fermentations [19]. Hence, soybean meal was selected as the organic nitrogen source in the medium.
Effect of micronutrients and amino acids
Micronutrients (trace elements) are known to affect the production of some secondary metabolites produced by actinomycetes. The concentration range of a trace element is much narrower for permitting secondary metabolism than the concentration range that permits superb vegetative growth [20].
For the estimation of the optimal concentration of each micronutrient for maximum antimicrobial production, the influence of one factor (one micronutrient) was determined at three concentration levels [Fig. 1(C)]. Each micronutrient was added to the modified GSS medium containing 2% glucose, 1% soluble starch, 2.5% soybean meal. Among the micronutrients examined, the mycelial growth and antimicrobial activity reached the highest levels in the media containing 0.01% MgCl2, 0.1% MnSO4, 0.01% FeCl3 and 0.1% NaHCO3. The results indicated that an increase in MnSO4 concentrations in the liquid medium significantly increased antimicrobial activity, while an increase in MgCl2, FeSO4, FeCl3 and NaHCO3 concentrations decreased antimicrobial production. The effects of Mg2+ may be due to its requirements in protein synthesis and its depletion may restrict enzyme synthesis and activity [21]. Also, Mg2+ is known as a growth-stimulating factor in submerged mycelia of Streptomyces azureus [22]. Fe3+ is the most important micronutrient used by bacteria as is required as a cofactor for multiple enzymes and Fe-containing proteins. Lubbe et al. [23, 24] reported that the complete cephamycin pathway benefited from an increase in Fe concentration. Mahmood [25] reported that Mn2+ had a marked effect on growth and bulbiformin production by Bacillus subtilis, and Liu et al. [26] reported that Mn2+ stimulated polyene production in Streptomyces.
Glutamate is the nitrogen donor for the biosynthesis of 85% of the nitrogenous compounds in the cells, while glutamine can transfer its nitrogen to 15% of the nitrogenous molecules, by the action of aminotransferases and amidotransferases, respectively [27]. An increase in glutamate and glutamine concentrations in the liquid medium significantly increased antimicrobial activity.
Optimization of fermentation medium using a statistical method
Screening of essential medium components using the PBD
PBD is an authentic method to evaluate the relative importance of various variables or medium components for specific output. Use of PBD decreases the total number of experiments, tremendously, as the interaction effects of the variables not consider and only those variables that actually affect the production of desired metabolite are screened [16]. PBD was used for initial screening of the medium components. Eleven different variables (corn starch, potato starch, MgCl2, glutamate, soybean meal, NaHCO3, fructose, pH, FeCl3, glutamine and MnSO4) were evaluated for their suitability to sustain increased antimicrobial activity of S. rimosus AG-P1441. Glucose was supplied as the carbon source in all of the media investigated. The nine components (corn starch, potato starch, MgCl2, glutamate, soybean meal, NaHCO3, fructose, pH, FeCl3) were also determined to be the most significant in antimicrobial production, based on the low P-values (< 0.05). Moreover, the regression coefficient (R2) was 0.967, meaning that 96.7% of the total variations could be explained by the model. On the Pareto chart of the standardized effects (Supplementary Fig. 3), the minimal effects were presented towards lower fields, near zero, and the maximal effects towards upper fields. NaHCO3, fructose, pH and FeCl3 were excluded because their effects were relatively weak. As a complex carbon source, corn starch was preferred over potato starch because of its positive effect. Mg2+ is by far the most frequently found metal ion cofactor in enzymatic systems [28]. Mn2+ is very similar to Mg2+ in terms of its chemical properties. Mn2+ effectively binds ATP and allows hydrolysis of the energy molecule by most ATPases. Mn2+ can also replace Mg2+ as the activating ion for a number of Mg2+-dependent enzymes [29]. Mg2+ is more essential for energy metabolism than Mn2+ and has been chosen to simplify experimental design. Further studies are needed to clarify the effect on Mn2+. Therefore, corn starch, soybean meal, MgCl2 and glutamate were determined as important factors for the antimicrobial activity of S. rimosus AG-P1441.
Rsm
In the optimization of significant media components, RSM has been proven to be a powerful tool. RSM is a sturdy, robust and efficient mathematical approach which includes statistical experimental designs and multiple regression analysis, for seeking the best formulation under a set of constrained equations. RSM is used to determine the factor levels which can simultaneously satisfy a set of desired specifications. This method helps us to determine, how a specific response is affected by changes in the level of the factors over the specified levels of interest and to achieve a quantitative understanding of the system behavior over the region tested. With the help of RSM we can predict the product properties throughout the region, even at factor combinations not actually run and to find conditions for the process stability [16]. Furthermore, CCD is a widely used statistical design for optimizing media components using a small number of experiments. CCD was first described by Box and Wilson [30]. Nowadays it is widely used in RSM for building a second order (quadratic) model for the response variable without using a complete three-level factorial experiment in terms of cost and effort [16]. For instance, RSM with the CCD has been adopted to improve antibacterial compound production by several Streptomyces species [24, 31–33].
The optimal concentration of media components was determined using CCD with the four variables, corn starch, soybean meal, MgCl2 and glutamate, in 30 experimental runs. Table 1 summarizes the observed responses of the RSM experiments for studying the effect of the four variables on the antimicrobial production. The regression coefficient of each variable, in terms of linear and quadratic response and interaction, along with t and P-values are provided in Table 2. The P values were used as a tool to check the significance of each of the coefficients, which, in turn, are necessary to understand the pattern of the mutual interactions between the test variables. The smaller the magnitude of P, the more significant is the corresponding coefficient. A level of P < 0.050 indicates the model terms are significant. In this instance, corn starch, soybean meal, MgCl2 and glutamate had a significant effect on antimicrobial acitivity (P < 0.05). A regression model having an R2 > 0.9 was considered as having a very high correlation [34]. It also meant that the model could explain 95.9% of the total variations. It was found that the response of the antimicrobial activity could be expressed by the following regression equation:
where Y is the antimicrobial activity (mm) and A, B, C and D were corn starch, soybean meal, MgCl2 and glutamate, respectively.
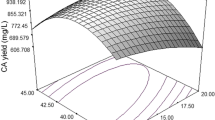
Furthermore, an analysis of variance (ANOVA) for the response surface quadratic model is presented in Table 3, which also proved that this regression was statistically significant at a 95% confidence level. The 3D surface plots were obtained to understand the interaction of the medium components and the optimum concentration of each component required for maximum antimicrobial activity. Because the 3D surface plots can only show response values for two variables, they illustrated the interaction of the two variables, keeping the other variables constant at the middle value. Figure 2(A) shows the effect of corn starch, and soybean meal on antimicrobial activity. With moderate concentration of corn starch, the antimicrobial activity increased with increase in soybean meal, and there after antimicrobial activity decreased with higher concentration of corn starch. The same trend was observed in Fig. 2(B)–(E). Figure 2(F) shows the effect of MgCl2 and glutamate on antimicrobial activity. The antimicrobial activity increased with increasing concentration of MgCl2 and decreasing concentration of glutamate. The 3D plots clearly showed that the maximum antimicrobial activity should occur with middle level of both corn starch and glutamate, behind middle level of soybean meal and higher levels of MgCl2. On the basis of numerical optimization, the quadratic model predicted that the maximum antimicrobial activity was 24.8 mm (inhibition zone diameter) with corn starch, soybean meal, MgCl2 and glutamate at 3.5, 2.5%, 1.2 and 5.9 mM, respectively.
3D surface plot of antimicrobial activity of S. rimosus AG-P1441: (A) the effect of corn starch and soybean meal on antimicrobial activity; (B) the effect of corn starch and MgCl2; (C) the effect of corn starch and glutamate; (D) the effect of soybean meal and MgCl2; (E) the effect of soybean meal and glutamate; (F) the effect of MgCl2 and glutamate
Verification in a 5L bioreactor
Figure 3(A) shows the typical time courses of antimicrobial production in a 5 L bioreactor. Under the non-optimized culture condition (10 g soluble starch, 20 g glucose, 25 g soybean meal, 1 g beef extract, 4 g yeast extract, 2 g NaCl, 0.25 g K2HPO4 and 2 g CaCO3 per liter; pH 7.2), the maximum antimicrobial activity (inhibition zone diameter in mm) indicated 15 mm at 40 h of the fermentation while the reducing sugar was completely depleted in 48 h. The pH of the medium decreased slightly to pH 5.15 at 40 h followed by a slow increase to pH 6.3. Meanwhile, under the optimal culture condition (3% glucose, 3.5% corn starch, 2.5% soybean meal, 1.2 mM MgCl2 and 5.9 mM glutamate), the maximum antimicrobial activity indicated 29 mm at 48 h of the fermentation while the reducing sugar was completely depleted in 96 h [Figure 3(B)]. The pH of the optimal medium decreased to pH 5 at 20 h and then increased to pH 6.5 at the end of fermentation. Antimicrobial production was high in the vicinity of pH 6. Optimization of operating parameters (e.g. effects of agitation, aeration, and dissolved oxygen level) in a bioreactor fermentation deserves further investigation, which is being studied in our laboratory. As a result, the model developed was considered to be accurate and reliable for predicting the production of antibiotics by S. rimosus AG-P1441. After optimization, the antimicrobial activity was improved from an initial 15 to a final 29 mm. Furthermore, the information presented is considered fundamental and useful for developing a cultivation process for S. rimosus AG-P1441 to allow efficient large-scale antimicrobial production.
References
Hausbeck MK, Lamour KH. Phytophthora capsici on vegetable crops: research progress and management challenges. Plant Dis. 88: 1292–1300 (2014)
Chen YY, Chen PC, Tsay TT. The biocontrol efficacy and antibiotic activity of Streptomyces plicatus on the oomycete Phytophthora capsici. Biol. Control. 98: 34–42 (2016)
Gavino PD, Smart CD, Sandrock RW, Miller JS, Hamm PB, Lee TY, Davis RM, Fry WE. Implications of sexual reproduction for Phytophthora infestans in the United States: generation of an aggressive lineage. Plant Dis. 84: 731–735 (2000)
Zohara F, Akanda AM, Paul NC, Rahman M, Islam T. Inhibitory effects of Pseudomonas spp. on plant pathogen Phytophthora capsici in vitro and in planta. Biocatal. Agric. Biotechnol. 5: 69–77 (2016)
Lee HB, Kim Y, Kim JC, Choi GJ, Park SH, Kim CJ, Jung HS. Activity of some aminoglycoside antibiotics against true fungi, Phytophthora and Pythium species. J. Appl. Microbiol. 99: 836–843 (2005)
Rothrock CS and Gottlieb D. Importance of antibiotic production in antagonism of Streptomyces species to two soil-borne plant pathogens. J. Antibiot. 34: 830–835 (1981)
Crawford DL, Linch JM, Whipps JM and Ousley MA. Isolation and characterization of actinomycete antagonists of a fungal root pathogen. Appl. Environ. Microbiol. 59: 3899–3905 (1993)
Mari M, Guizzardi M and Pratella GC. Biological control of gray mold in pears by antagonistic bacteria. Biol. Control. 7: 30–37 (1996)
Georgakopoulos DG, Fiddaman P, Leifert C and Malathrakis NE. Biological control of cucumber and sugar beet damping off caused by Pythium ultimum with bacterial and fungal antagonists. J. Appl. Microbiol. 92: 1078–1086 (2002)
Kim CJ, Park DJ, Lee JC, Ju YJ, Yoon BS. Development of biocontrol agent from bioactive compounds of microbial origin. Annual report of Rural Development Administration. (2013)
Rathi P, Goswami VK, Sahai V, Gupta R. Statistical medium optimization and production of a hyperthermostable lipase from Burkholderia cepacia in a bioreactor. J. Appl. Microbiol. 93(6): 930–936 (2002)
Grove and Randall. Assay methods of antibiotics medical encyclopedia. Inc. New York, NY, USA. (1955)
Singh AK, Mehta G, Chhatpar HS. Optimization of medium constituents for improved chitinase production by Paenibacillus sp. D1 using statistical approach. Lett. Appl. Microbiol. 49(6): 708–714 (2009)
Plackett RL and Burman JP. The design of optimum multifactorial experiments. Biometrika. 33(4): 305–325 (1946)
Box G, Hunter W, Hunter J. Statistics for experiments. Wiley, New York, USA. (1958)
Singh V, Haque S, Niwas R, Srivastava A, Pasupuleti M, Tripathi CK. Strategies for Fermentation Medium Optimization: An In-Depth Review. Front. Microbiol. Jan 6;7:2087 (2017)
Sánchez S, Chávez A, Forero A, García-Huante Y, Romero A, Sánchez M, Rocha D, Sánchez B, Avalos M, Guzmán-Trampe S, Rodríguez-Sanoja R, Langley E, Ruiz B. Carbon source regulation of antibiotic production. J. Antibiot. 63: 442–459 (2010)
Aharonowitz, Y. Nitrogen metabolite regulation of antibiotic biosynthesis. Ann. Rev. Microbiol. 34: 209–233 (1980)
Porter MA, Jones AM. Variability in soy flour composition. J. Am. Oil Chem. Soc. 80(6): 557–562 (2003)
Schrader KK, Blevins WT. Effects of carbon source, phosphorus concentration, and several micronutrients on biomass and geosmin production by Streptomyces halstedii. J. Ind. Microbiol. Biotechnol. 26(4): 241–247 (2001)
Natsume M, Kamo Y, Hirayama M, Adachi T. Isolation and characterization of alginate-derived oligosaccharides with roof growth-promoting activities. Carbohydr. Res. 258: 187–197 (1994)
Okba AK, Ogata T, Matsubara H, Matsuo S, Doi K, Ogata S. Effects of bacitracin and excess Mg2+ on submerged mycelial growth of Streptomyces azureus. J. Ferment. Bioeng. 86: 28–33 (1998)
Lubbe C, Jensen SE, Demain AL. Prevention of phosphate inhibition of cephalosporin synthetases by ferrous ion. FEMS Microbiol. Lett. 25: 75–79 (1984)
Raza W, Yang XM, Wu HS, Huang QW, Xu YC, Shen QR. Evaluation of metal ions (Zn2+, Fe3+, Mg2+) effect on production of fusaricidin-type antifungal compounds by Paenibacillus polymyxa SQR-2. Bioresour. Technol. 101: 9264–9271 (2010)
Mahmood M. Trace elements for growth and bulbiformin production by Bacillus subtilis. J. Appl. Bacteriol. 35: 1–5 (1972)
Liu CM, McDaniel LE, Schaffner CP. Factors affecting the production of Candicidin. Antimicrob. Agents. Chemother. 7: 196–202 (1975)
Voelker F, Altaba S. Nitrogen source governs the patterns of growth and pristinamycin production in ‘Streptomyces pristinaespiralis’ Microbiology. 147: 2447–2459 (2001)
Sissi C, Palumbo M. Effects of magnesium and related divalent metal ions in topoisomerase structure and function. Nucleic Acids Res. Feb;37(3): 702–711 (2009)
Cowan JA. Introduction to the biological chemistry of magnesium ion. The biological chemistry of magnesium. VCH. New York, 1–23 (1995)
Box, GEP, Wilson, KB. On the experimental attainment of optimum conditions. J. Roy. Stat. Soc. (Ser. B) 13: 1–45 (1951)
Rajeswari P, Jose PA, Amiya R, Jebakumar SR. Characterization of saltern based Streptomyces sp. and statistical media optimization for its improved antibacterial activity. Front. Microbiol. 5: 753 (2015)
Gao H, Liu M, Liu J, Dai H, Zhou X, Liu X, Zhuo Y, Zhang W, Zhang L. Medium optimization for the production of avermectin B1a by Streptomyces avermitilis 14-12A using response surface methodology. Bioresour. Technol. 100: 4012–4016 (2009)
Vineeta Singh, C.K.M. Tripathi. Production and statistical optimization of a novel olivanic acid by Streptomyces olivaceus MTCC 6820. Process biochem. 43: 1313–1317 (2008)
Chen XC, Bai JX, Cao JM, Li ZJ, Xiong J, Zhang L, Hong Y, Ying HJ. Medium optimization for the production of cyclic adenosine 3′,5′-monophosphate by Microbacterium sp. no. 205 using response surface methodology. Bioresour. Technol. 100: 919–924 (2009)
Acknowledgements
This research was supported by a Grant (10045326) from the R&D Program of MOTIE/KEIT of Republic of Korea, the KRIBB Research Initiative Program, Republic of Korea and a Grant (NRF-2013M3A9A5076601) from a study on the strategies of improving the value of microbial resources funded by Ministry of Science, ICT and Future Planning of the Korea Government.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ju, Y., Son, KH., Jin, C. et al. Statistical optimization of culture medium for improved production of antimicrobial compound by Streptomyces rimosus AG-P1441. Food Sci Biotechnol 27, 581–590 (2018). https://doi.org/10.1007/s10068-017-0257-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10068-017-0257-1