Abstract
Objective
There are inconclusive data connecting single-nucleotide polymorphisms (SNPs) of TNF-α (rs361525) and TNF-β (rs909253) to potential malignant oral disorder (PMOD) such as lichen planus and oral fibrosis. Here, we have investigated the risk of oral squamous cell carcinoma as well as oral pre-cancerous lesions in North Indian population with the polymorphism of the TNFα/ β genes.
Material and methods
A total 500 patients with oral pre-cancer and OSCC and 500 healthy volunteers were genotypes for the TNF-α (-238) G/A (rs361525) and TNF-β (252) A/G (rs909253) gene polymorphism. Genotypes were identified by polymerase chain reaction (PCR) restriction fragment length polymorphism (RFLP). Genotype frequencies were evaluated by Chi-square test.
Results
Compared to the GG genotype, the GA genotype of TNF-α (G238A) polymorphism (rs361525) has been found to significantly increase the risk of oral disease (OR = 1.99) and especially the risk of lichen planus and OSCC (OR = 2.805 and 5.790, respectively). Similarly, the risk of oral disease was also more in the heterozygote (AG) than the common allele homozygote (AA) of TNF-β (A252G) polymorphism (rs909253) (OR = 1.483).
Conclusion
We conclude that the SNPs rs361525 and rs909253 were significantly associated with oral pre-cancer and OSCC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Oral cancer represents an important problem worldwide. The incidence and prevalence rates for these tumors are double in men than in women. Cancers of the oral cavity rank as the eighth most common cancer among men, being responsible for 3% of the cancers diagnosed in this gender. Oral pre-cancerous lesion, a benign morphologically altered tissue that has a greater than normal risk of malignant transformation, is also very common. Leukoplakia, lichen planus, and oral submucous fibrosis are early indicators of damage to the oral mucosa with a transformation rate of 2–12% to frank malignancies [1]. These lesions are termed as potential malignant oral disorder (PMOD). Many environmental factors like tobacco, smoking, and alcohol consumption and genetic factors (oncogenes and suppressor genes) are implicated in the development of oral cancer [2] which may sometimes predispose from a clinically evident PMOD, the most common being the oral leukoplakia [3]. Tobacco and alcohol usage are strong risk factors with the development of oral cancer [4]. Oral diseases are highly prevalent in Indian subcontinent, and oral cancer ranked 3rd in Indian scenario [4].
Several salivary tumor markers are found to be significantly increased in the saliva of oral cancer patients [5, 6] such as 8-OHdG and MDA biomarkers.
Few biomarkers are needed to predict the evolution of oral cancer from an PMOD and also to screen the high-risk patients with smoking and tobacco habits. It is important that the combined early detection and the effective treatment strategies could help in the prevention and the good prognostic outcomes of oral cancer [7].
Several factors related to angiogenesis, inflammation, and thrombosis have been associated with oral cancer [2, 4, 8]. Previous studies have shown that polymorphism in extracellular matrix MMP1 (1607GG) is highly associated with colorectal cancer, renal cancer, and head and neck carcinoma [9,10,11]. The vascular endothelial growth factor (VEGF) family of proteins known to involve in vasculogenesis and angiogenesis is the prime requirement for tumor growth. Polymorphisms of VEGF-C such as rs7664413 (− 33 nt upstream of the VEGF-C gene) and rs2046463 (downstream 5008 nt) have been shown to be strongly associated with OSCC growth [12]. Similarly, polymorphism of VEGF-A is also known to regulate the malignancy [13]. Polymorphisms of different inflammatory genes such as IL-2 (−330A > C) [14], COX-2 gene −1195 G > A (rs689466), +837 T > C (rs5275), and − 765 G > C (rs20417) have also been shown to strongly correlated with oral cancer progression [15]. Predisposition of individuals to different carcinogens is also associated with genetic polymorphisms, as ALDH2 (aldehyde dehydrogenase-2) Rs671G > A is strongly correlated with cancer progression [16]. Sensitivity toward tobacco consumption also varies with genetic polymorphism in genes such as CYP1A1, GSTM1, and GSTT1 [17]. Susceptibility of pre-cancer oral diseases to genetic polymorphism is also well documented as SNPs in GSTM1 (null) [18], CCND1 (G870A) [19], XPD (codon 751) [20], and MMP3 (-1171; promotor region) [21] are well correlated across the majority of populations (Asians, Caucasians, Brazilians, and others). Risk associated with a SNP in p53 (codon 72) was reported in Indian populations only [22].
The progression of cytokines as biomarkers for detecting malignancies has helped in realizing the importance of inflammation-mediated carcinogenesis in PMODs and in oral cancer. Inflammatory genes as tumor necrosis factor alpha (TNF-α) and beta (TNF-β) are, respectively, encoded by TNF-α and TNF-β genes [23]. TNF-α gene is located on chromosome 6p21.231 in the polymorphic region of MHC III. The promoter polymorphisms of TNF-α have been intensively studied as a potential determinant of disease susceptibility [24, 25]. There is also an increasing evidence that TNF-α may promote the development and spread of cancer [26,27,28]. Commonly described variants of TNF-α gene polymorphisms consist of G to A transitions in the promoter region at positions _238 and _308 [24]. G to A transition at position 238 in promoter region is important because it results into higher expression of TNF-α protein [29]. So far, TNF-α promoter polymorphism has been related to numerous cancers, such as bladder cancer [30], renal cell carcinoma [31], non-small cell lung carcinoma [32], cervical cancer [33], and breast carcinoma [34]. Importance of this polymorphism in oral cancer has been shown in western population [35], but significance in Indian population is still unknown. TNF-β is a proinflammatory cytokine produced by lymphocytes and is structurally related to TNF-α [35]. The two factors have 30% amino acid sequence identity, are recognized by the same widely distributed cellular receptors, and, therefore, share many of their functions [35]. The TNF-β gene is adjacent to the TNF-α gene within a 7 kb locus in the major histocompatibility complex [35]. An A/G polymorphism located at position 252 within the first intron of the TNF-β gene affects expression of both genes and concentration of TNF-α and TNF- β proteins in plasma [36, 37]. The less common allele B1 (252G) has been correlated with a higher TNF-β expression both at the mRNA and the protein level [37]. On the other hand, the common allele B2 (252A) is associated with increased TNF-α gene expression [36]. The TNF-β (A252G) polymorphism has been associated with a risk of development of breast, esophageal, gastric, and colorectal cancer [38,39,40]. The reported allele frequency of the high expression B1 homozygotes is about 16% in Europeans and 13% in Asians [38, 41]. Interestingly, when the combined TNF-α and TNF-β polymorphisms were examined together in patients with breast or esophageal cancer, certain genotypes were found to be significantly overrepresented and were associated with an increased risk of cancer development [38, 42]. Although the role of TNF-alpha (-308 G/A) and TNF-beta (252 G/A) polymorphism in oral cancer was reported earlier, the study was based upon European population [35]. There is one report of the study of TNF α polymorphism in Indian population. Singh et al. [43] showed TNF-α (-238) G/A is highly but TNF-α (-308) G/A is not associated with incidence of oral cancer. Singh et al. had not analyzed the impact of TNF-β (+252A/G) or TNF-α (-238) G/A on oral cancer or pre-cancer lesions. There was one study in Taiwan where TNF-α (-308) G/A polymorphism was found associated with oral submucous fibrosis [44]. Another study conducted on Saudi Arabian population the polymorphism TNF-α (-308 G/A), and TNF-β (+252A/G) was found significantly associated with oral lichen planus [45]. These information strongly indicate that there is no thorough study of TNF polymorphism in connection with oral pre-cancer and cancer condition in Indian population. Habit of taking pan masala, gutka, areca nut, betel nut, and tobacco by North Indian population makes them highly vulnerable to oral diseases such as oral submucous fibrosis, oral lichen planus, oral leukoplakia, and oral cancer. In this study, the possible correlation of TNF-α (-308 G/A) and TNF-β (+252A/G) with OPMD and OSCC has been evaluated in North Indian population.
Materials and methods
Study subjects
The study was undertaken on a total of 500 patients with previously treated and pathologically confirmed oral pre-cancer and OSCC who were registered at Department of Oral Pathology and Microbiology, King George’s Medical University and 500 healthy controls. This study was approved by the Institutional Ethics Committee of the King George’s Medical University, Lucknow. An informed written consent was obtained from all subjects. Ethical clearance was obtained from institutional ethical committee. Venous blood samples were collected in EDTA tubes and stored at − 80 °C, until DNA extraction. Genomic DNA extraction from blood samples was carried out by salting out method [46].
Genotyping by RFLP
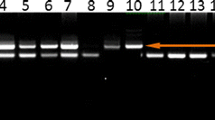
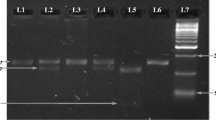
Genotypes for polymorphisms in TNF-α (-238) G/A and TNF-β (252) A/G were detected using PCR—RFLP technique. PCR products are generated by using 10 ng of genomic DNA in 10 μl volume reactions containing 20 mMTris- HCl, 50 mMKCl, 2.0 mM MgCl2, 0.11 mMeachdNTP, 0.3 mM each primer (Table 1), and 0.3 U Taq DNApolymerase (Invitrogen, Paisley, UK). PCR primer used to genotype the TNF-α and TNF-β polymorphisms is detailed in Table 1. PCR products were digested with the BamHI enzyme to screen the TNF alpha (-238G/A) polymorphism (Fig. 1) and NcoI enzyme to screen for the TNF-β (252) A/G polymorphism (Fig. 2) that recognized and cut wild-type, homozygous, heterozygous sequences at 37 °C for overnight. The digested PCR products were resolved on a 2% agarose gel and stained with ethidium bromide for visualization under UV light. Most of the assays were carried out including samples with known genotypes as controls.
Statistical analysis
The significance in this study was evaluated by Chi-square test. Odds ratio (OR) was calculated as an estimate of relative risk of having disease according to the relative frequency of different genotypes among the cases as well as the controls. P value was considered significant at < 0.05. Odd ratio (OR) is given with 95% confidence interval (CI).
Results
In this study, we have evaluated 500 individuals of oral pre-cancer and OSCC (300 males and 200 females) and their age ranging from 20 to 70 years. Oral pre-cancer and OSCC cases were further categorized as oral submucous fibrosis (n = 200), lichen planus (n = 100), leukoplakia (n = 100), and OSCC (n = 100). TNM staging and grades of OSCC patients are detailed in Table 2. Alcohol consumption, smoking, masala, and tobacco chewing all are found to increase the risk of oral pre-cancer as well as OSCC (Table 2).
Genotype frequencies among the patients with oral disease and control are shown in Table 3. Compared to GG genotype, the frequency of GA genotype for TNF-α G238A polymorphism was significantly more in cases than in control. Similarly, the frequency of allele A for the same polymorphism is significantly higher in cases than in control (Table 3). The genotype GA and allele A both confer high risk for developing oral diseases (OR = 1.99 and 1.88, respectively). Similarly, the risk of oral disease was also more in the heterozygote (AG) than the common allele homozygote (AA) of TNF-β (A252G) polymorphism (OR = 1.483).
Risk of disease with different genotypes of TNF-α (G238A) and TNF-β (A252G) polymorphisms is further evaluated in different disease categories of oral diseases including oral submucous fibrosis, lichen planus, Leukoplakia, and OSCC (Table 4). Compared to the GG genotype, the GA genotype of TNF-α (G238A) polymorphism has been found to significantly increase the risk of lichen planus and OSCC (OR = 2.805 and 5.790, respectively). The risk of lichen planus and OSCC is also high with allele A compared to allele G of TNF-α (G238A) polymorphism (Table 4). For the TNF-β (A252G) polymorphism compared to A allele, the G allele was associated with lower risk of OSMF and Leukoplakia (OR = 0.45 and 0.63, respectively) and higher risk of OSCC (OR = 2.81). Compared to the AA genotype, the AG genotype of TNF-β (A252G) polymorphism increased the risk of lichen planus and OSCC (OR = 2.65 and 3.77, respectively). However, the GG genotype is associated with lower risk of OSMF (OR = 0.42) while compared to AA genotype of TNF-β (A252G) polymorphism (Table 4).
In a stratified analysis, we have correlated the TNF-α G238A and TNF-β A252G genotypes with the risk of oral disease among the smokers, masala/tobacco chewers, alcohol consumers, and areca nut/pan masala chewers (Table 5). Among the smokers, compared to GG genotype, the risk of oral disease was lower in the individual with AA genotype for TNF-α (G238A) polymorphism. The genotype GA for the same TNF-α polymorphism was also found to lower the risk of oral diseases among the areca nut/pan masala chewers. In this group of patients, the AG genotype and G allele of TNF-β A252G polymorphism were also associated with reduced risk of oral diseases. However, the AG genotype of TNF-β A252G polymorphism was found to increase the risk of oral diseases among the masala/tobacco chewers. Interaction between alcohol consumption and TNF-α G238A and TNF-β A252G genotypes is not found to modulate the risk of diseases (Table 5).
Influence of genotypes with the disease outcome/clinical parameters of OSCC patients is analyzed and documented on Table 6. None of the genotypes from TNF-α G238A and TNF-β A252G polymorphisms is found to be associated with the tumor stage, grade, or metastasis. However, the risk for developing larger sized tumor (tumor T status) was significantly lower with A and G allele for TNF-α G238A and TNF-β A252G polymorphisms, respectively. The A allele of TNF-α G238A polymorphism also reduces the risk of lymph node metastasis among the patients of OSCC (Table 6).
Since each person can express TNFα as well as TNFβ, it was important to analyze impact of combined polymorphism on disease progression. Four different genotype combinations of TNFα: TNFβ are analyzed, namely GG: AA, GA: AA, GG:AG, and GA:AG (Table 7). We found that the combine genotype GG:AA was more frequent in healthy control subjects where as other combined genotypes where more frequent in oral disease patients. Whenever TNFα G238A hetero genotype is present, it plays dominant role than TNFβ G252A homo or hetero genotype. Comparison between TNFα GA and TNFβ AA genotype was not possible as there was no healthy control available with this genotypes, although trend shows more diseased type phenotype. More samples are needed for comparison.
Discussion
The present study indicates association of TNF-α G238A and TNF-β A252G polymorphisms with the risk of oral diseases including pre-oral cancer lesions and OSCC. Boyle et al. [47] establish the fact that 71% of pre-oral cancer or cancer salivary sample shows tumor-specific p53 mutation.
Heterozygotes from both the polymorphism were associated with increased risk for development of lichen planus as well as OSCC. However, among the smokers and areca nut/pan masala chewers, these polymorphisms seem to reduce the risk of OSCC. Similarly, variant alleles for both the polymorphisms (A and G, respectively, for TNF-α G238A and TNF-β A252G polymorphisms) were found to reduce the risk of developing larger sized tumor in patients with OSCC. These results indicate that susceptibility of person to pan masala, areca nut, and smoking is independent of tumor or pre-lesions growth.
TNF-α gene is located on chromosome 6p21.231, and its promoter polymorphisms have been explored as a potential determinant of disease susceptibility [24, 25]. TNF-α not only alters disease susceptibility but may also promote the development and spread of cancer [26,27,28]. Commonly described variants of TNF-α gene polymorphisms consist of G to A transitions in the promoter region at positions _238 and _308 [24]. So far, many studies have reported the association of TNF-α promoter polymorphism with different cancers, including bladder cancer [29], renal cell carcinoma [30], non-small cell lung carcinoma [31], cervical cancer [28, 32], and breast carcinoma [33]. Some previous studies reported that the TNF-α (-238) G/A gene polymorphism was significantly associated with oral cancer and non-small cell lung carcinoma (NSCLC) [48, 49]. Our report confirms these previous findings. Although TNF-α level is necessary to maintain cell in-setting and to prevent certain diseases through regulating immune response [50] however, excessive TNF-α is associated with autoimmune diseases and malignant tumors.
The G allele of the 252A/G polymorphism of TNF-β gene has repeatedly been correlated with a higher expression of TNF-β gene both at the level of mRNA and the protein [36]. The G allele has been detected to be significantly increased in patients with osteosarcoma, as well as breast, colorectal, and bladder cancer [32, 51,52,53]. Similar to that, we have also observed a significantly higher frequency of the G allele of TNF-β 252A/G polymorphism among the subjects of OSCC and lichen planus. It seems that TNF-β contributes in certain malignancies due to its high expression level which is imparted by allele G of 252A/G polymorphism of TNF-β gene.
This study not only confirmed previous finding [35] but also shows the importance of TNF-α and TNF-β in the growth of pre-oral cancer lesions and OSCC in North Indian population. This also points that susceptibility to carcinogen and growth of pre-cancer or OSCC lesions are differently regulated by polymorphic genes of TNF-α and TNF-β. This study indicates that the susceptibility of different pre-oral cancer lesions to these polymorphisms is not the same too. We do not claim that analysis of these polymorphisms would predict the OSCC of pre-oral lesions as the cohorts in our study are quite heterogeneous. Further, subcategorization of different pre-oral cancer lesions is needed to strongly determine the correlation between TNF-α/ TNF-β polymorphism and predisposition of North Indian population to the OSCC. The study confirmed that TNFα genotype has stronger impact compare to TNF.β on oral disease progression.
In conclusion, two common functional polymorphisms in TNF-αand TNF-β genes seem to increase the risk of development of OSCC as well as pre-oral cancer lesions in North Indian population. The detection of such genetic predisposition may be of considerable importance for safeguarding the life and health status of certain individuals at risk in the general population through early prevention measures. Future studies with larger sample sizes and validation at protein level are needed. .Investigation to find correlation among different polymorphism is required to understand the interplay of TNFα and TNFβ.
Results of association between TNF-α, TNF-β polymorphisms, and OSCC as well as pre-oral cancer lesions are contradictory. However, due to small sample size in different disease group, our findings are rather suggestive than conclusive and warn larger studies.
References
Speight PM, Khurram SA, Kujan O (2018) Oral potentially malignant disorders: risk of progression to malignancy. Oral Surg Oral Med Oral Pathol Oral Radiol 125:612–627
Williams HK (2000) Molecular pathogenesis of oral carcinoma. J ClinPathol 53:165–172
Choi S, Myers JN (2008) Molecular pathogenesis of oral squamous cell carcinoma: implications for therapy. J Dent Res. 87:14–32
Kumar M, Nanavati R, Modi TG, Dobariya C (2017) Oral cancer: etiology and risk factors: a review. Journal Of Cancer Research And Theraputics. 2(12):458–463
Nagler R, Bahar G, Shpitzer T, Feinmesser R (2006) Concomitant analysis of salivary tumor markers: a new diagnostic tool for oral cancer. Clin Cancer Res. 12:3979–3984
Rajkumar K, Kumar AR, Ramyamalini V, Nandhini G, Kumar TD, Ashwini BK et al (2010) Estimation of serological and salivary biomarkers in patients with oral squamous cell carcinoma, premalignant lesions and conditions. SRM Univ J Dent Sci. 1:14–19
Juretic M, Cerovic R, Belusic-Gobic M et al (2013) Salivary levels of TNF-[alpha] and IL-6 in patients with oral premalignant and malignant lesions. Folia Biol. 59:99–102
Vairaktaris E, Yapijakis C, Vylliotis A, Kessler P, Vylliotis A, Ries J, Wiltfang J, Vassiliou Derka S, Neukam FW (2006) Methylenetetrahydrofolate reductase polymorphism and minor increase of risk for oral cancer. J Cancer Res ClinOncol 132:219–222
Hinoda Y, Okayama N, Takano N, Fujimura K, Suehiro Y, Hamanaka Y (2002) Association of functional polymorphisms of matrix metalloproteinase (MMP)-1 and MMP-3 genes with colorectal cancer. Int J Cancer 102:526–529
Hirata H, Okayama N, Naito K, Inoue R, Yoshihiro S, Matsuyama H (2004) Association of a haplotype of matrix metalloproteinase (MMP)-1 and MMP-3 polymorphisms with renal cell carcinoma. Carcinogenesis 25:2379–2384
Hashimoto T, Uchida K, Okayama N, Imate Y, Suehiro Y, Hamanaka Y (2004) Association of matrix metalloproteinase (MMP)-1 promoter polymorphism with head and neck squamous cell carcinoma. Cancer Lett. 211:19–24
Chien M-H, Liu Y-F, Hsin C-H, Lin C-H, Shih C-H, Yang SF, Cheng C-W, Lin C-W (2013) Impact of VEGF-C gene polymorphisms and environmental factors on oral cancer susceptibility in Taiwan. PLoS One. 8(4):e60283
Supic G, Jovic N, Zeljic K, Kozomara R, Magic Z (2012) Association of VEGF-A genetic polymorphisms with cancer risk and survival in advanced-stage oral squamous cell carcinoma patients. Oral oncology vol. 48(11):1171–1177. https://doi.org/10.1016/j.oraloncology.2012.05.023
Singh PK, Kumar V, Ahmad MK, Gupta R, Mahdi AA, Jain A, Bogra J, Chandra G (2017) Association of -330 interleukin-2 gene polymorphism with oral cancer. The Indian journal of medical research. 146(6):730–737
Dong L, Hao S-H, Sun Y, Hu C-M, Ma Z-H, Wang ZM, Liu J, Liu HB, Ye M, Zhang YF, Yang DS, Shi G (2015) Functional polymorphisms in COX-2 gene are correlated with the risk of oral cancer. BioMed Res Int. 2015:1–12. https://doi.org/10.1155/2015/580652
Zuo W, Zhan Z, Lin M, Bai W, Zeng S (2019) Effect of ALDH2 polymorphism on cancer risk in Asians. Medicine (Baltimore). 98(13):e14855
Anantharaman D, Chaubal PM, Kannan S, Bhisey RA, Mahimkar MB (2007) Susceptibility to oral cancer by genetic polymorphisms at CYP1A1, GSTM1 and GSTT1 loci among Indians: tobacco exposure as a risk modulator. Carcinogenesis. 28(7):1455–1462
Li YF, Sung FC, Tsai MH, Hua CH, Liu CS, Huang YT (2013) Interactions between cigarette smoking and polymorphisms of xenobiotic-metabolizing genes: the risk of oral leukoplakia. Dis Markers. 34(4):247–255
Yadav BK, Kaur J, Srivastava A, Ralhan R (2009) Effect of polymorphisms in XRCC1, CCND1 and GSTM1 and tobacco exposure as risk modifier for oral leukoplakia. Int J Biol Markers. 24(2):90–98
Wang Y, Spitz MR, Lee JJ, Huang M, Lippman SM, Wu X (2007) Nucleotide excision repair pathway genes and oral premalignant lesions. Clin Cancer Res. 13(12):3753–3758
Tu HF, Liu CJ, Chang CS, Lui MT, Kao SY, Chang C (2006) The functional (-1171 5A–>6A) polymorphisms of matrix metalloproteinase 3 gene as a risk factor for oral submucous fibrosis among male areca users. J Oral Pathol Med. 35(2):99–103
Sikka S, Sikka P (2014) Association of human papilloma virus 16 infection and p53 polymorphism among tobacco using oral leukoplakia patients: a clinicopathologic and genotypic study. Int J Prev Med. 5(4):430–438
Aggarwal BB, Shishodia S, Sandur SK, Pandey MK, Sethi G (2006) Inflammation and cancer: how hot is the link? BiochemPharmacol 72:1605–1621
Kaluza W, Reuss E, Grossmann S, Hug R, Schopf RE, Galle PR, Maerker-Hermann E, Hoehler T (2000) Different transcriptional activity and in vitro TNF-alpha production in psoriasis patients carrying the TNF-alpha 238A promoter polymorphism. J Invest Dermatol 114:1180–1183
Gupta R, Sharma SC, Das SN (2008) Association of TNF-α and TNFR1 promoters and 30 UTR region of TNFR2 gene polymorphisms with genetic susceptibility to tobacco-related oral carcinoma in Asian Indians. Oral Oncol 44:455–463
Liu C-J, Wong Y-K, Chang K-W, Chang H-C, Liu H-F, Lee Y-J (2005) Tumor necrosis factor-α promoter polymorphism is associated with susceptibility to oral squamous cell carcinoma. J Oral Pathol Med 34:608–612
Hohberger L, Wuertz BR, Xie H, Griffin T, Ondrey F (2008) TNF-alpha drives matrix metalloproteinase-9 in squamous oral carcinogenesis. Laryngoscope 118:1395–1399
Liu L, Yang X, Chen X, Kan T, Shen Y, Chen Z, Hu Z (2012) Association between TNF-α polymorphisms and cervical cancer risk: a meta-analysis. MolBiol Rep 39:2683–2688
Dutta D, Choudhuri S, Mondal SA, Maisnam I, Reza AHH, Ghosh S, Chowdhury S, Bhattacharya B, Mukhopadhyay S (2015) Tumor necrosis factor alpha -238G/A (Rs 361525) gene polymorphism predicts progression to type-2 diabetes in an Eastern Indian population with prediabetes. Diabetes Res Clin Pract 99(3):e37–e41
Marsh HP, Haldar NA, Bunce M, Marshall SE, le Monier K, Winsey SL, Christodoulos K, Cranston D, Welsh KI, Harris AL (2003) Polymorphisms in tumor necrosis factor (TNF) are associated with risk of bladder cancer and grade of tumor at presentation. Br J Cancer 89:1096–1101
Nakajima K, Sasaki M, Nojima D, Oh BR, Ishii N, Miura K, Dahiya R (2001) Tumor necrosis factor-alpha gene mutations and genotype changes in renal cell carcinoma. J Urol 165:612–615
Shih C-M, Lee Y-L, Chiou H-L, Chen W, Chang G-C, Chou M-C, Lin L-Y (2006) Association of TNF-alpha polymorphism with susceptibility to and severity of non-small cell lung cancer. Lung Cancer 52:15–20
Govan VA, Constant D, Hoffman M, Williamson A-L (2006) The allelic distribution of -308 tumor necrosis factor-alpha gene polymorphism in South African women with cervical cancer and control women. BMC Cancer 6:24
Mestiri S, Bouaouina N, Ahmed SB, Khedhaier A, Jrad BB, Remadi S, Chouchane L (2001) Genetic variation in the tumor necrosis factor alpha promoter region and in the stress protein hsp70-2: susceptibility and prognostic implications in breast carcinoma. Cancer 91:672–678
Yapijakis C, Serefoglou Z, Vylliotis A, Nkenke E, Derka S, Vassiliou S, Avgoustidis D, Neukam FW, Patsouris E, Vairaktaris E (2006) Association of polymorphisms in tumor necrosis factor alpha and beta genes with increased risk for oral cancer. Anticancer Res. 29(6):2379–2386
Mestiri S, Bouaouina N, Ahmed SB, Khedhaier A, Jrad BB, Remadi S, Chouchane L (2001) Genetic variation in the tumor necrosis factoralpha promoter region and in the stress protein hsp70-2: susceptibility and prognostic implications in breast carcinoma. Cancer. 91:672–678
Bazzoni F, Beutler B (1996) The tumor necrosis factor ligand and receptor families. N Engl J Med 334:1717–1725
Messer G, Spengler U, Jung MC, Honold G, Blomer K, Pape GR, Riethmuller G, Weiss EH (1991) Polymorphic structure of the tumor necrosis factor (TNF) locus: an NcoI polymorphism in the first intron of the human TNF-beta gene correlates with a variant aminoacid in position 26 and a reduced level of TNF-beta production. J Exp Med 173:209–219
Pociot F, Briant L, Jongeneeel CV, Molvig J, Worsaae H, Abbal M, Thomsen M, Nerup J, Cambon-Thomsen A (1993) Association of tumor necrosis factor (TNF) and class II major histocompatibility complex alleles with the secretion of TNF-α and TNF-β by human mononuclear cells: a possible link to insulin-dependent diabetes mellitus. Eur J Immunol 23:224–231
Guo W, Wang N, Li Y, Zhang JH (2005) Polymorphisms in tumor necrosis factor genes and susceptibility to esophageal squamous cell carcinoma and gastric cardiac adenocarcinoma in a population of high incidence region of North China. Chin Med J 118:1870–1878
Azmy IA, Balasubramanian SP, Wilson AG, Stephenson TJ, Cox A, Brown NJ, Reed MW (2004) Role of tumor necrosis factor gene polymorphism (-308 and -238) in breast cancer susceptibility and severity. Breast Cancer Res 6:395–400
Saito S, Kasai Y, Nomoto S, Fujiwara M, Akiyama S, Ito K, Nakao A (2001) Polymorphism of tumor necrosis factor in esophageal, gastric or colorectal carcinoma. Hepatogastroenterology l48:468–470
Singha PK, Bograa J, Chandraa G, Ahmadb MK, Guptaa R, Kumarc V, Jaind A, Mahdib AA (2015) Association of TNF-α (-238 and -308) promoter polymorphisms with susceptibility of oral squamous cell carcinoma in North Indian population. Cancer Biomarkers 15:125–131
Chiu CJ, Chiang CP, Chang ML, Chen HM, Hahn LJ, Hsieh LL, Kuo YS, Chen CJ (2001) Association between Genetic Polymorphism of Tumor Necrosis Factor-a and Risk of Oral Submucous Fibrosis, a Pre-cancerous Condition of Oral Cancer. J Dent Res. 80(12):2055–2059
Al-Mohaya MA, Al-Harthi F, Arfin M, Al-Asmari A (2015) TNF-α, TNF-β and IL-10 Gene Polymorphism and Association With Oral Lichen Planus Risk in Saudi Patients. J Appl Oral Sci 23(3):295–301
Suguna S, Nandal DH, Kamble S, Bharatha A, Kunkulol R (2014) Genomic DNA isolation from human whole blood samples by non enzymatic salting out method. Int J Pharm Pharm Sci 6(6):198–199
Boyle JO, Hakim J, Koch W, Van der Riet P, Hruban RH, Roa RA et al (1993) The incidence of p53 mutations increases with progression of head and neck cancer. Cancer Res. 53:4477–4480
Trabace S, Brioli G, Lulli P, Morellini M, Giacovazzo M, Cicciarelli G, Martelletti P (2002) Tumor necrosis factor gene polymorphism in migraine. Headache 42:341–345
Kamali-Sarvestani E, Gharesi-Fard B, Sarvari J, Talei AR (2005) Association of TNF-α and TNF-β gene polymorphism with steroid receptor expression in breast cancer patients. Pathol Oncol Res. 11:99–102
Liu CJ, Wong YK, Chang KW, Chang HC, Liu HF, Lee YJ (2005) Tumor necrosis factor-alpha promoter polymorphism is associated with susceptibility to oral squamous cell carcinoma. J Oral Pathol Med. 34:608–612
Chen LW, Huang HL, Lee IT (2006) Thermal injury-induced priming effect of neutrophil is TNF-α and P38. Depedent Shock 26:69–76
Nonomura N, Tokizane T, Nakayama M, Inoue H, Nishimura K, Muramatsu M, Okuyama (2006) A: Possible correlation between polymorphism in the tumor necrosis factor-betagene and the clinicopathological features of bladder cancer in Japanese patients. Int J Urol 13:971–976
Oliveira ID, Petrilli AS, Tavela MH, Zago MA, de Toledo SR (2007) TNF-alpha, TNF-beta, IL-6, IL-10, PECAM-1 and the MPO inflammatory gene polymorphisms in osteosarcoma. J Pediatr Hematol Oncol 29:293–297
Availability of data and material
All the data and material are presented in this article.
Funding
This study was supported by Council of Science & Technology U.P. (UPCST), India.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
Authors declare that they have no conflict of interest.
Code availability
Not applicable.
Ethics approval
Ethical clearance was obtained from ethical committee of King George’s Medical University. The study was performed in accordance with the Declaration of Helsinki.
Consent to participate
An informed written consent was obtained from all subjects involved in this study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gupta, S., Nigam, K., Srivastav, R.K. et al. Genetic polymorphism of tumor necrosis factor alpha (TNF-α) and tumor necrosis factor beta (TNF-β) genes and risk of oral pre-cancer and cancer in North Indian population. Oral Maxillofac Surg 26, 33–43 (2022). https://doi.org/10.1007/s10006-020-00929-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10006-020-00929-5