Abstract
The waters of Lake Magic in Western Australia are among the most geochemically extreme on Earth. This ephemeral saline lake is characterized by pH as low as 1.6 salinity as high as 32% total dissolved solids, and unusually complex geochemistry, including extremely high concentrations of aluminum, silica, and iron. We examined the microbial composition and putative function in this extreme acid brine environment by analyzing lake water, groundwater, and sediment samples collected during the austral summer near peak evapoconcentration. Our results reveal that the lake water metagenome, surprisingly, was comprised of mostly eukaryote sequences, particularly fungi and to a lesser extent, green algae. Groundwater and sediment samples were dominated by acidophilic Firmicutes, with eukaryotic community members only detected at low abundances. The lake water bacterial community was less diverse than that in groundwater and sediment, and was overwhelmingly represented by a single OTU affiliated with Salinisphaera. Pathways associated with halotolerance were found in the metagenomes, as were genes associated with biosynthesis of protective carotenoids. During periods of complete desiccation of the lake, we hypothesize that dormancy and entrapment in fluid inclusions in halite crystals may increase long-term survival, leading to the resilience of complex eukaryotes in this extreme environment.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Studying how life adapts and diversifies in extreme environments is crucial to understanding the range of physical, chemical, and biological conditions that exist on Earth and possibly on other planets. Acid saline environments host organisms living in them not only with extreme hydrogen ion concentration gradients, but often with accompanying challenges, such as low nutrient and high metal concentrations (Brake and Hasiotis 2010; Ruecker et al. 2016). Moreover, these lakes undergo repeated cycles of flooding, evapoconcentration, and complete desiccation, giving rise to critical questions about the survival mechanisms utilized by microbial cells under these extreme conditions.
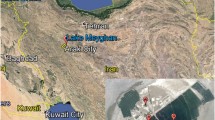
Although the modern chemical and sedimentological processes of these acid lakes have recently been documented (Benison et al. 2007; Bowen and Benison 2009; Benison and Bowen 2013, 2015; Ruecker et al. 2016), the biology of these extreme environments remains enigmatic. Lake Magic is located 4 km northeast of the town of Hyden in Western Australia (Fig. 1), and stands out for having amongst the lowest pH, highest salinity, and highest dissolved aluminum, iron, and silica known from any surface water. Lake Magic waters have a pH low of 1.6, high salinity (32% TDS), and concentrations of aluminum, iron, and silica reaching 1774, 331, and 510 mg/L, respectively (Wave Rock Lake 2 in Bowen and Benison 2009; Conner and Benison 2013). During evapoconcentration stage, the lake bed itself is rich in halite with moderate amounts of gypsum. The site also precipitates small amounts of hematite, jarosite, and alunite from its groundwaters. Similar to other acid salt lakes in the Yilgarn Craton, Lake Magic an ephemeral lake, to which water is supplied both via a regional acid brine groundwater system and by infrequent meteoric precipitation (Benison et al. 2007). The oxidation of sulfides has been suggested as the primary source of acidity, with ferrolysis (a combined process of oxidation and hydrolysis that occurs in waters enriched with dissolved iron), high rates of evaporation, and potentially acidophilic microorganisms serving as secondary acidification mechanisms (Benison and Bowen 2015).
Location of Lake Magic, Western Australia. a Google Earth image of area near town of Hyden and tourist attraction Wave Rock. Satellite image by DigitalGlobe 2017. b Google Earth image of Lake Magic with approximate locations of the four sample sites: LW lake water, GW groundwater, MFS mudflat sediment, SS subaqueous sediment. Satellite image by DigitalGlobe 2017. c Photograph, looking north from the lake toward brown mudflat and tan sandflat. In foreground, halite raft floating on water surface cast shadows on lake bottom, and (inset) photograph of brown mud under halite crust at lake bottom
Microbiological studies of acid brine lakes in the region have revealed a surprising level of bacterial diversity within the lake water, groundwater, and sediment, with acidophilic and acidotolerant, as well as halophilic and halotolerant, community members (Mormile et al. 2007, 2009; Johnson et al. 2015; Weigold et al. 2016). Metagenomic analysis of sediment in a nearby lake, Lake Gneiss, has indicated that microbes inhabiting these acid, saline depositional environments may be involved in sulfur transformations, possibly including direct formation of sulfur minerals (Johnson et al. 2015). It has also been hypothesized that metabolic activities of these microbial communities could alter sediment chemistry by generating acidity and facilitating mineral precipitation (Johnson et al. 2015; Benison and Bowen 2013; Mormile et al. 2009). Further, micro-algae, prokaryotes and organic compounds have been documented in fluid inclusions within halite crystals precipitated by the lake water (Conner and Benison 2013). To date, no metagenomic studies have been published on the Lake Magic system or on the waters of any acid brine lake in the region. To better understand population structure, biogeochemical roles and cell survival strategies in this extreme environment, we use high-throughput sequencing of bacterial 16S rRNA gene amplicons and metagenomes to investigate the diversity and putative function of the communities inhabiting Lake Magic, including the lake’s pelagic zone, sediments, and groundwater.
Materials and methods
Sample collection
Samples were collected from Lake Magic during the austral summer during a stage of evapoconcentration with large amounts of halite and gypsum precipitation (Fig. 1). Samples of lake water, groundwater, and sediments (subaqueous and mudflat) were collected in January 2015 (Table 1). Approximate sample locations are shown in Fig. 1b. The temperature, pH, and salinity of water at sampling sites were measured and are presented in Table 1. Temperatures were taken with plastic-encased glass and alcohol thermometers at various depths within the water column, and no thermal stratification was found. Oakton triple junction pH test 2 portable pH meters with automatic temperature compensation, a range from − 1.0 to 15.0 pH, and accuracy to 0.1 pH unit were used. Orion pH buffers at pH 10, 7, 4, and 1.38 were used to calibrate the pH meters at the start and middle of each field day. All pHs were measured in the field with two pH meters and confirmed later in the lab with an Orion 230A pH meter. Hoake optical salinity refractometers, models HRS-28, were used to measure total dissolved solids in the field. Salinity was also tested at both bottom and top of water column to check for salinity stratification; none was found.
Lake water was collected in sterile high-density polyethylene bottles and subsequently filtered in 100 mL aliquots onto 0.2 µm polyethersulfone filters. The filters were then placed into sterile 2.0 mL Nalgene Cryovial tubes and suspended in LifeGuard Soil Preservation Solution (Mo Bio, Carlsbad, CA, USA).
Groundwater was sampled by trenching the sandflat on the north side of the lake and collecting 20 mL of groundwater from a depth of 34 cm in sterile 50 mL polypropylene centrifuge tubes (Fisher Scientific, Hampton, NH, USA). The groundwater from the trench was subsequently filtered with the same process described for lake water filtration. It was subsampled into five vials and suspended in LifeGuard Soil Preservation Solution (Mo Bio, Carlsbad, CA, USA). The vials containing lake water and groundwater samples were maintained at 4 °C from sample collection through shipment to Mormile’s laboratory. Once in Mormile’s lab, the vials were kept at − 70 °C until the DNA was extracted.
Three types of sediment samples were taken from the vicinity of the groundwater trench, collected in 1 mL aliquots using sterile spatulas. The sediment was placed into sterile 2.0 mL Nalgene Cryovial tubes and subsequently suspended in LifeGuard Soil Preservation Solution (Mo Bio, Carlsbad, CA, USA). Subaqueous sediment (SS, see Fig. 1) was collected near the northern edge of the lake where there was no halite and gypsum bottom-growth crust. Additional sediment samples were collected below a crust of bottom-growth halite and gypsum crystals at the lake bottom, but these samples failed to yield enough DNA for sequencing, and therefore were excluded from further analyses. Subaerially exposed sediment was also collected on the mudflat between the groundwater trench and the shoreline. This mudflat sediment (MFS, see Fig. 1) was slightly greenish in hue and dry, as it was situated above the water line. All sediment was collected from ~ 1 to 2 cm from below the surface.
DNA extraction and library preparation
Total genomic DNA was prepared and pooled from replicate DNA extractions of Lake Magic lake water (LW), groundwater (GW) and sediment (MFS and SS) samples, according to standard protocols using PowerSoil, PowerWater, and Powerlyzer DNA Isolation Kits (MO Bio Laboratories, Carlsbad, CA, USA). The retrieved DNA was shipped overnight on dry ice to Washington, DC, where the DNA quality and quantity were estimated by UV–Vis spectrophotometry with a ND-1000 spectrophotometer (Thermo Fisher NanoDrop) and fluorometry using the Qubit dsDNA HS Assay Kit on the Qubit 2.0 fluorometer (Life Technologies), respectively. Indexed, paired-end sequencing libraries were prepared using the Illumina MiSeq platform with the MiSeq Reagent Kit v3 (600 cycle).
Bacterial 16S rRNA gene amplicon sequencing
To gauge the diversity of the bacterial communities present at Lake Magic, DNA extracted from lake water (LW), groundwater (GW), and sediment (SS) was amplified using bacteria-specific primers targeting the V6–V8 hypervariable region of the 16S rRNA gene. Amplicons were then sequenced on an Illumina MiSeq platform. The resultant amplicon sequences were analyzed using mothur software (Schloss 2009). Forward and reverse reads were assembled, and the resulting contigs were screened for appropriate length, homopolymer content (< 6) and sequences containing any ambiguous bases were removed from further analysis. Sequences passing initial quality control were aligned to a reference database (Silva release 123) for operational taxonomic unit (OTU) clustering and downstream analyses. Prior to clustering, chimeric sequences were identified using the VSEARCH algorithm and were excluded from the dataset. The remaining sequences were clustered at 97% identity by first calculating uncorrected pairwise distances between aligned DNA sequenced, and then clustering the distance matrix. Alpha diversity metrics and rarefaction curves were calculated using mothur. The taxonomic identity of OTUs and OTU abundances were used to construct an OTU table in R. Sequences were subsampled in mothur for multivariate statistics carried out in R using vegan and hclust packages. Singleton OTUs consisting of a single read were removed from comparative analyses. Krona and R ggplot2 and default packages were used to visualize analyses outputs.
Metagenomic sequencing
To assess non-bacterial community composition as well as investigate community function, whole genome shotgun libraries from Lake Magic lake water (LW) and groundwater (GW) and mudflat sediment (MFS) were constructed and also sequenced on an Illumina MiSeq platform. Sequences were filtered for length and quality scores > 20, and where compared to the RefSeq database using LAST. LAST hits were filtered for a minimum e-score of 120 and were subsequently analyzed using MEGAN (version 7.6.10). Reads with hits to genes encoding proteins were assigned to KEGG, COG and SEED database categories. Taxonomic affiliation of the genes in the metagenomic dataset was determined using the taxonomic LCA placement and minimum taxon cover algorithms. Taxonomic composition of metagenomes and distribution of gene assignments are based on normalized data to ensure even sampling effort between samples. Metagenomic and SSU rRNA amplicon sequences reported in this study were deposited in the GenBank Sequence Read Archive, with sequence accession numbers SRR5990693–SRR5990698.
Results
Physicochemical parameters of Lake Magic
At time of sampling in early January 2015, the daytime air temperature at Lake Magic ranged from 21.5 to 35 °C. The lake was only 20 cm deep at its deepest point and lake water was bright yellow in color (Fig. 1). All water samples were highly saline and had low pH (Table 1). Over the course of the day, as evaporation increased, lake water pH decreased from 2.3 to 1.7, and salinity increased from 26 to 30% TDS. Lake water temperatures remained within 4 °C of local air temperature throughout the day. However, no differences were detected in salinity, pH, or temperature from bottom to top of water column, indicating unstratified lake water. Numerous halite crystal rafts with diameters up to approximately 6.0 cm had formed on the surface of the water, indicating that the water was at saturated conditions.
At the time of sampling, the groundwater was warmer than lake water, but was considerably cooler than the water surrounding the SS sample (Table 1). Groundwater was also less saline and higher in pH than the lake water and water above the subaqueous sediment. Although the SS water was less saline and lower in pH than lake water at the time of sampling, it fell within the range observed in lake water throughout the day.
Bacterial community composition and structure in Lake Magic based on 16S rRNA gene amplicon reads
To characterize the identities and relative abundance of bacteria in Lake Magic, a total of 1.6 million 16S rRNA gene amplicon reads were generated for lake water, ground water, and sediment samples. The most abundant and diverse bacterial phyla recovered from Lake Magic were Proteobacteria and Firmicutes (Fig. 2). Although, the same phyla were present in the different lake compartments, the composition and OTU distribution varied among them. Each lake compartment had more OTUs that were unique to it and not shared with the other samples and lake water shared a larger proportion of its OTUs with sediment than with groundwater (Fig. 2b). Of the three lake habitats, the highest bacterial diversity was observed in the sediment (SS) sample, with > 1000 non-singleton OTUs (Table 2, Fig. 2). The GW bacterial community was of a similar diversity, with ~ 980 OTUs. Conversely, the bacterial community of the lake water exhibited much lower diversity, consisting of 11 phyla (Table 2, Fig. 3).
Bacterial SSU rRNA gene OTU (97% ID) diversity and distribution. a Observed species richness rarefaction curve, with 250,000 sequences subsampled per sample. b Venn diagram showing bacterial OTU distribution among samples. Circle sizes are scaled to total number of OTUs within each sample. LW lake water (green), GW groundwater (blue), and SS subaqueous sediment (purple)
Bacterial community composition and structure. a Barplot of OTU-level diversity of bacterial phyla within the samples. Bars represent the proportion of OTUs assigned to each phylum. b Barplot of OTU-level abundance of bacterial phyla within the samples, where bars represent the proportion of reads assigned to each phylum. LW lake water, GW groundwater, SS subaqueous sediment
Lake Magic contained bacterial phyla that were diverse (percent OTU), but low abundance (percent reads), as well as phyla that were abundant, but not very diverse (Fig. 3). Moreover, a single OTU dominated the lake water bacterial community, with 90% of all reads belonging to a single OTU affiliated with the genus Salinisphaera (Supplementary Fig. 1). This OTU was rare in groundwater (0.03%), but represented 27% of the sediment community, where it was the second-most abundant OTU. Salinisphaera species are halotolerant, mesophilic, and slightly acidophilic bacteria, with pH optima of 5.0–7.5 (Crespo-Medina et al. 2009; Shimane et al. 2013; Antunes et al. 2003). The pH tolerance ranges differ between species belonging to this genus, for example, pH 5.0–7.5 for S. hydrothermalis and pH 3.8–9.5 for S. japonica (Crespo-Medina et al. 2009; Shimane et al. 2013). The second-most abundant OTU in lake water (5.7%) was assigned to the alphaproteobacterial genus Methylobacterium. Species belonging to this genus are aerobic facultative methylotrophs that are capable of forming biofilms and exhibit slow growth and resistance to desiccation and high temperatures (Fredrickson et al. 2008; and reviewed in Kovaleva et al. 2014).
In the sediment and groundwater, the most abundant phylum was Firmicutes; however, the taxonomic composition of this phylum differed noticeably between these two samples (Fig. 3). In the sediment, the most abundant OTU (33%) was rare in the water samples (0.02% in LW and 0.2% in GW) and belonged to a thermoacidophilic, endospore-forming, sulfur-oxidizing Firmicutes genus, Alicyclobacillus, as did its third most abundant OTU. Members of this extremophilic genus occur in soils, mining sites, acid mine drainage, and geothermal sites (Méndez-García et al. 2015; Jiang et al. 2008; Baker and Banfield 2003; Simbahan et al. 2004; Kim et al. 2014). These bacteria are also known agents of fruit juice spoilage due to the ability of the Alicyclobacillus spp. spores to survive the high temperatures of typical pasteurization (Walls and Chuyate 2000; Chang and Kang 2004). The fourth-most abundant OTU in the sediment was an unclassified Cytophagia bacterium and represented 3.6% of the SS bacterial community. Additional Bacilli lineages, Planctomycetes, and the deeply branching halophilic Mollicutes (Tenericutes) represented most of the remaining SS bacterial diversity (Fig. 3, Supplementary Fig. 1), suggesting a primarily heterotrophic salt-tolerant community (Antunes et al. 2008).
The groundwater was also dominated (~ 43%) by a single OTU, belonging to the halophilic Firmicutes genus Oceanobacillus. This OTU was just 0.001% of SS bacteria and 0.08% of the LW bacterial community. Oceanobacillus species are halophilic, and typically alkaliphilic, and have been isolated from a variety of environments including fermented foods, deep-sea sediments, and the human gut (Lu et al. 2001; Whon et al. 2010; Jang et al. 2014; Lagier et al. 2015; Amoozegar et al. 2016). The second and third most abundant OTU in groundwater belonged to Paenibacillus, a genus of extremophilic, facultatively anaerobic, endospore-forming Firmicutes (La Duc et al. 2007; Stieglmeier et al. 2009). These two OTUs combined were 0.08 and 0.004% of the lake water and sediment bacteria, respectively. Another notable (3.5%) member of the groundwater community was related to a putative sulfide-oxidizing Acidithiobacillales uncultured bacterium clone 9M32. These organisms were not present in the sediment and were represented by just one read in the lake water.
Eukaryotes comprise most of the pelagic metagenome
To extend beyond our 16S rRNA gene-based bacterial community characterization and identify other members of the Lake Magic community, over 1.8 million high quality whole genome shotgun reads were generated for lake water, mudflat sediment and groundwater samples (Table 3). Taxonomic analysis of genomic reads revealed that although 2–11% of reads with good LAST hits could not be assigned to a specific lineage, most of the metagenomic sequences recovered from MFS and GW were bacterial, 92 and 85%, respectively (Fig. 4). Lake water sequences, surprisingly, were primarily eukaryotic, with 73% of the reads assigned to this domain (Fig. 4). Groundwater contained archaeal reads at a higher proportion (2.4%) than either lake water (0.030%) or the dry mudflat sediment (0.046%). Most of the archaea belonged to Euryarchaeota, more specifically the extremely acid-tolerant order of iron-oxidizers, Thermoplasmatales (Schleper et al. 1995; Edwards et al. 2000; Ruepp et al. 2000). Other archaeal members were methanogenic Euryarchaeota species and Sulfolobus species (Crenarchaeota). All known members of the Sulfolobus genus are thermophilic and acidophilic aerobic sulfur oxidizers that are capable of CO2 fixation, and are typically found in areas of volcanic or geothermal activity (Chen et al. 2005; Mao and Grogan 2012; Dai et al. 2016).
The taxonomic distribution of reads assigned to bacteria was consistent with observations in the 16S rRNA gene dataset. The LW bacterial diversity was low, with bacterial reads assigned to the phyla Proteobacteria (Salinisphaera), Fibrobacteres, Chlorobi, and Bacteroidetes (FCB) group, and Terrabacteria (Supplementary Fig. 2). Similar to the 16S rRNA gene dataset, a large proportion of GW sequences were assigned to Oceanobacillus (~ 15% of all assigned reads) and Paenibacillus (~ 17%) (Supplementary Fig. 2). Metagenomic sequences also revealed the presence of bacteria of the extremophilic phylum Deinococcus–Thermus, particularly in the groundwater. Sequences assigned to photoautotrophic phylum Cyanobacteria, with representatives from sub-taxa Nostocales, Oscillatoriophycideae, and Synechococcales, were present in small proportions in all three samples (0.12–0.17%), possibly indicating that autotrophic carbon fixation may be carried out by other taxa, for example, eukaryotic microbes. The distribution of bacterial taxa among the different lake compartments indicates possible exchange between the sediment and lake water and less input of bacterial groups from groundwater into either the sediment or lake water.
Lake water eukaryotes consisted of, in order of decreasing abundance, Opisthokonta (primarily Fungi), Viridiplantae, Alveolata, and Euglenozoa as well as some more rare members (Fig. 4). A small percentage (0.51%) of the fungi were Basidiomycota, whereas ~ 98.5% fungi present were Ascomycota, the vast majority of which were Leotiomyceta. Two Aspergillaceae genera, namely Aspergillus and Penicillium, comprised the majority of lake water Leotiomyceta. These genera contain known halophiles and acidophiles (Leitão et al. 2012; Nazareth and Gonsalves 2014). For example, Aspergillus fungi can grow on organic carbon-rich sources, but can also grow in oligotrophic environments and at high salt concentrations (Nazareth and Gonsalves 2014).
In addition to fungal sequences, sequences assigned to Viridiplantae (7.5% total) were abundant in lake water. More specifically, 4.6% of LW reads were assigned to the green algae division Chlorophyta. These included members of the Chlorophyceae (Dunaliella, Volvox, Chlamydomonas, Oedogonium and Monoraphidium species, in decreasing order) as well as Trebouxiophyceae, Mamiellophyceae and Ulvophyceae families. Red algae, Rhodophyta, were rare in lake water (0.02% total reads) and were absent in GW and MFS metagenomes. The GW sample possessed far fewer eukaryotic sequences than LW (9% assigned reads), and these were primarily fungal (Fig. 4, Supplementary Fig. 2). The mudflat sediment sample contained only ~ 5.7% eukaryote-assigned reads, most of which were diatoms (3.7% total reads). Green algae were not detected in groundwater and comprised just 0.01% of MFS reads, with none assigned to Chlorophyceae. No diatom reads were detected in LW, and diatoms represented 0.05% of the groundwater metagenome.
Distribution of putative metabolic genes of Lake Magic microbial communities
Most of the genes that had good LAST hits were assigned to the “unclassified” category of the KEGG hierarchy and SEED subsystem (Fig. 5, Fig. S3). Many of these genes are involved in multiple functions, including transcription regulators, as well as regulatory peptidases and permeases, suggesting that organisms inhabit Lake Magic actively sense and respond to changes in environmental conditions. Along similar lines, the presence of chemotaxis, two-component signaling and regulatory pathways indicate that Lake Magic bacteria likely sense and respond (by moving away or toward) to environmental cues. Cellular motility in lake water was based on actin and related proteins, suggestive of eukaryotic cells, while groundwater contained genes for flagellar synthesis and bacterial chemotaxis. Most chemotaxis-associated reads were from taxa within five bacterial phyla—Verrucomicrobia, Proteobacteria, Firmicutes, Actinobacteria and Acidobacteria.
Distribution of functional assignments of Lake Magic metagenomic reads. a Bubble plot showing the number of reads mapping to each COG category. The COG categories, from top to bottom, are: [A] RNA processing and modification, [B] Chromatin structure and dynamics, [J] Translation, ribosomal structure and biogenesis, [K] Transcription, [L] Replication, recombination and repair, [D] Cell cycle control, cell division, chromosome partitioning, [M] Cell wall/membrane/envelope biogenesis, [N] Cell motility, [O] Posttranslational modification, protein turnover, chaperones, [T] Signal transduction mechanisms, [U] Intracellular trafficking, secretion and vesicular transport, [V] Defense mechanisms, [W] Extracellular structures, [Z] Cytoskeleton, [C] Energy production and conversion, [E] Amino acid transport and metabolism, [F] Nucleotide transport and metabolism, [G] Carbohydrate transport and metabolism, [H] Coenzyme transport and metabolism, [I] Lipid transport and metabolism, [P] Inorganic ion transport and metabolism, and [Q] Secondary metabolites biosynthesis, transport and catabolism. b Bubble plot showing the number of reads mapping to each KEGG category identified in Lake Magic metagenomes. Total number of reads assigned to the highest-level categories are indicated by bars. LW lake water (green), GW groundwater (blue), MFS mudflat sediment (purple). Bubble sizes represent the number of reads assigned to each category
The majority of predicted functional genes in Lake Magic are implicated in amino acid, carbohydrate, lipid as well as cofactor, pigment and vitamin metabolism (Fig. 5). Membrane transport and active uptake genes, including those encoding ABC transporters and components of bacterial secretion systems were detected in the pelagic, groundwater, and sediment compartments of Lake Magic. Putative genes with roles in the phosphotransferase system (PTS), a major carbohydrate uptake mechanism, were represented by fewer reads in LW than MFS and GW. MFS and GW metagenomes also contained genes associated with iron acquisition and transport, which were less prevalent in lake water (Fig. 5, Supplementary Fig. 3). Most of the iron acquisition reads were from Proteobacteria, with a third of all Fe acquisition reads assigned to Alphaproteobacteria. Other phyla with iron uptake reads include Firmicutes (Bacilli), Acidobacteria, and Actinobacteria. Different transport systems were prevalent in the lake water metagenome, which contained a higher proportion of genes involved in vesicular transport, secretion, and intracellular trafficking. These differences likely reflect the taxonomic composition of the communities. In this vein, differences in types of genes and pathways involved in cellular motility between lake water and the groundwater/sediment samples are also likely due to differences in community composition.
Photosynthesis in Lake Magic was associated with two bacterial groups—Cyanobacteria and Alphaproteobacteria—as well as Viridiplantae and diatoms. Pelagic photosynthesis-associated reads were from mostly green algae, including Dunaliella, although a number were from Synechococcus. Chemosynthetic pathways also appear to be present. For example, although sulfur oxidation reads were not abundant, they were detected in Lake Magic, and were primarily assigned to proteobacterial classes—Alphaproteobacteria, Gammaproteobacteria, and Acidithiobacillia (formerly classified within the Gammaproteobacteria).
Evidence of a variety of survival strategies can be found in the metagenomes, including sporulation, accumulation of non-toxic osmolytes, and protein adaptations to function under high solute ion concentrations. For example, sporulation, dormancy, and spore germination genes, all from Bacillus Firmicutes bacteria, were detected.
Reactive oxygen species (ROS) increase when algal cells are exposed to stresses, and are an important factor in the cellular response in algae. In the green alga, Dunaliella salina, increase in ROS appears to stimulate lipid overproduction and accumulation (Yilancioglu et al. 2014). Sequences associated with sterol and steroid biosynthesis in the lake water community were mostly assigned to fungi, including Aspergillus, although a smaller proportion was from Chlorophyta. In the sediment, in addition to fungal and green algae, sterol biosynthesis genes were also associated with diatoms (Bacillariophyta) and Proteobacteria.
Sequences associated with genes involved in biosynthesis of osmoprotectants were present in Lake Magic. Choline and betaine biosynthesis is carried out by bacterial taxa that were differentially distributed between the different lake compartments, and mainly consisted of Alphaproteobacteria, Gammaproteobacteria, Betaproteobacteria, and Firmicutes. In the pelagic zone, choline biosynthesis was mainly associated with Salinisphaera, as were sequences with possible roles in osmoregulation, stress response, ectoine and betaine biosynthesis. Fungi can accumulate glycerol in response to salt stress. Biosynthesis pathways for glycerol, an important osmolyte, were detected in Lake Magic, and were primarily assigned to Fungi including Aspergillus spp, and secondarily—with many fewer reads—to green algae and other Viridiplantae, diatoms, and Proteobacteria. The enzyme glycerol dehydrogenase is necessary for osmotolerance in Aspergillus fungi, and results in glycerol accumulation in salt-stressed A. nidulans (Redkar et al. 1995; De Vries et al. 2003). In addition to these, pigment biosynthesis pathways were also detected in the metagenomes. In Dunaliella, the chloroplast accumulates large quantities of beta-carotene, making cells look orange-red rather than green under sub-optimal conditions, including high salinity, high temperature, and high light (Heidelberg et al. 2013). Reads with possible roles in metabolism of phytoene, an intermediate in carotenoid biosynthesis, were assigned to green algae, Pleosporineae fungi, and to a lesser extent cyanobacteria and Alphaproteobacteria in Lake Magic. Specifically, carotenoid biosynthesis in lake water was identified by Chlorophyta reads and some Rhodobacteriaceae (Alphaproteobacteria) reads. In the mudflat sediment, diatoms and the aerobic heterotrophic Alphaproteobacteria Acidiphilium also had reads associated with carotenoid biosynthesis.
Discussion
Previous research on community composition of nearby ephemeral lakes in Western Australian, with a pH range of 2.7–8.2, indicated a high level of bacterial diversity, although with varying microbial communities (Hong et al. 2006; Mormile et al. 2009). We note that the microbial communities of lake water, sediment, and groundwater of Lake Magic were each dominated by a single OTU dominated the bacterial community (33–90%). This suggests niche partitioning and differences in the physicochemical parameters and redox gradients of this lake are reflected in the microbial composition.
The extreme pH and salinity of Lake Magic lake water seems to allow the expansion of a single bacterial Salinisphaera species. The genus, Salinisphaera, was first described by Antunes et al. (2003). These organisms have been isolated from environments that range from seawater, the slime of the surface of a marine fish, hydrothermal vents, solar salterns, and brine from a salt well (Crespo-Medina et al. 2009; Gi et al. 2010; Antunes et al. 2011; Park et al. 2012; Zhang et al. 2012; Shimane et al. 2013). For the most part, these are acidotolerant and halotolerant and can thrive under a range of moderately acid and saline conditions. In addition to the characterized Salinisphaera isolates, there have been other reports of this genus, including two from saline environments of Australia. Mormile et al. (2007) isolated a Salinisphaera from Lake Brown, a slightly acid saline lake in Western Australia. This genus of bacteria possesses the ability to metabolize autotrophically and heterotrophically (Antunes et al. 2003; Crespo-Medina et al. 2009). Furthermore, analysis of the genome sequence of S. shabanensis indicated that it possesses the genetic versatility to thrive under different environmental stresses (Antunes et al. 2011). Of these genetic capabilities, heavy metal resistance and detoxification are of interest due to the high concentration of metallic ions in Lake Magic.
The presence of facultative aerobes and anaerobes suggests tolerance to changing O2 conditions. Indeed, work in two other lakes, Lake Orr and Lake Whurr, in Western Australia indicate that lake sediment redox conditions, amount of halite crust formed, as well as potentially oxygen penetration are strongly affected by weather conditions (Ruecker et al. 2016). The dominant bacterial species detected in this study have been noted to have a number of survival strategies, such as the formation of spores and organization into desiccation-resistant biofilms. These organisms appear to be adapted to the multiple extremes that characterize their habitats, including osmotic tension and temperature gradients. Glycerol pathways associated with halotolerance were found in the Lake Magic metagenome dataset, as were genes associated with carotenoid biosynthesis, indicating that the eukaryotic and bacterial organisms in Lake Magic may use these strategies for stress tolerance.
Iron cycling and metal mobilization may be important functions of the dominant members in Lake Magic sediment. Organisms assigned to the most abundant bacterial genus in Lake Magic sediment, Alicyclobacillus, have also been reported in acidic, hypersaline river sediments form the same region of Western Australia, where they along with Aplasma Archaea potentially function as Fe(II)-oxidizers and may contribute to a drop in pH (Lu et al. 2016). In addition to oxidizing Fe(II), most isolated iron-oxidizing Alicyclobacillus are capable of reducing Fe(III) (Lu et al. 2010; Yahya et al. 2008). Therefore, it is possible that members of this genus are similarly involved in iron transformations in Lake Magic. Fe(III)-reducer genera Acidocella and Acidiphilium were dominant bacterial groups in these river sediments (Lu et al. 2010, 2016; Jones et al. 2013). In Lake Magic, Acidiphilium was an important community member in the sediment, with OTUs assigned to this genus representing 11% of the reads. Members of this heterotrophic genus have been documented in other extremely acidic environments, including acid mine drainage and Rio Tinto, where they may couple iron reduction to organic carbon oxidation (Johnson and Bridge 2002; Weber et al. 2006; Sánchez-Andrea et al. 2011). In Lake Magic groundwater, Thermoplasmatales archaea are likely involved in metal oxidation, and there appears to be active iron uptake in groundwater and sediment, whereas little appears to occur in lake water, indicating that different processes may be prevalent in different lake compartments. Curiously, Acidiphilium spp. grown under anaerobic, highly acidic and saline conditions form long filaments, possibly a coping mechanism, and are unable to reduce Fe(III) (Lu et al. 2016). This observation suggests that conditions present in the Western Australian sediments may prevent iron reducing activity by dominant community members, and invites further investigations into in situ gene expression. Lake Strawbridge sediments are covered by a salt crust of several cm thick, and host a fairly abundant 108 16S rRNA gene/g sediment (Weigold et al. 2016). Sequencing genomic and transcribed 16S rRNA genes revealed that some rare taxa were highly active at time of sampling (Weigold et al. 2016). Future temporal studies targeting genomes and transcriptomes during different stages of evapoconcentration would help delineate community responses and ecological impact of changing environmental conditions in extreme environments.
Intriguingly, eukaryotes, primarily fungi and green algae, were far more abundant than prokaryotes in the pelagic zone of Lake Magic. This finding mirrors the discovery of a eukaryote-dominated ecosystem in the highly acidic, iron-rich waters of the Rio Tinto, Spain’s “River of Fire” (Amaral Zettler et al. 2002; Amaral-Zettler 2012). The heterotrophic Lake Magic eukaryotes were extremotolerant fungi and were more abundant than the photoautotrophic algae. The fungi may have a role in decreasing the lake’s pH, as Aspergillus sp. fungi produce and secrete organic acids, rendering them industrially and biotechnologically important (Yang et al. 2017). The secretion of these acidic compounds can lower the pH of surrounding milieu. The ability of fungi to lower or elevate environmental (host) pH has been observed in a number of pathogenic fungal species (Prusky et al. 2001; Naglik et al. 2003; Prusky and Yakoby 2003; Bi et al. 2016). Viridiplantae, including the green algal genus Dunaliella, was the most abundant autotrophic eukaryotic group. Similar algae have been detected in Lake Tyrrell, yet no fungi have been reported there (Heidelberg et al. 2013). The presence of diatoms in the sediments suggests that different algae fix CO2 in lake water versus the sediment.
During evaporation, lake waters precipitate abundant halite and gypsum. Microbes, algae, and organic compounds have been documented within fluid inclusions in halite and gypsum from acid saline lakes (Benison 2013; Conner and Benison 2013; Benison and Karmanocky 2014). Petrography and laser Raman spectrometry found evidence for 5–7 μm dimpled, yellow spherules—interpreted to be Dunaliella—in Lake Magic halite samples (Conner and Benison 2013). Approximately 20% of primary fluid inclusions were suspected of containing at least one eukaryote (Conner and Benison 2013). Dunaliella represented ~ 35% of the lake water community and over 80% of the microbial community isolated from halite crusts in Lake Tyrrell (Heidelberg et al. 2013). We hypothesize that encasing cells in inclusion fluids within precipitate minerals may shield organisms from desiccation and enable survival. Coatings of iron oxides and clays on the halite and gypsum may further protect entrapped microorganisms from dilute water and UV-radiation (Farmer et al. 2009).
Long-term preservation and survival of archaea and bacteria in fluid inclusions in neutral-alkaline halite have been investigated for decades (Reiser and Tasch 1960; Dombrowski 1966; Norton and Grant 1988; Fredrickson et al. 1997; Grant et al. 1998; Stan-Lotter et al. 1999; McGenity et al. 2000; Vreeland et al. 2000; Mormile et al. 2003), yet, with a few notable exceptions, little work has been done on eukaryotic survival. In addition to Lake Magic halite fluid inclusions, Dunaliella has been reported in halite fluid inclusions up to 30 kyr in age from Death Valley and Saline Valley in California (Lowenstein and Brennan 2001; Schubert et al. 2009). Within these inclusions, Dunaliella cells co-occur with archaea, which may utilize glycerol derived from Dunaliella cells as a carbon source (Schubert et al. 2010a, b). A similar relationship may exist in acid saline environments, as Dunaliella, beta-carotene, and bacteria and archaea have been documented in fluid inclusions from halite in Lake Magic (Conner and Benison 2013) and necessary genes have been detected in our study.
As Lake Magic goes through repeated cycles of evapoconcentration and desiccation, the microbial eukaryotes present in the lake water may be entombed in halite crystals. As flooding occurs and dissolves halite, the eukaryotes may be released back into lake water (Conner and Benison 2013). Following complete desiccation, our data suggest that repopulation of green algae into the lake water is unlikely to proceed via input from the microbial communities in the surrounding sediment or groundwater. It is possible that algal cells could be delivered to the lake by aeolian processes, though what appears more likely is that these eukaryotes are preserved through time within fluid inclusions in Lake Magic’s precipitated halite deposits.
Our results call for additional studies into eukaryotic cell survival under these challenging conditions, with implications for our understanding of cell physiology and polyextremophile adaptation on Earth as well as microbial survival through changing conditions on Mars, where desiccating acid brine lakes are thought to have been once common (Squyres et al. 2004; Benison and Bowen 2006).
Conclusion
Lake Magic, Australia, hosts some of the most geochemically extreme waters on Earth, with extremely low pH (as low as 1.6), high salinity (reaching 32% total dissolved solids), many heavy metals, and periods of extreme evapoconcentration leading to the onset of complete desiccation. This work provides new insight into how microorganisms may cope with polyextremophilic conditions. In this study, we used high-throughput sequencing of bacterial 16S rRNA gene amplicons and metagenomics to investigate the diversity and putative function in lake water, groundwater, and sediment communities.
The bacterial diversity and genomic abundance in lake water was much lower than that in groundwater and subaqueous sediment. Furthermore, the distribution of bacterial OTUs, especially those affiliated with Salinisphaera spp., indicated possible exchanges between sediment and lake water bacteria, but little input to/from the groundwater. The lake water community was primarily composed of eukaryotes, predominantly fungi, and secondarily, green algae. Genes associated with halotolerance, osmoregulation metal transformations and thermal tolerance provide insights into survival mechanisms employed by polyextremophilic community members. Further, we surmise that entrapment in halite crystals may be a desiccation-survival strategy for lake water-dwelling microbes, first being entombed in salt structures during times of evapoconcentration, then released back into lake water upon flooding. Our findings call for additional studies into eukaryotic cell survival under these challenging conditions, with implications for our understanding of cell biology, extremophile physiology, and astrobiology.
References
Amaral Zettler LA, Gómez F, Zettler E et al (2002) Microbiology: eukaryotic diversity in Spain’s River of Fire. Nature 417:137. https://doi.org/10.1038/417137a
Amaral-Zettler LA (2012) Eukaryotic diversity at pH extremes. Front Microbiol 3:1–17. https://doi.org/10.3389/fmicb.2012.00441
Amoozegar MA, Bagheri M, Makhdoumi A et al (2016) Oceanobacillus longus sp. nov., a moderately halophilic bacterium isolated from a salt lake. Int J Syst Evol Microbiol. https://doi.org/10.1099/ijsem.0.001339
Antunes A, Eder W, Fareleira P, Santos H, Huber R (2003) Salinisphaera shabanensis gen. nov., sp. nov., a novel, moderately halophilic bacterium from the brine-seawater interface of the Shaban Deep, Red Sea. Extremophiles 7(1):29–34. https://doi.org/10.1007/s00792-002-0292-5
Antunes A, Rainey FA, Wanner G et al (2008) A new lineage of halophilic, wall-less, contractile bacteria from a brine-filled deep of the Red Sea. J Bacteriol 190:3580–3587. https://doi.org/10.1128/JB.01860-07
Antunes A, Alam I, Bajic VB, Stingl U (2011) Genome sequence of Salinisphaera shabanensis, a gammaproteobacterium from the harsh, variable environment of the brine-seawater interface of the Shaban Deep in the Red Sea. J Bacteriol 193:4555–4556. https://doi.org/10.1128/JB.05459-11
Baker BJ, Banfield JF (2003) Microbial communities in acid mine drainage. FEMS Microbiol Ecol 44:139–152. https://doi.org/10.1016/S0168-6496(03)00028-X
Benison KC (2013) Acid saline fluid inclusions: examples from modern and Permian extreme lake systems. Geofluids 13:579–593. https://doi.org/10.1111/gfl.12053
Benison KC, Bowen BB (2006) Acid saline lake systems give clues about past environments and the search for life on Mars. Icarus 183:225–229. https://doi.org/10.1016/j.icarus.2006.02.018
Benison KC, Bowen BB (2013) Extreme sulfur-cycling in acid brine lake environments of Western Australia. Chem Geol 351:154–167. https://doi.org/10.1016/j.chemgeo.2013.05.018
Benison KC, Bowen BB (2015) The evolution of end-member continental waters: the origin of acidity in southern Western Australia. GSA Today 25:4–10. https://doi.org/10.1130/GSATG231A.1
Benison KC, Karmanocky FJ (2014) Could microorganisms be preserved in Mars gypsum? Insights from terrestrial examples. Geology 42:615–617. https://doi.org/10.1130/G35542.1
Benison KC, Bowen BB, Oboh-Ikuenobe FE et al (2007) Sedimentology of acid saline lakes in southern Western Australia: newly described processes and products of an extreme environment. J Sedim Res 77:366–388. https://doi.org/10.2110/jsr.2007.038
Bi F, Barad S, Ment D et al (2016) Carbon regulation of environmental pH by secreted small molecules that modulate pathogenicity in phytopathogenic fungi. Mol Plant Pathol 17:1178–1195. https://doi.org/10.1111/mpp.12355
Bowen BB, Benison KC (2009) Geochemical characteristics of naturally acid and alkaline saline lakes in southern Western Australia. Appl Geochem 24:268–284. https://doi.org/10.1016/j.apgeochem.2008.11.013
Brake SS, Hasiotis ST (2010) Eukaryote-dominated biofilms and their significance in acidic environments. Geomicrobiol J 27:534–558. https://doi.org/10.1080/01490451003702966
Chang SS, Kang DH (2004) Alicyclobacillus spp. in the fruit juice industry: history, characteristics, and current isolation/detection procedures. Crit Rev Microbiol 30:55–74. https://doi.org/10.1080/10408410490435089
Chen L, Brügger K, Skovgaard M et al (2005) The genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota. J Bacteriol 187:4992–4999. https://doi.org/10.1128/JB.187.14.4992
Conner AJ, Benison KC (2013) Acidophilic halophilic microorganisms in fluid inclusions in halite from Lake Magic, Western Australia. Astrobiology 13:850–860. https://doi.org/10.1089/ast.2012.0956
Crespo-Medina M, Chatziefthimiou A, Cruz-Matos R et al (2009) Salinisphaera hydrothermalis sp. nov., a mesophilic, halotolerant, facultatively autotrophic, thiosulfate-oxidizing gammaproteobacterium from deep-sea hydrothermal vents, and emended description of the genus Salinisphaera. Int J Syst Evol Microbiol 59:1497–1503. https://doi.org/10.1099/ijs.0.005058-0
Dai X, Wang H, Zhang Z et al (2016) Genome sequencing of Sulfolobus sp. A20 from Costa Rica and comparative analyses of the putative pathways of carbon, nitrogen, and sulfur metabolism in various Sulfolobus strains. Front Microbiol. https://doi.org/10.3389/fmicb.2016.01902
De Vries RP, Flitter SJ, Van De Vondervoort PJI et al (2003) Glycerol dehydrogenase, encoded by gldB is essential for osmotolerance in Aspergillus nidulans. Mol Microbiol 49:131–141. https://doi.org/10.1046/j.1365-2958.2003.03554.x
Dombrowski H (1966) Geological problems in the question of living bacteria in Paleozoic salt deposits. Second Symp Salt 1:215–220
Edwards KJ, Bond PL, Gihring TM et al (2000) An archaeal iron-oxidizing extreme acidophile important in acid mine drainage. Science 287:1796–1799. https://doi.org/10.1126/science.287.5459.1796
Farmer J, Bell JFI, Benison KC et al (2009) Assessment of planetary protection requirements for Mars sample return missions. The National Academies Press, Washington, DC
Fredrickson JK, Chandler DP, Onstott TC (1997) Potential for preservation of halobacteria and their macromolecular constituents in brine inclusions from bedded salt deposits, pp 318–329
Fredrickson JK, Li SW, Gaidamakova EK et al (2008) Protein oxidation: key to bacterial desiccation resistance? ISME J 2:393–403. https://doi.org/10.1038/ismej.2007.116
Gi DB, Chung YH, Hye MK, Byung CC (2010) Salinisphaera dokdonensis sp. nov., isolated from surface seawater. Int J Syst Evol Microbiol 60:680–685. https://doi.org/10.1099/ijs.0.010058-0
Grant WD, Gemmell RT, McGenity TJ (1998) Halobacteria: the evidence for longevity. Extremophiles 2:279–287
Heidelberg KB, Nelson WC, Holm JB et al (2013) Characterization of eukaryotic microbial diversity in hypersaline Lake Tyrrell, Australia. Front Microbiol 4:1–14. https://doi.org/10.3389/fmicb.2013.00115
Hong B, Christiansen JM, Oboh-Ikuenobe FE, Bowen BB, Benison KC, Mormile MR (2006) Microbial diversity found in the acid saline lakes of Australia. In: American society for microbiology 106th general meeting, Orlando, FL, N-089, p 388
Jang SJ, Kim YJ, Lee SH et al (2014) Oceanobacillus gochujangensis sp. nov., isolated from gochujang a traditional Korean fermented food. J Microbiol 52:1050–1055. https://doi.org/10.1007/s12275-014-4220-z
Jiang CY, Liu Y, Liu YY, You XY, Guo X, Liu SJ (2008) Alicyclobacillus ferrooxydans sp. nov., a ferrous-oxidizing bacterium from solfataric soil. Int J Syst Evol Microbiol 58:2898–2903
Johnson DB, Bridge TAM (2002) Reduction of ferric iron by acidophilic heterotrophic bacteria: evidence for constitutive and inducible enzyme systems in Acidiphilium spp. J Appl Microbiol 92:315–321. https://doi.org/10.1046/j.1365-2672.2002.01535.x
Johnson SS, Chevrette MG, Ehlmann BL, Benison KC (2015) Insights from the metagenome of an acid salt lake: the role of biology in an extreme depositional environment. PLoS One 10:1–19. https://doi.org/10.1371/journal.pone.0122869
Jones RM, Hedrich S, Johnson DB (2013) Acidocella aromatica sp. nov.: an acidophilic heterotrophic alphaproteobacterium with unusual phenotypic traits. Extremophiles 17:841. https://doi.org/10.1007/s00792-013-0566-0
Kim MG, Lee JC, Park DJ, Li WJ, Kim CJ (2014) Alicyclobacillus tengchongensis sp. nov., a thermo-acidophilic bacterium isolated from hot spring soil. J Microbiol 52:884–889
Kovaleva J, Degener JE, Van Der Mei HC (2014) Methylobacterium and its role in health care-associated infection. J Clin Microbiol 52:1317–1321. https://doi.org/10.1128/JCM.03561-13
La Duc MT, Dekas A, Osman S et al (2007) Isolation and characterization of bacteria capable of tolerating the extreme conditions of clean room environments. Appl Environ Microbiol 73:2600–2611. https://doi.org/10.1128/AEM.03007-06
Lagier J-C, Khelaifia S, Azhar EI et al (2015) Genome sequence of Oceanobacillus picturae strain S1, an halophilic bacterium first isolated in human gut. Stand Genom Sci 10:91. https://doi.org/10.1186/s40793-015-0081-2
Leitão AL, García-Estrada C, Ullán RV et al (2012) Penicillium chrysogenum var. halophenolicum, a new halotolerant strain with potential in the remediation of aromatic compounds in high salt environments. Microbiol Res 167:79–89. https://doi.org/10.1016/j.micres.2011.03.004
Lowenstein T, Brennan ST (2001) Fluid inclusions in paleolimnological studies of chemical sediments. In: Last WM, Smol JP (eds) Tracking environmental change using lake sediments: physical and geochemical methods. Springer, Dordrecht, pp 189–216
Lu J, Nogi Y, Takami H (2001) Oceanobacillus iheyensis gen. nov., sp. nov., a deep-sea extremely halotolerant and alkaliphilic species isolated from a depth of 1050 m on the Iheya Ridge. FEMS Microbiol Lett 205:291–297. https://doi.org/10.1016/S0378-1097(01)00493-1
Lu S, Gischkat S, Reiche M, Akob DM, Hallberg KB, Küsel K (2010) Ecophysiology of Fe-cycling bacteria in acidic sediments. Appl Environ Microbiol 76:8174–8183. https://doi.org/10.1128/AEM.01931-10
Lu S, Peiffer S, Lazar CS et al (2016) Extremophile microbiomes in acidic and hypersaline river sediments of Western Australia. Environ Microbiol Rep 8:58–67. https://doi.org/10.1111/1758-2229.12351
Mao D, Grogan D (2012) Genomic evidence of rapid, global-scale gene flow in a Sulfolobus species. ISME J 6:1613–1616. https://doi.org/10.1038/ismej.2012.20
McGenity TJ, Gemmell RT, Grant WD, Stan-Lotter H (2000) Origins of halophilic microorganisms in ancient salt deposits. Environ Microbiol 2:243–250
Méndez-García C, Peláez AI, Mesa V, Sánchez J, Golyshina OV, Ferrer M (2015) Microbial diversity and metabolic networks in acid mine drainage habitats. Front Microbiol 6:475. https://doi.org/10.3389/fmicb.2015.00475
Mormile MR, Biesen MA, Gutierrez MC et al (2003) Isolation of Halobacterium salinarum retrieved directly from halite brine inclusions. Environ Microbiol 5:1094–1102. https://doi.org/10.1046/j.1462-2920.2003.00509.x
Mormile MR, Hong B, Adams NT, et al (2007) Characterization of a moderately halo-acidophilic bacterium isolated from Lake Brown, Western Australia. In: Instruments, methods, and missions for astrobiology X. https://doi.org/10.1117/12.734741
Mormile MR, Hong B-Y, Benison KC (2009) Molecular analysis of the microbial communities of Mars analog lakes in Western Australia. Astrobiology 9:919–930. https://doi.org/10.1089/ast.2008.0293
Naglik JR, Challacombe SJ, Hube B (2003) Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev 67:400–428. https://doi.org/10.1128/mmbr.67.3.400
Nazareth SW, Gonsalves V (2014) Halophilic Aspergillus penicillioides from athalassohaline, thalassohaline, and polyhaline environments. Front Microbiol 5:1–5. https://doi.org/10.3389/fmicb.2014.00412
Norton C, Grant W (1988) Survival of halobacteria within fluid inclusions in salt crystals. J Gen Microbiol 134:1365–1371. https://doi.org/10.1099/00221287-134-5-1365
Park SJ, Cha IT, Kim SJ et al (2012) Salinisphaera orenii sp. nov., isolated from a solar saltern. Int J Syst Evol Microbiol 62:1877–1883. https://doi.org/10.1099/ijs.0.028647-0
Prusky D, Yakoby N (2003) Pathogenic fungi: leading or led by ambient pH? Mol. Plant Pathol. 4:509–516
Prusky D, McEvoy JL, Leverentz B, Conway WS (2001) Local modulation of host pH by Colletotrichum species as a mechanism to increase virulence. Mol Plant Microb Interact 14:1105–1113. https://doi.org/10.1094/MPMI.2001.14.9.1105
Redkar RJ, Locy RD, Singh NK (1995) Biosynthetic pathways of glycerol accumulation under salt stress in Aspergillus nidulans. Exp Mycol 19:241–246
Reiser R, Tasch P (1960) Investigation of the viability of osmophile bacteria of great geological age. Trans Kans Acad Sci 63:31–34. https://doi.org/10.2307/3626919
Ruecker A, Schröder C, Byrne J et al (2016) Geochemistry and mineralogy of Western Australian salt lake sediments: implications for Meridiani Planum on Mars. Astrobiology 16:525–538. https://doi.org/10.1089/ast.2015.1429
Ruepp A, Graml W, Santos-Martinez ML et al (2000) The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature 407:508–513. https://doi.org/10.1038/35035069
Sánchez-Andrea I, Rodríguez N, Amils R, Sanz JL (2011) Microbial diversity in anaerobic sediments at Rio Tinto, a naturally acidic environment with a high heavy metal content. Appl Environ Microbiol 77:6085–6093. https://doi.org/10.1128/AEM.00654-11
Schleper C, Piihler G, Kuhlmorgen B, Zillig W (1995) Life at extremely low pH. Nature 375:741–742. https://doi.org/10.1038/375741b0
Schubert BA, Lowenstein TK, Timofeeff MN, Parker MA (2009) How do prokaryotes survive in fluid inclusions in halite for 30 k.y.? Geology 37:1059–1062. https://doi.org/10.1130/G30448A.1
Schubert BA, Lowenstein TK, Timofeeff MN, Parker MA (2010a) Halophilic archaea cultured from ancient halite, Death Valley, California. Environ Microbiol 12:440–454. https://doi.org/10.1111/j.1462-2920.2009.02086.x
Schubert BA, Timofeeff MN, Lowenstein TK, Polle JEW (2010b) Dunaliella cells in fluid inclusions in halite: significance for long-term survival of prokaryotes. Geomicrobiol J 27:61–75. https://doi.org/10.1080/01490450903232207
Shimane Y, Tsuruwaka Y, Miyazaki M et al (2013) Salinisphaera japonica sp. nov., a moderately halophilic bacterium isolated from the surface of a deep-sea fish, malacocottus gibber, and emended description of the genus Salinisphaera. Int J Syst Evol Microbiol 63:2180–2185. https://doi.org/10.1099/ijs.0.047845-0
Simbahan J, Drijber R, Blum P (2004) Alicyclobacillus vulcanalis sp. nov., a thermophilic, acidophilic bacterium isolated from Coso Hot Springs, California, USA. Int J Syst Evol Microbiol 54:1703–1707
Squyres SW, Grotzinger JP, Arvidson RE et al (2004) In situ evidence for an ancient aqueous environment at Meridian Planum. Science 306(80):1709–1714. https://doi.org/10.1126/science.1104559
Stan-Lotter H, McGenity TJ, Legat A et al (1999) Very similar strains of Halococcus salifodinae are found in geographically separated Permo-Triassic salt deposits. Microbiology 145:3565–3574. https://doi.org/10.1099/00221287-145-12-3565
Stieglmeier M, Wirth R, Kminek G, Moissl-Eichinger C (2009) Cultivation of anaerobic and facultatively anaerobic bacteria from spacecraft-associated clean rooms. Appl Environ Microbiol 75:3484–3491. https://doi.org/10.1128/AEM.02565-08
Vreeland RH, Rosenzweig WD, Powers DW (2000) Isolation of a 250 million-year-old halotolerant bacterium from a primary salt crystal. Nature 407:897–900. https://doi.org/10.1038/35038060
Walls I, Chuyate R (2000) Spoilage of fruit juices by Alicyclobacillus acidoterrestris: Alicyclobacillus in the food industry. Food Aust 52:286–288
Weber KA, Achenbach LA, Coates JD (2006) Microorganisms pumping iron: anaerobic microbial iron oxidation and reduction. Nat Rev Microbiol 4:752–764. https://doi.org/10.1038/nrmicro1490
Weigold P, Ruecker A, Loesekann-Behrens T et al (2016) Ribosomal tag pyrosequencing of DNA and RNA reveals “rare” taxa with high protein synthesis potential in the sediment of a hypersaline lake in Western Australia. Geomicrobiol J 33:426–440. https://doi.org/10.1080/01490451.2015.1049304
Whon TW, Jung M-J, Roh SW et al (2010) Oceanobacillus kimchii sp. nov. isolated from a traditional Korean fermented food. J Microbiol 48:862–866. https://doi.org/10.1007/s12275-010-0214-7
Yahya A, Hallberg KB, Johnson DB (2008) Iron and carbon metabolism by a mineral-oxidizing Alicyclobacillus-like bacterium. Arch Microbiol 189:305–312
Yang L, Lübeck M, Lübeck PS (2017) Aspergillus as a versatile cell factory for organic acid production. Fungal Biol Rev 31:33–49
Yilancioglu K, Cokol M, Pastirmaci I et al (2014) Oxidative stress is a mediator for increased lipid accumulation in a newly isolated Dunaliella salina strain. PLoS One 9:e91957. https://doi.org/10.1371/journal.pone.0091957
Zhang YJ, Tang SK, Shi R et al (2012) Salinisphaera halophila sp. nov., a moderately halophilic bacterium isolated from brine of a salt well. Int J Syst Evol Microbiol 62:2174–2179. https://doi.org/10.1099/ijs.0.035584-0
Acknowledgements
We thank Yi Cui (Missouri S&T) for assistance with DNA extraction. This work was supported by an Experimental Program to Stimulate Competitive Research at the National Aeronautics and Space Administration, Missouri Research Infrastructure Development award (M.M.), a Georgetown University Main Campus Research Fellowship (S.S.J. and E.Z.), and West Virginia University (K.B.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by A. Oren.
Electronic supplementary material
Below is the link to the electronic supplementary material.
792_2018_1000_MOESM1_ESM.pdf
Supplementary Fig. 1. Taxonomic composition and diversity of bacterial SSU rRNA gene OTUs (97% ID). Krona charts showing the relative abundance of each bacterial taxon identified in the Lake Magic samples. Within each chart, circles are arranged hierarchically, with the innermost circle corresponding to phylum and outermost circle representing genus. LW = lake water, GW = groundwater, and SS = subaqueous sediment (PDF 923 kb)
792_2018_1000_MOESM2_ESM.pdf
Supplementary Fig. 2. Taxonomic composition and diversity of Lake Magic metagenomic reads. (a) Krona charts showing the relative abundance of bacterial phyla identified in the Lake Magic samples. The PVC clade includes the phyla Chlamydiaea, Lentisphaerae, Planctomycetes, and Verrucomicrobia, the FCB group encompasses the Chlorobi, Bacteroidetes and Fibrobacteres phyla, and the Terrabacteria group includes the Actinobacteria, Deinococcus–Thermus, Cyanobacteria, Chloroflexi, and Firmicutes. (b) Krona charts showing the relative abundance of eukaryotic phyla identified in the Lake Magic samples. LW = lake water, GW = groundwater, and MFS = mudflat sediment (PDF 866 kb)
792_2018_1000_MOESM3_ESM.pdf
Supplementary Fig. 3. Distribution of Lake Magic metagenomic read assignments to SEED subsystems. Bars represent the total number of reads assigned to each subsystem. LW = lake water (green), GW = groundwater (blue), and MFS = mudflat sediment (purple) (PDF 872 kb)
Rights and permissions
About this article
Cite this article
Zaikova, E., Benison, K.C., Mormile, M.R. et al. Microbial communities and their predicted metabolic functions in a desiccating acid salt lake. Extremophiles 22, 367–379 (2018). https://doi.org/10.1007/s00792-018-1000-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-018-1000-4