Abstract
Objective
Evaluate if automatic segmentation of mandibular three-dimensional (3D) models is reliable and accurate.
Materials and methods
Eight dry mandibles with eight silica markers were scanned in the i-CAT Classic device (Imaging Sciences International). Automatic segmentation was performed using nine standard preset thresholds in the Dolphin software (Dolphin Imaging & Management Solutions). Three observers individually made twice eight linear measurements on the mandibular 3D models. Another observer made physical measurements, twice as well, on the dry mandibles. Reliability and accuracy were evaluated with intraclass correlation coefficients (ICCs), Dahlberg’s formula, Bland-Altman analyses, and changing bias with regression analyses.
Results
Inter-observer and intra-observer ICCs and Dahlberg’s error were ≥ 0.75 and ≤ 1.0 mm, respectively, for all measurements. Inter-observer agreement between mandibular 3D models and physical measurements ranged from −0.37 to 0.91 mm.
Conclusions
Linear measurements made on mandibular 3D models obtained using standard preset thresholds are reliable and accurate. However, additional studies are necessary to confirm this hypothesis for clinical applications.
Clinical relevance
Since the 3D models are useful for diagnostics and surgical planning, it is necessary to determinate whether the linear measurements made on 3D models obtained by automatic segmentation are sufficiently reliable and accurate.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Three-dimensional (3D) modeling from cone-beam computed tomography (CBCT) has been the imaging modality of choice in oral and maxillofacial radiology when the conventional radiological techniques are insufficient to reach the diagnostic and treatment planning needs in dentistry [1, 2]. For this, 3D model segmentation, e.g., to distinguish bone and soft tissue of the mandible and maxilla, is an important step in software programs [3,4,5].
From digital imaging and communications in medicine (DICOM) CBCT data sets, 3D models can be rendered using various software programs. Rendering is the process by which an object is made to appear as it does in the real world. Each rendering program has a unique algorithm for the transformation of CBCT data to vector data, i.e., to construct the surface of a triangulated mesh covering the selected surface of interest [2].
On the other hand, segmentation of a 3D model requires the operator to input a threshold value specifying the structure of interest [2]. Segmentation can be performed completely manually (with prior knowledge of the grayscale range of interest) [3,4,5], automatically (using a standard preset threshold defined by the software) [2,3,4,5,6], or semi-automatically (with manual checking and refinement of the threshold before segmentation) [7]. Manual and semi-automatic segmentation are operator-dependent and time-consuming tasks [5, 7].
Due to the growing clinical acceptance of the digital workflow in dentistry, much software with automatic segmentation tools for 3D models is commercially available. Dolphin software (Dolphin Imaging & Management Solutions, Patterson Technology, Chatsworth, USA) is a software widely used to perform diagnostics and planning in orthodontics and surgery that allows the professional to choose which parameter preset by the software will be used to segment the 3D model. This software has already been used in other studies to evaluate craniometric measurements using 3D models. However, no study has been conducted to evaluate the reliability and accuracy of the different options of 3D models automatically segmented by the Dolphin software.
Since 3D models are useful for diagnostics and surgical planning, it is necessary to determinate whether the linear measurements made on 3D models obtained by automatic segmentation are sufficiently reliable and accurate. Therefore, the scope of this reliability and agreement study was to evaluate if automatic segmentation of 3D models of dry adult human mandibles with software’s standard preset thresholds in the Dolphin software is reliable and accurate.
Methodology
This study was reported according to Guidelines for Reporting Reliability and Agreement Studies [8, 9]. Eight dry adult human mandibles with preserved bone cortices were selected from the Department of Anatomy of the University of Sao Paulo with the approval of the Research Ethics Committee of the institution (process number: 006). Sample size calculation had been performed based on the Co-Co measurements (Dolphin Solid-1) of a previous study of our research group [6] with an alpha level of 0.5 and beta value of 0.2% to attain 80% test power. Furthermore, 1 mm was determined as being the clinically acceptable error for linear measurements, and eight mandibles were necessary.
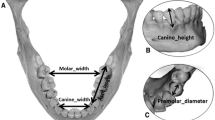
Eight mandibular points, which were used in a previous study [6], were selected (Table 1). Silica-based hyperdense cylindrical markers (2 × 2 mm each) with a density similar to that of bone cortex and central orifices (0.5 mm diameter) were glued with ethylene-vinyl acetate polymer [6].
Eight linear measurements (in millimeters) were made on the mandibles directly and on mandibular 3D models (Table 1 and Fig. 1). The measurements were selected based on a previous study [6] and represent linear measurements in the three dimensions (height, width, and depth). A single observer performed the physical measurements (PMs) twice at an interval of 30 days using a digital caliper (Mitutoyo Sul Americana Ltda., Suzano, Brazil) with a 0.1-mm-thick edge [6, 10, 11].
After PM, each mandible was positioned with wax on the acrylic table of an i-CAT Classic instrument (Imaging Sciences International, Hatfield, USA) [6, 11]. The median sagittal plane was positioned perpendicular to the ground, with the occlusal plane parallel to the ground, to reproduce the position used for patient examination [6, 10, 11]. The mandibles were scanned using the following parameters: 0.3-mm voxels, 8-cm field of view, 20 s, 18.45 mAs, and 120 kv [6]. The original CBCT data were stored in DICOM format and transferred to an independent workstation with a 21.5″ screen and a resolution of 1920 × 1080 pixels (Samsung, São Paulo, Brazil). Mandibular 3D models were rendered at the workstation using the Dolphin 11.5 software. Automatic segmentation was performed using nine of the software’s standard preset thresholds (translucent default (TD), translucent grayscale (TG), solid default (SD), solid smooth (SS), and solid-1–5 (S1–S5) (Fig. 2)). Due to the fact that measurements on mandibular 3D models are distinct sources of error, three other observers familiarized with the software who did not know the physical measures performed measurements on all mandibular 3D models twice at an interval of 30 days [6].
The measurement conditions were close to the clinical routine of the oral radiologist, as the observer randomly selected a mandible and predefined standard thresholds and followed an analysis sequence (Co-Co, Co-Me, Co-Go, Go-Go, Go-Me, MF-AC, MF-MF, and MF-Go). All the measurements were directly typed into a pre-validated Excel spreadsheet, used to perform the standardization of the data for all the outcomes [12, 13].
The medians of the data were calculated to avoid the influence by the extremes of the dataset and were used for statistical analyses [12, 13]. To measure intra-observer and inter-observer reliability, the intraclass correlation coefficient (ICC) and Dahlberg’s formula were used [12, 13].
To determine the accuracy, Bland-Altman analyses were used with limits of agreement (LOA) and 95% confidence intervals and changing bias with regression analyses [12, 13]. Normality test was performed with Shapiro-Wilk and histograms. For data interpretation, an error up to 1 mm [14] was considered to be clinically acceptable for Dahlberg’s error and systematic bias, and 0.75 for ICCs [12]. All statistical analyses were performed with Microsoft Excel™ 2010 (Microsoft Corp., Redmond, USA) and IBM SPSS™ Statistics software version 23.0 (IBM Corp., Armonk, USA).
Results
Intra-observer reliability (Table 2) in mandibular 3D models was greater than 0.75 for 100% of the variables and standard preset thresholds (0.96 ≤ ICC ≤ 1.0). Dahlberg’s error was less than 1 mm for 100% of the variables and standard preset thresholds and ranged from 0.02 to 0.78 mm. The outcomes measured on mandibular 3D models with the largest errors were observed for Go-Me measured in the SS model (observer 2).
Inter-observer reliability (Table 3) in mandibular 3D models was greater than 0.75 for 100% of the variables and standard preset thresholds (0.97 ≤ ICC ≤ 1.0). Dahlberg’s error was less than 1 mm for 100% of the variables and standard preset thresholds and ranged from 0.02 to 0.78 mm. The outcomes measured on mandibular 3D models with the largest errors were observed for M-F-MF measured in the TD model (observers 1 × 3).
The PMs and measurements performed on the mandibular 3D models are compared in Table 4. Systematic bias ranged from −0.37 mm ≤ Δ ≤ 0.91 mm. Although all measurements are below 1 mm, there was a tendency to underestimate the measurement in mandibular 3D models in 70.83% of the variables. Finally, no signs of changing bias were detected.
Discussion
As mentioned before, manual and semi-automatic segmentation are operator-dependent and time-consuming tasks [5, 7]. Thus, knowledge of the reliability and accuracy of linear measurements made on 3D models obtained by automatic segmentation is very important to determine whether the professional time saved by using this method is justifiable. The results of this study are clinically important because the predictability of errors and the diagnosis, treatment plan, and realistic prognosis must be established [15]. The current study revealed excellent reliability and accuracy of linear measurements made after automatic segmentation of mandibular 3D models with the Dolphin software, comparable to those obtained with manual and semi-automatic segmentation in previous studies [2, 7, 16, 17].
Our findings of high intra-observer and inter-observer reliability of the mandibular 3D model measurements are similar to results obtained with manual segmentation using the Dolphin software (ICC > 0.96) [16], CB Works software (CyberMed, Seoul, Korea; ICC > 0.95) [17], and SimPlant Ortho Pro 2.1 software (Materialize Dental, Leven, Belgium; ICC > 0.97) [2]. For semi-automatic segmentation with the Matlab software (Mathworks Inc., Natick, Massachusetts USA), ICCs >0.97 have been reported [7]. Despite the high accuracy observed, we found significant differences between PMs and mandibular 3D models measures of Co-Me and Go-Me, independent of the standard preset threshold used. We believe these differences were due to the difficulty of marking the Me point on the mandibular 3D models, as reported by the three examiners. All these measurements were underestimated compared with PMs, as also found in a previous study [17].
Systematic bias (absolute error) is one of the most important clinical parameters in studies of 3D models. In this study, the largest absolute error was observed for Co-Me measured in the TG model (0.91 mm). This error is smaller than that reported by Periago et al. [16] (−3.32 mm) for manual segmentation using the Dolphin software. The difference may be attributable to the use of hyperdense markers in our study. Hassan et al. [18] reported that more time and care are needed to perform measurements when hyperdense markers are not used. All mandibular 3D models obtained by automatic segmentation with the Dolphin software showed errors below 0.91 mm. This result is very important because as proposed by Damstra et al. [14], ≤ 1 mm is a clinically acceptable error for linear measurements.
The reliability and accuracy of linear measurements are influenced by factors such as image quality, accuracy of measuring instruments (caliper and software), and anatomical landmarks, when markers are not used, and even the size and material from landmarks [19,20,21,22]. The scanner used in this study was the i-CAT CBCT device, which produces images with good resolution and has been used in other craniometric studies [6, 16, 23]. Our protocol generated intermediate spatial resolution [24]. This device is considered accurate and presents images with good resolution. The 0.3-mm voxel protocol chosen was used with an intermediate spatial resolution size in the i-CAT instrument [14, 21]. Most recent studies of this nature have involved the use of hyperdense markers made of stainless steel [19], gutta-percha [25], titanium [26] and glass [22], or bone references. In this study, as in two previous studies [6, 21], we used beads with a density similar to that of bone cortex. PMs also showed excellent reliability in this study, as in other studies [6, 16, 22]. Finally, the Dolphin software was chosen because it was designed to assist with cephalometric tasks and is commonly used by dentists worldwide [27].
The major advantage of the automatic segmentation process used in this study relative to manual or semi-automatic segmentation is a significant reduction of operator time; manual segmentation requires about 1 h and semi-automatic segmentation requires about 10–15 min [7].
Limitations of this study include the experimental condition, as accuracy may vary, e.g., in the presence of soft tissue [2]. The thresholding is easier without soft tissue; dry mandibles do not move during imaging, which makes the image sharper and easier to segment; and anatomical structures with cortical bone influence imaging. In addition, this study assessed only measurements made on the mandible; the threshold values used for segmentation of the maxilla and mandible are different [18]. Therefore, additional studies including measurement of the maxilla are needed.
Conclusion
Linear measurements made on mandibular 3D models obtained using standard preset thresholds of the Dolphin software are reliable and accurate compared with PMs. However, additional studies are necessary to confirm this hypothesis for clinical applications.
References
Scarfe WC, Farman AG, Sukovic P (2006) Clinical applications of cone-beam computed tomography in dental practice. J Can Dent Assoc 72:75–80
Engelbrecht WP, Fourie Z, Damstra J, Gerrits PO, Ren Y (2013) The influence of the segmentation process on 3D measurements from cone beam computed tomography-derived surface models. Clin Oral Investig 17:1919–1927
Wang L, Chen KC, Gao Y, Shi F, Liao S, Li G, Shen SG, Yan J, Lee PK, Chow B, Liu NX, Xia JJ, Shen D (2014) Automated bone segmentation from dental CBCT images using patch-based sparse representation and convex optimization. Med Phys 41:043503
Abdolali F, Zoroofi RA, Otake Y, Sato Y (2016) Automatic segmentation of maxillofacial cysts in cone beam CT images. Comput Biol Med 72:108–119
Abdolali F, Zoroofi RA, Abdolali M, Yokota F, Otake Y, Sato Y (2017) Automatic segmentation of mandibular canal in cone beam CT images using conditional statistical shape model and fast marching. Int J Comput Assist Radiol Surg 12:581–593
Poleti ML, Fernandes TMF, Pagin O, Moretti MR, Rubira-Bullen IRF (2016) Analysis of linear measurements on 3D surface models using CBCT data segmentation obtained by automatic standard pre-set thresholds in two segmentation software programs: an in vitro study. Clin Oral Investig 20:179–185
Xi T, Schreurs R, Heerink WJ, Bergé SJ, Maal TJ (2014) A novel region-growing based semi-automatic segmentation protocol for three-dimensional condylar reconstruction using cone beam computed tomography (CBCT). PLoS One 9:e111126
Cronin P, Rawson JV (2016) Review of research reporting guidelines for radiology researchers. Acad Radiol 23:537–558
Kottner J, Audigé L, Brorson S, Donner A, Gajewski BJ, Hróbjartsson A, Roberts C, Shoukri M, Streiner DL (2011) Guidelines for reporting reliability and agreement studies (GRRAS) were proposed. J Clin Epidemiol 64:96–106
Lopes PM, Moreira CR, Perrella A, Antunes JL, Cavalcanti MG (2008) 3-D volume rendering maxillofacial analysis of angular measurements by multislice CT. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 105:224–230
Fernandes TMF, Julie Adamczyk J, Poleti ML et al (2015) Comparison between 3D volumetric rendering and multiplanar slices on the reliability of linear measurements on CBCT images: an in vitro study. J Appl Oral Sci 23:56–63
Koretsi V, Kirschbauer C, Proff P, Kirschneck C (2019) Reliability and intra-examiner agreement of orthodontic model analysis with a digital caliper on plaster and printed dental models. Clin Oral Investig 23:3387–3396
Koretsi V, Tingelhoff L, Proff P, Kirschneck C (2018) Intra-observer reliability and agreement of manual and digital orthodontic model analysis. Eur J Orthod 40:52–57
Damstra J, Fourie Z, Huddleston Slater JJ, Ren Y (2011) Reliability and the smallest detectable difference of measurements on 3-dimensional cone-beam computed tomography images. Am J Orthod Dentofac Orthop 140:e107–e114
Kapila S, Conley RS, Harrell WE Jr (2011) The current status of cone beam computed tomography imaging in orthodontics. Dentomaxillofac Radiol 40:24–34
Periago DR, Scarfe WC, Moshiri M, Scheetz JP, Silveira AM, Farman AG (2008) Linear accuracy and reliability of cone beam CT derived 3-dimensional images constructed using an orthodontic volumetric rendering program. Angle Orthod 78:387–395
Baumgaertel S, Palomo JM, Palomo L, Hans MG (2009) Reliability and accuracy of cone-beam computed tomography dental measurements. Am J Orthod Dentofac Orthop 136:19–25
Hassan B, van der Stelt P, Sanderink G (2009) Accuracy of three-dimensional measurements obtained from cone beam computed tomography surface-rendered images for cephalometric analysis: influence of patient scanning position. Eur J Orthod 31:129–134
Berco M, Rigali PH Jr, Miner RM, DeLuca S, Anderson NK, Will LA (2009) Accuracy and reliability of linear cephalometric measurements from cone-beam computed tomography scans of a dry human skull. Am J Orthod Dentofac Orthop 136:17e1–17e9
Choi JY, Choi JH, Kim NK, Kim Y, Lee JK, Kim MK, Leea JH, Kima MJ (2002) Analysis of errors in medical rapid prototyping models. Int J Oral Maxillofac Surg 31:23–32
Gribel BF, Gribel MN, Frazao DC, McNamara JA Jr, Manzi FR (2011) Accuracy and reliability of craniometric measurements on lateral cephalometry and 3D measurements on CBCT scans. Angle Orthod 81:26–35
Damstra J, Fourie Z, Huddleston Slater JJ, Ren Y (2010) Accuracy of linear measurements from cone-beam computed tomography-derived surface models of different voxel sizes. Am J Orthod Dentofac Orthop 137:16 e1–16 e6
Brown AA, Scarfe WC, Scheetz JP, Silveira AM, Farman AG (2009) Linear accuracy of cone beam CT derived 3D images. Angle Orthod 79:150–157
Ballrick JW, Palomo JM, Ruch E, Amberman BD, Hans MG (2008) Image distortion and spatial resolution of a commercially available cone-beam computed tomography machine. Am J Orthod Dentofac Orthop 134:573–582
Mischkowski RA, Pulsfort R, Ritter L, Neugebauer J, Brochhagen HG, Keeve E, Zölle JE (2007) Geometric accuracy of a newly developed cone-beam device for maxillofacial imaging. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 104:551–559
Lagravere MO, Carey J, Toogood RW, Major PW (2008) Three-dimensional accuracy of measurements made with software on cone-beam computed tomography images. Am J Orthod Dentofac Orthop 134:112–116
Nouri M, Hamidiaval S, Akbarzadeh Baghban A, Basafa M, Fahim M (2015) Efficacy of a newly designed cephalometric analysis software for McNamara analysis in comparison with Dolphin software. J Dent 12:60–69
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
For this type of study, formal consent is not required.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 1118 kb)
Rights and permissions
About this article
Cite this article
Poleti, M.L., Fernandes, T.M.F., Moretti, M.R. et al. Reliability and accuracy of automatic segmentation of mandibular 3D models on linear measurements. Clin Oral Invest 25, 6335–6346 (2021). https://doi.org/10.1007/s00784-021-03934-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00784-021-03934-4