Abstract
The relationship between the expression of the SATB2 and CDX2 proteins and common molecular changes and clinical prognosis in colorectal cancer (CRC) still needs further clarification. We collected 1180 cases of CRC and explored the association between the expression of SATB2 and CDX2 and clinicopathological characteristics, molecular alterations, and overall survival of CRC using whole-slide immunohistochemistry. Our results showed that negative expression of SATB2 and CDX2 was more common in MMR-protein-deficient CRC than in MMR-protein-proficient CRC (15.8% vs. 6.0%, P = 0.001; 14.5% vs. 4.0%, P = 0.000, respectively). Negative expression of SATB2 and CDX2 was more common in BRAF-mutant CRC than in BRAF wild-type CRC (17.2% vs. 6.1%, P = 0.003; 13.8% vs. 4. 2%; P = 0.004, respectively). There was no relationship between SATB2 and/or CDX2 negative expression and KRAS, NRAS, and PIK3CA mutations. The lack of expression of SATB2 and CDX2 was associated with poor histopathological features of CRC. In multivariate analysis, negative expression of SATB2 (P = 0.030), negative expression of CDX2 (P = 0.043) and late clinical stage (P = 0.000) were associated with decreased overall survival of CRC. In conclusion, the lack of SATB2 and CDX2 expression in CRC was associated with MMR protein deficiency and BRAF mutation, but not with KRAS, NRAS and PIK3CA mutation. SATB2 and CDX2 are prognostic biomarkers in patients with CRC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is a common malignant tumor of the digestive tract worldwide [1], and its occurrence is a complex process involving multiple genes and factors. At present, it is considered that sporadic CRC is primarily formed through two pathways. One is chromosomal instability, which is characterized by loss of heterozygosity and DNA aneuploidy, and is associated with APC, KRAS, and TP53 mutations; the other is the microsatellite instability pathway, which is related to methylation of the promoter of the DNA mismatch repair gene MLH1, BRAF mutation, and CpG island methylation phenotype [2,3,4]. Molecular changes in CRC often potentially affect protein expression [4,5,6,7,8]. The AT-rich sequence-binding protein 2 (SATB2) and the caudal type homeobox transcription factor 2 (CDX2) are considered as specific immune protein markers of CRC [9,10,11]. SATB2 is a transcription factor that regulates chromatin remodeling and transcription. It is highly expressed in the lower gastrointestinal epithelium (including the appendix, colon, and rectum), specific neurons (cerebral cortex and hippocampus), osteoblast differentiated tumors, and the ductal epithelium of the testis and epididymis [12,13,14]. SATB2 exhibits high sensitivity to colorectal tumors, as >93% of the tumors are positive for this protein, although its level of expression and distribution vary [15]. Thus far, few studies have addressed the relationship between SATB2 protein expression and CRC-associated molecules. A limited number of studies have shown that SATB2 expression in CRC is associated with MMR protein deficiency and BRAF mutation [5, 6, 16]. Moreover, loss of SATB2 expression often occurs in MMR-deficient and BRAF-mutant colon cancer [5, 6]. In colitis-associated colorectal adenocarcinoma, the loss of SATB2 expression is not related to KRAS or BRAF mutation, or MMR protein deficiency [17, 18]. One possible explanation for this finding is that the formation of colitis-associated colorectal adenocarcinoma and sporadic CRC are based on different molecular changes [17]. Research has shown that SATB2 suppresses the progression of CRC both in vitro and in vivo [19]. SATB2-negative expression is associated with reduced survival in CRC patients [20,21,22]. The relationship between the expression of SATB2 protein and the prognosis of CRC still needs further clarification.
During the process of clinicopathological diagnosis, SATB2 is often used in combination with CDX2 to diagnose the origin of colorectal adenocarcinoma or metastatic adenocarcinoma [9,10,11]. CDX2 is a Drosophila tail related homeobox gene that encodes a transcription factor and plays an important role in intestinal development by inhibiting the proliferation and promoting the differentiation and expression of intestine-specific genes [23]. 90 percent of CRC cases show strong CDX2 nuclear positivity. In a study of 713 cases of CRC, two different clones of an anti-CDX2 antibody were used for immunohistochemical detection, which each revealed that CDX2 expression was lost in 5.9% and 6.0% of cases, respectively [8]. Loss of CDX2 expression is closely related to molecular changes in CRC, such as the CpG island methylation phenotype (CIMP), microsatellite instability, and BRAF mutation [4,5,6,7,8]. Concomitantly, it was also found that the loss of CDX2 expression is associated with an aggressive tumor behavior and poor clinical outcomes [8]. CDX2 has been identified as a prognostic biomarker for colon cancer. Loss of CDX2 expression is associated with reduced survival in CRC patients [24,25,26].
Although previous studies have shown that the loss of SATB2 and CDX2 expression is associated with BRAF mutation and MMR protein deficiency in colon cancer [5, 6, 16], the relationship between SATB2 and the status of other molecules (such as KRAS, NRAS, and PIK3CA mutation) remains poorly understood. Moreover, most of those studies were based on the data collected from the Western population, and few reports have examined the Chinese population. To clarify the relationship between the expression of the SATB2 and CDX2 proteins and CRC-associated molecules, we collected tissue samples from 1180 patients with CRC at the Fujian Provincial Hospital, China. Furthermore, to avoid the heterogeneity associated with tumor cell immune protein expression, we used whole-slide sections to evaluate the relationship between SATB2 and CDX2 immunohistochemical expression, and MMR protein status, as well as BRAF, KRAS, NRAS, and PIK3CA mutations. In addition, we evaluated the relationship between the expression of SATB2 and CDX2 proteins and the overall survival of colorectal cancer. This was the largest study in this field to use whole-slide immunohistochemistry and the simultaneous evaluation of the relationship between SATB2 and CDX2 protein expression and CRC-associated molecules. In addition, this study is a supplement to the current lack of research data on the Chinese population.
Lastly, we further confirmed whether the loss of SATB2 and CDX2 expression was associated with MMR protein deficiency and BRAF mutation, but not with KRAS, NRAS, and PIK3CA mutation. In addition, loss of SATB2 and CDX2 expression is associated with decreased overall survival in CRC.
Materials and methods
Patient selection and case review
The clinicopathological data of 1180 cases of colorectal adenocarcinoma diagnosed at the Department of Pathology of the Fujian Provincial Hospital from January 2017 to January 2022 were retrospectively collected. The inclusion criteria were as follows: (1) all patients underwent surgical resection of CRC and were diagnosed with primary colorectal adenocarcinoma; (2) all cases had relatively complete clinical and pathological data; and (3) all cases were diagnosed by two gastrointestinal pathologists. The cohort included 752 males and 428 females and encompassed 231 cases of right colon cancer and 949 cases of left colon and rectum cancer. All tissue samples were fixed in 10% neutral formalin solution (pH 7.2) for 24 h, dehydrated routinely, embedded in paraffin, sectioned at 4 µm, stained with hematoxylin and eosin, and observed. The trial was approved by the ethics committee of the hospital, and the patients voluntarily participated in the trial. All patients signed the informed consent form themselves. The initial operation type, demographic information, and clinical data were obtained from the electronic case system.
Histopathological evaluation
A histopathological evaluation was performed for all cases, and the histological characteristics of each case, including histological grade, pathological stage, lymphatic invasion, venous invasion, and nerve invasion, were then re-evaluated. All histological features were evaluated via routine hematoxylin and eosin staining. The tumor sites were divided into two types: right colon (including the ascending colon, hepatic flexure, and transverse colon), and left colon (including the splenic flexure, descending colon, and sigmoid colon) and rectum. A two-tier tumor grading scheme was employed. A low grade was defined as gland formation >50%, and a high grade was defined as gland formation <50% and/or signet ring cell differentiation. Combined with the clinical data, all cases were classified according to clinical stage.
Immunohistochemical analysis
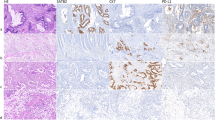
Immunohistochemical staining was performed using the EnVision two-step method. Antibodies for CDX2 (EPR2764Y, Fuzhou Maixin Biotech Co., Ltd, China), SATB2 (EP281, Fuzhou Maixin Biotech Co., Ltd, China), MLH1 (EPR3894, Abcam, UK), PMS2 (EPR3947, Abcam, UK), MSH2 (EPR21017123, Abcam, UK), and MSH6 (EPR3945, Abcam, UK) were used in this experiment. On-slide tissue positive and negative controls were used for all cases. The immunohistochemical expression levels of SATB2 and CDX2 (Fig. 1) were scored as reported previously [6]. For a negative score, two staining patterns were considered: (a) complete loss of expression in tumor cells (score, 0); and (b) a few tumor cells showed scattered and fuzzy nuclear expression (score, 1). For a positive score, two staining patterns were considered: (a) strong staining in most tumor cells (score, 2); and (b) strong staining in all tumor cells (score, 3). For MLH1, PMS2, MSH2, and MSH6, the expression was defined as nuclear staining of tumor cells, using infiltrating lymphocytes and surrounding nontumor intestinal mucosa as internal positive controls. Loss of expression of the MLH1, PMS2, MSH2, and MSH6 proteins was defined as complete absence of nuclear staining within tumor cells, whereas positive signals were detected in the non-neoplastic area of the intestinal mucosa. The absence of MMR protein expression was defined as the nuclear expression of all four MMR proteins. Deficient MMR protein expression was defined as the loss of expression of at least one of the four MMR proteins. Proficient MMR protein expression was defined as preserved nuclear expression of all four MMR proteins.
Molecular mutation analysis
BRAF, KRAS, NRAS, and PIK3CA mutation analysis was performed in paraffin-embedded tissue samples. Typical paraffin blocks were selected and cut into 10 pieces of 10 μm. The cut tissue was placed in a 1.5 mL centrifuge tube. DNA was extracted from paraffin sections according to the instructions of a nucleic acid extraction or purification kit (Xiamen Eide Biomedical Technology Co., Ltd., China). The collected DNA samples were stored at −20 ℃. According to the steps shown in the manual of the human KRAS/NRAS/BRAF/PIK3CA multi gene mutation joint detection kit (Xiamen Eide Biomedical Technology Co., Ltd., China), samples were prepared and detected via real-time fluorescent quantitative PCR. The amplification procedure was as follows: first stage: one cycle of 95 ℃ for 5 min; second stage: 15 cycles of 95 ℃ for 25 s, 64 ℃ for 20 s, and 72 ℃ for 20 s; and third stage: 31 cycles of 93 ℃ for 25 s, 60 ℃ for 35 s, and 72 ℃ for 20 s. The mutation detection sites included KRAS exon 2 (G12D, G12A, G12V, G12S, and G12C), KRAS exon 3 (Q16H), NRAS exon 2 (G12D), NRAS exon 3 (Q61R, Q61K), BRAF exon 15 (V600E), and PIK3CA exon 21 (H1047). Molecular detection in resected tumors is part of the routine evaluation performed at the Fujian Provincial Hospital, and is carried out at the initial pathological evaluation.
Statistical analysis
The Excel software was used to screen, classify, and summarize the data. Statistical analyses were performed using SPSS for Windows 22 (IBM, Armonk, NY). The χ2 or Fisher’s exact tests were used to characterize the relationship between categorical variables. The Kruskal–Wallis test was used to characterize the relationship between continuous variables. The disordered variables were evaluated by two-sided tests, and the ordered variables were evaluated by one-sided tests. P-values < 0.05 were considered statistically significant. Overall survival (OS) is defined as the time (in months) from the date of initial diagnosis to the date of recorded death or the last clinical follow-up. Survival rates were determined by the Kaplan–Meier method and differences between groups were evaluated by log-rank test. Univariate analyses were performed using the binary logistic regression model. Multivariate analyses were performed using the Cox proportional hazard model. Data from univariate and multivariate analyses were reported as hazard ratios with 95% confidence intervals. All statistical tests were performed two-sided, p-values < 0.05 were considered significant.
Results
Negative expression of SATB2 and CDX2 was correlated with poor histopathological features of CRC
Among the 1180 cases of CRC, 78 cases (6.6%) were negative for SATB2 immunohistochemical expression. Compared with SATB2-positive tumors, SATB2-negative tumors exhibited more adverse histological features (Table 1), including a high tumor grade (28.2% vs. 15.5%, P = 0.003), neural invasion (47.4% vs. 33.1%, P = 0.010), vascular invasion (51.3% vs. 39.5%, P = 0.040), lymphatic invasion (51.3% vs. 35.1%, P = 0.004), pN2 stage (24.4% vs. 12.7%, P = 0.004), pM1 stage (15.4% vs. 6.5%, P = 0.004), 003), and later clinical stage (stage III–IV) (52.6% vs. 38.1%, P = 0.011). There was no significant difference in age, tumor diameter, gender, tumor location, and pT stage among these two types of tumor (P > 0.05).
Among the 1180 cases of CRC, 55 cases (4.7%) were negative for CDX2 immunohistochemical expression. Similar to that observed for SATB2, CDX2-negative expression was associated with poor histological features (Table 1). Compared with CDX2-positive tumors, CDX2-negative tumors exhibited a high tumor grade (27.3% vs. 15.8%, P = 0.025), vascular invasion (56.4% vs. 39.5%, P = 0.013), lymphatic invasion (52.7% vs. 35.4%, P = 0.009), pM1 stage (18.2% vs. 6.6%, P = 0.004), and later clinical stage (III–IV) (54.5% vs. 38.3%, P = 0.016) more often. There was no significant difference in age, tumor diameter, gender, nerve invasion, tumor location, pT stage, and pN stage between these two types of tumor (P > 0.05).
The SATB2 and CDX2 expression pattern was associated with MMR protein deficiency and the BRAF V600E mutation
SATB2 and CDX2 immunohistochemical expression was significantly affected in MMR-protein-deficient and BRAF-mutant CRC (Table 2). Negative SATB2 and CDX2 expression was observed significantly more often in BRAF V600E mutated tumors compared with BRAF wild-type tumors (17.2% vs. 6.1%, P = 0.003; 13.8% vs. 4.2%, P = 0.004, respectively) (Fig. 2A). Negative SATB2 and CDX2 expression was observed significantly more often in MMR-protein-deficient CRC compared with MMR-protein-proficient CRC (15.8% vs. 6.0%, P = 0.001; 14.5% vs. 4.0%, P < 0.001, respectively) (Fig. 2B). Immunohistochemical expression of SATB2 and CDX2 was not altered in KRAS, NRAS, and PIK3CA mutated CRC.
A The proportion of SATB2- and CDX2-negative cases in BRAF-mutant colorectal cancer (CRC) (17.2% and 13.8%, respectively) was higher than that detected in wild-type BRAF CRC (6.1% and 4.2%, respectively). B The proportion of SATB2- and CDX2-negative cases in mismatch repair (MMR)-protein-deficient CRC (15.8% and 14.5%, respectively) was higher than that detected in MMR-protein-proficient CRC (6.0% and 4.0%, respectively)
Effects of MMR protein status and BRAF mutation on the combined expression profile of SATB2 and CDX2 in CRC
SATB2 and CDX2 combined immunohistochemical expression profiles were also affected by MMR protein and BRAF mutation status (Table 3). In fact, 27.6% of SATB2-negative and/or CDX2-negative tumors (including 13.2% SATB2−/CDX2+, 11.8% SATB2+/CDX2−, and 2.6% SATB2−/CDX2−) were found among MMR-protein-deficient CRC samples. Only 9.2% of the tumors that were proficient for MMR proteins were SATB2-negative and/or CDX2-negative tumors (P < 0.001). Moreover, 25.9% of BRAF V600E mutated CRC cases were found to be SATB2-negative and/or CDX2-negative tumors, (including 12.1% SATB2−/CDX2+, 8.6% SATB2+/CDX2−, and 5.2% SATB2−/CDX2−). Only 9.5% of BRAF wild-type tumors were SATB2-negative and/or CDX2-negative tumors (P < 0.001).
Negative expression of SATB2 and CDX2 was correlated with poor prognosis in CRC
A total of 993 (84%) out of 1180 CRC patients with known SATB2 and CDX2 expression status underwent clinical follow-up data for overall survival analysis, including 78 SATB2-negative patients and 52 CDX2-negative patients. The median follow-up interval was 42 months (range: 3–66 months) from the time of the initial diagnosis. There were 194 deaths.
In the full cohort (n = 993), Kaplan Meier analysis showed a correlation between negative expression of SATB2 and a decrease in overall survival rate of CRC compared to positive expression of SATB2 (P < 0.05) (Fig. 3a). Similarly, compared to CDX2-positive expression, CDX2-negative expression was associated with a decrease in overall survival in CRC (P < 0.05) (Fig. 3b). In addition, in the univariate survival analysis, lymphatic invasion and the later clinical stage (stage III–IV) were significantly associated with the reduction of the overall survival of CRC (P < 0.001) (Table 4). In multivariate analysis, only negative expression of SATB2 (multivariate risk ratio 1.58, 95% confidence interval 1.05–2.39; P = 0.030), negative expression of CDX2 (multivariate risk ratio 1.64, 95% confidence interval 1.02–2.66; P = 0.043), and later clinical stage (multivariate risk ratio 3.73, 95% confidence interval 1.94–7.16; P = 0.000) were associated with decreased overall survival of CRC (Table 4).
a Kaplan Meier survival curve was used to compare the overall survival of CRC patients through SATB2 expression stratification. Patients with SATB2-negative tumors had reduced overall survival compared with patients with SATB2-positive tumors (p < 0.001). b Kaplan Meier survival curve was used to compare the overall survival of CRC patients through CDX2 expression stratification. Patients with CDX2-negative tumors had reduced overall survival compared with patients with CDX2-positive tumors (p = 0.003)
Discussion
In this study, whole-slide immunohistochemistry was used to evaluate the association between SATB2 and CDX2 immunohistochemical expression in cases of CRC and MMR protein deficiency, as well as BRAF, KRAS, NRAS, and PIK3CA mutations. Our results showed that the immunohistochemical expression of SATB2 and CDX2 was affected by molecular changes in CRC, and that negative SATB2 and CDX2 expression was more common in MMR-protein-deficient CRC and BRAF-mutant CRC, but not in KRAS-, NRAS-, and PIK3CA-mutant CRC. In addition, we observed that negative expression of SATB2 and CDX2 was associated with poor histopathological features and decreased overall survival of CRC.
Thus far, few studies have addressed the relationship between SATB2 protein expression and molecules commonly associated with CRC. Ma et al. [5] analyzed 499 cases of colon cancer and observed negative SATB2 and/or CDX2 expression in 33% of MMR-protein-deficient tumors and 36% of BRAF V600E-mutant tumors. This result is similar to that obtained in our study (Table 2). In addition, those authors found that the negative expression of SATB2 was associated with a low disease-specific survival rate among MMR-protein-deficient cases of colon cancer [5]. In their study, the negative expression rate of SATB2 (67/499, 13%) was higher than that detected in our study (78/1180, 6.6%). Eberhardt et al. [21] analyzed 527 cases of colon cancer and also observed that SATB2 expression was often absent in MMR-protein-deficient tumors; moreover, negative expression of SATB2 can be used as an independent predictor of a decreased disease-specific survival rate among patients with colon cancer. In their study, the negative expression rate of SATB2 was 28.8% (152/527), which was much higher than that detected here. We propose the following explanations for this discrepancy: first, compared with Ma et al. (499 cases) and Eberhardt et al. (527 cases), our study included a larger sample (1180 cases) because it included cases of rectal cancer. Second, different antibody clones were used in each study, which may explain the differences in SATB2 expression detected in the samples of CRC. In addition, the definition of negative expression of SATB2 was slightly different. Further, we used the whole-slide immunohistochemistry method, rather than the tissue microarray method, to analyze the expression of SATB2. Cigerova et al. [16] observed that the SATB2 protein was absent only in 7.2% of CRC cases. This is similar to our results. In addition, our study found that negative SATB2 expression was not associated with KRAS, NRAS, and PIK3CA mutation status. This is rarely mentioned in the remaining pertinent literature. It was recently reported that expression of SATB2 is also frequently absent in colitis-associated colorectal adenocarcinoma; however, the loss of SATB2 is not related to BRAF mutation and MMR protein deficiency [17, 18]. One possible explanation for this finding is that the formation of colitis-associated colorectal adenocarcinoma may be triggered by a continuous inflammatory environment, which induces epithelial DNA mutation [17] and is different from the mechanism underlying sporadic CRC. Eberhardt et al. [21] found that high expression of SATB2 is associated with a good prognosis in colon cancer, and SATB2 expression is an independent prognostic factor for overall survival and cancer specific survival in colon cancer. Wang et al. [22] found that low expression of SATB2 is closely related to tumor invasion, lymph node metastasis, distant metastasis, and Dukes' classification of CRC. Low expression of SATB2 is closely related to decreased overall survival and disease-free survival. Schmitt et al. [20] found that SATB2 showed particularly high prognostic relevance in univariate and multivariate analysis of high-risk clinicopathological subgroups. In our study, we observed that SATB2 negative expression was associated with poor histopathological features in patients with CRC, including a high tumor grade, neural invasion, vascular invasion, lymphatic invasion, and later pathological and clinical stage. In addition, we also found that negative expression of SATB2 was associated with decreased overall survival of CRC. These results are consistent with the literature [5, 6, 20,21,22].
Previous studies have shown that the loss of CDX2 expression is closely related to the molecular changes of colorectal cancer [4,5,6,7,8, 27, 28]. Lugli et al. [4] observed that CDX2 was more likely to be lost in MMR-protein-deficient colorectal cancer than in MMR-protein-proficient CRC. Subsequently, Ma et al. [6] further confirmed that CDX2 expression is often absent in MMR-protein-deficient and BRAF-mutant colon cancer. Most of those studies did not mention the relationship between CDX2 and KRAS, NRAS, and PIK3CA mutation. We observed that the expression of CDX2 was not affected by the mutation status of KRAS, NRAS, and PIK3CA in patients with CRC. In a study of 713 cases of CRC, Bae et al. [8] used two different clones of an anti-CDX2 antibody (CDX2-88 and EPR2764Y) and found that the CDX2 negative expression rates were 5.9% (CDX-88) and 6.0% (EPR2764Y). This was similar to our study (55/1180, 4.7%). Olsen et al. [27] conducted a qualitative systematic review of 52 studies of CDX2 expression in CRC. They observed that the loss of CDX2 expression was related to tumor grade, tumor stage, right tumor location, MMR deficiency, high CIMP, and BRAF mutation. Similar results were observed in our study. Some studies have also pointed out that the lack of CDX2 expression is an independent risk factor for low disease-specific survival in CRC, and can serve as a prognostic biomarker for stage II and III colon cancer [25]. Chen et al. [29] found that loss of CDX2 expression in CRC is associated with poor overall survival and disease-free survival in a recent meta-analysis. Similarly, our study found that negative expression of CDX2 is associated with a decreased overall survival of CRC patients.
Although SATB2 and CDX2 are often negatively expressed in CRC cases with MMR protein deficiency and BRAF mutation, their expression is different. Concurrent negative SATB2 and CDX2 expression (SATB2−/CDX2−) was only rarely observed as it was identified in 2.6% of MMR-protein-deficient and 5.2% of BRAF V600E-mutant CRC samples. Therefore, the losses of SATB2 and/or CDX2 expression are independent from each other in most cases of CRC with MMR protein deficiency and BRAF mutation. This is similar to that reported by Ma et al. [5, 6]. Therefore, when the morphology of CRC is not typical, the combination of SATB2 and CDX2 can help establish a correct diagnosis and avoid misdiagnosis, especially in MMR-protein-deficient and BRAF-mutant tumors. The reason for the loss of SATB2 and CDX2 expression in MMR-protein-deficient and BRAF-mutant CRC remains unclear. Some studies have suggested that epigenetic silencing caused by a high level of CpG island promoter methylation may be a mechanism of CDX2 expression reduction [7, 25, 26]. In addition, CDX2 plays an important role in the regulation of the polarity of epithelial cells, and the loss of CDX2 may be related to the interruption of epithelial tight junction and epithelial mesenchymal transition (EMT) [25, 30]. There are also studies that suggest that reduced CDX2 expression is caused by a passenger mutation in the simple repeat sequence of the CDX2 gene [31]. It is not clear whether the reduced SATB2 expression occurs via the same mechanism as CDX2. Ma et al. [6] proposed that alternative mechanisms lead to the loss of SATB2 expression, as only 14% of MMR-protein-deficient tumors exhibited concurrent loss of CDX2 and SATB2 expression. Our results also support this view.
This study has some limitations, e.g., the inclusion and screening of cases using a retrospective design and the heterogeneity of protein expression in tissue sections. However, the advantage of this study is that partial heterogeneity was solved using whole-slide immunohistochemistry, which also conferred considerable reliability to the negative expression of SATB2, and CDX2. In addition, our study is one of the largest studies thus far that investigated the negative expression of SATB2 and CDX2 and molecular changes in CRC using whole-slide immunohistochemistry. Our study is a representative of CRC resected at a large academic medical center in Fujian Province, China; therefore it has an inherent referral bias.
In conclusion, our results suggest that negative SATB2 and CDX2 expression is associated with MMR protein deficiency and BRAF mutation, but not with KRAS, NRAS, and PIK3CA mutation, in patients with CRC. Negative expression of SATB2 and CDX2 is an independent marker for poor prognosis in CRC.
Availability of data and materials
The datasets generated and/or analyzed in the present study are available from the corresponding author on reasonable request.
References
Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F (2017) Global patterns and trends in colorectal cancer incidence and mortality. Gut 66:683–691. https://doi.org/10.1136/gutjnl-2015-310912
Kambara T, Simms LA, Whitehall VL, Spring KJ, Wynter CV, Walsh MD, Barker MA et al (2004) BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut 53:1137–1144. https://doi.org/10.1136/gut.2003.037671
Jass JR (2007) Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology 50:113–130. https://doi.org/10.1111/j.1365-2559.2006.02549.x
Lugli A, Tzankov A, Zlobec I, Terracciano LM (2008) Differential diagnostic and functional role of the multi-marker phenotype CDX2/CK20/CK7 in colorectal cancer stratified by mismatch repair status. Mod Pathol 21:1403–1412. https://doi.org/10.1038/modpathol.2008.117
Ma C, Olevian D, Miller C, Herbst C, Jayachandran P, Kozak MM et al (2019) SATB2 and CDX2 are prognostic biomarkers in DNA mismatch repair protein deficient colon cancer. Mod Pathol 32:1217–1231. https://doi.org/10.1038/s41379-019-0265-1
Ma C, Olevian DC, Lowenthal BM, Jayachandran P, Kozak MM, Chang DT et al (2018) Loss of SATB2 expression in colorectal carcinoma is associated with DNA mismatch repair protein deficiency and BRAF mutation. Am J Surg Pathol 42:1409–1417. https://doi.org/10.1097/PAS.0000000000001116
Kim JH, Rhee YY, Bae JM, Kwon HJ, Cho NY, Kim MJ et al (2013) Subsets of microsatellite-unstable colorectal cancers exhibit discordance between the CpG island methylator phenotype and MLH1 methylation status. Mod Pathol 26:1013–1022. https://doi.org/10.1038/modpathol.2012.241
Bae JM (2015) Loss of CDX2 expression is associated with poor prognosis in colorectal cancer patients. World J Gastroenterol 21:1457. https://doi.org/10.3748/wjg.v21.i5.1457
Lin F, Shi J, Zhu S, Chen Z, Li A, Chen T et al (2014) Cadherin-17 and SATB2 are sensitive and specific immunomarkers for medullary carcinoma of the large intestine. Arch Pathol Lab Med 138:1015–1026. https://doi.org/10.5858/arpa.2013-0452-OA
Dragomir A, de Wit M, Johansson C, Uhlen M, Ponten F (2014) The role of SATB2 as a diagnostic marker for tumors of colorectal origin: results of a pathology-based clinical prospective study. Am J Clin Pathol 141:630–638. https://doi.org/10.1309/AJCPWW2URZ9JKQJU
Kaimaktchiev V, Terracciano L, Tornillo L, Spichtin H, Stoios D, Bundi M et al (2004) The homeobox intestinal differentiation factor CDX2 is selectively expressed in gastrointestinal adenocarcinomas. Mod Pathol 17: 1392–1399. https://doi.org/10.1038/modpathol.3800205
Berg KB, Schaeffer DF (2017) SATB2 as an immunohistochemical marker for colorectal adenocarcinoma: a concise review of benefits and pitfalls. Arch Pathol Lab Med 141:1428–1433. https://doi.org/10.5858/arpa.2016-0243-RS
Magnusson K, de Wit M, Brennan DJ, Johnson LB, McGee SF, Lundberg E et al (2011) SATB2 in combination with cytokeratin 20 identifies over 95% of all colorectal carcinomas. Am J Surg Pathol 35: 937–948. https://doi.org/10.1097/PAS.0b013e31821c3dae
FitzPatrick DR, Carr IM, McLaren L, Leek JP, Wightman P, Williamson K et al (2003) Identification of SATB2 as the cleft palate gene on 2q32-q33. Hum Mol Genet 12: 2491–2501. https://doi.org/10.1093/hmg/ddg248
Mezheyeuski A, Ponten F, Edqvist P-H, Sundström M, Thunberg U, Qvortrup C et al (2019) Metastatic colorectal carcinomas with high SATB2 expression are associated with better prognosis and response to chemotherapy: a population-based Scandinavian study. Acta Oncolog 59:284–290. https://doi.org/10.1080/0284186X.2019.1691258
Cigerova V, Adamkov M, Drahosova S, Grendar M (2021) Immunohistochemical expression and significance of SATB2 protein in colorectal cancer. Ann Diagn Pathol 52: 151731. https://doi.org/10.1016/j.anndiagpath.2021.151731
Ma C, Henn P, Miller C, Herbst C, Hartman DJ, Pai RK (2019) Loss of SATB2 expression is a biomarker of inflammatory bowel disease-associated colorectal dysplasia and adenocarcinoma. Am J Surg Pathol 43:1314–1322. https://doi.org/10.1097/PAS.0000000000001330
Iwaya M, Ota H, Tateishi Y, Nakajima T, Riddell R, Conner JR (2019) Colitis-associated colorectal adenocarcinomas are frequently associated with non-intestinal mucin profiles and loss of SATB2 expression. Mod Pathol 32:884–892. https://doi.org/10.1038/s41379-018-0198-0
Mansour MA, Hyodo T, Ito S, Kurita K, Kokuryo T, Uehara K et al (2015) SATB2 suppresses the progression of colorectal cancer cells via inactivation of MEK5/ERK5 signaling. FEBS J 282:1394–1405. https://doi.org/10.1111/febs.13227
Schmitt M, Silva M, Konukiewitz B et al (2021) Loss of SATB2 occurs more frequently than CDX2 loss in colorectal carcinoma and identifies particularly aggressive cancers in high-risk subgroups. Cancers (Basel) 13(24):6177. https://doi.org/10.3390/cancers13246177
Eberhard J, Gaber A, Wangefjord S, Nodin B, Uhlen M, Ericson Lindquist K et al (2012) A cohort study of the prognostic and treatment predictive value of SATB2 expression in colorectal cancer. Br J Cancer 106:931–938. https://doi.org/10.1038/bjc.2012.34
Wang S, Zhou J, Wang XY, Hao JM, Chen JZ, Zhang XM et al (2009) Down-regulated expression of SATB2 is associated with metastasis and poor prognosis in colorectal cancer. J Pathol 219:114–122. https://doi.org/10.1002/path.2575
Guo RJ, Suh ER, Lynch JP (2004) The role of Cdx proteins in intestinal development and cancer. Cancer Biol Ther 3:593–601. https://doi.org/10.4161/cbt.3.7.913
Dalerba P, Sahoo D, Paik S, Guo X, Yothers G, Song N et al (2016) CDX2 as a prognostic biomarker in stage II and stage III colon cancer. New Eng J Med 374: 211–222. https://doi.org/10.1056/NEJMoa1506597
Slik K, Turkki R, Carpen O, Kurki S, Korkeila E, Sundstrom J et al (2019) CDX2 loss with microsatellite stable phenotype predicts poor clinical outcome in stage II colorectal carcinoma. Am J Surg Pathol 43: 1473–1482. https://doi.org/10.1097/PAS.0000000000001356
Cecchini MJ, Walsh JC, Parfitt J, Chakrabarti S, Correa RJ, MacKenzie MJ et al (2019) CDX2 and Muc2 immunohistochemistry as prognostic markers in stage II colon cancer. Hum Pathol 90: 70–79. https://doi.org/10.1016/j.humpath.2019.05.005
Olsen J, Espersen ML, Jess P, Kirkeby LT, Troelsen JT (2014) The clinical perspectives of CDX2 expression in colorectal cancer: a qualitative systematic review. Surg Oncol 23:167–176. https://doi.org/10.1016/j.suronc.2014.07.003
Baba Y, Nosho K, Shima K, Freed E, Irahara N, Philips J et al (2009) Relationship of CDX2 loss with molecular features and prognosis in colorectal cancer. Clin Cancer Res 15: 4665–4673. https://doi.org/10.1158/1078-0432.CCR-09-0401
Chen K, Collins G, Wang H, Toh JWT (2021) Pathological features and prognostication in colorectal cancer. Curr Oncol 28:5356–5383. https://doi.org/10.3390/curroncol28060447
Zhang JF, Qu LS, Qian XF, Xia BL, Mao ZB, Chen WC (2015) Nuclear transcription factor CDX2 inhibits gastric cancer-cell growth and reverses epithelial-to-mesenchymal transition in vitro and in vivo. Mol Med Rep 12:5231–5238. https://doi.org/10.3892/mmr.2015.4114
Salari K, Spulak ME, Cuff J, Forster AD, Giacomini CP, Huang S et al (2012) CDX2 is an amplified lineage-survival oncogene in colorectal cancer. Proc Natl Acad Sci USA 109:E3196–E3205. https://doi.org/10.1073/pnas.1206004109
Acknowledgements
Not applicable.
Funding
Research works in this paper are financially supported by Qihang Fund General Project of Fujian Medical University (2022QH1301).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest. All authors participated sufficiently in the work to take public responsibility for the content, including participation in the concept, design, analysis, writing, or revision of the manuscript. In addition, each author certifies that this material or similar material has not been and will not be submitted to or published in any other publication.
Ethical approval
The study complied with the Helsinki Declaration, and the study protocol and exemption of informed consent were approved by the Institutional Ethics Committee of Fujian Provincial Hospital, Fuzhou, China (K2021-09-042).
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, J., Zeng, Q., Lin, J. et al. Loss of SATB2 and CDX2 expression is associated with DNA mismatch repair protein deficiency and BRAF mutation in colorectal cancer. Med Mol Morphol 57, 1–10 (2024). https://doi.org/10.1007/s00795-023-00366-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00795-023-00366-9