Abstract
Desulfovibrio gigas aldehyde oxidoreductase (DgAOR) is a mononuclear molybdenum-containing enzyme from the xanthine oxidase (XO) family, a group of enzymes capable of catalyzing the oxidative hydroxylation of aldehydes and heterocyclic compounds. The kinetic studies reported in this work showed that DgAOR catalyzes the oxidative hydroxylation of aromatic aldehydes, but not heterocyclic compounds. NMR spectroscopy studies using 13C-labeled benzaldehyde confirmed that DgAOR catalyzes the conversion of aldehydes to the respective carboxylic acids. Steady-state kinetics in solution showed that high concentrations of the aromatic aldehydes produce substrate inhibition and in the case of 3-phenyl propionaldehyde a suicide substrate behavior. Hydroxyl-substituted aromatic aldehydes present none of these behaviors but the kinetic parameters are largely affected by the position of the OH group. High-resolution crystallographic structures obtained from single crystals of active-DgAOR soaked with benzaldehyde showed that the side chains of Phe425 and Tyr535 are important for the stabilization of the substrate in the active site. On the other hand, the X-ray data of DgAOR soaked with trans-cinnamaldehyde showed a cinnamic acid molecule in the substrate channel. The X-ray data of DgAOR soaked with 3-phenyl propionaldehyde showed clearly how high substrate concentrations inactivate the enzyme by binding covalently at the surface of the enzyme and blocking the substrate channel. The different reactivity of DgAOR versus aldehyde oxidase and XO towards aromatic aldehydes and N-heterocyclic compounds is explained on the basis of the present kinetic and structural data.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
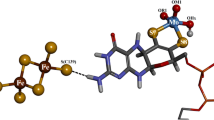
Molybdenum (42Mo) is a trace element that can be found in the active site of enzymes from both eukaryotic and prokaryotic cells catalyzing key reactions of the nitrogen (nitrate reductase, nitrogenase), sulfur (sulfite oxidase, polysulfide reductase), carbon (formate dehydrogenase, CO dehydrogenase) and chlorine [(per)chlorate reductase] cycles, among others [1]. The biological activity of this transition metal depends on the synthesis of complex cofactors such as the FeMoCo in nitrogenase and the pyranopterin in mononuclear Mo-enzymes. The latter cofactor is made up of a heterocyclic pterin structure fused to a pyran ring, and the latter is substituted by a dithiolene moiety responsible for Mo coordination (Fig. 1) [2]. Several enzymes are reported to have an identical cofactor but with tungsten (74W) instead of Mo, and for this reason both Mo- and W-containing mononuclear enzymes were grouped together. According to X-ray structural data, primary sequence alignments, and spectroscopic and biochemical features, mononuclear Mo- and W-enzymes have been classified in four broad families; xanthine oxidase (XO), sulfite oxidase (SO), dimethyl sulfoxide reductase (DMSOr), and W-aldehyde oxidoreductase (W-AOR) [1, 3–6].
Schematic representation of the active site and additional redox cofactors present in mononuclear Mo(W)-enzymes of the XO family. R=H pyranopterin monophosphate (PMP) cofactor found in XO, AO and other mammalian Mo-enzymes, R=CMP pyranopterin cytidine dinucleotide (PCD) cofactor which is found in the prokaryotic Mo(W)-enzymes of the XO family. An FAD cofactor is not present in DgAOR
Enzymes from the XO family are probably the most extensively studied mononuclear Mo-enzymes [3–5, 7, 8]. With the exception of CO dehydrogenase and 4-hydroxylbenzoyl-CoA reductase, they catalyze hydroxylation reactions according to
This oxidative half reaction takes place at the Mo site, and the two reducing equivalents generated upon hydroxylation of a carbon atom are transferred to an external electron acceptor by means of an electron transfer pathway made up of the Mo(W)-pyranopterin, two (2Fe-2S) clusters, and an FAD (Fig. 1).
In the as-isolated (oxidized) state, the active site of the XO family members comprises a Mo ion in a distorted square-pyramidal geometry coordinated by one dithiolene moiety from the pyranopterin molecule, one apical oxo-group, one hydroxyl ligand, and one equatorial sulfido ligand (Fig. 1). The latter can be removed by cyanide treatment (cyanolysis) to yield the inactive desulfo form of the enzyme [9]. Recently, we demonstrated that the Mo site of the catalytically competent aldehyde oxidoreductase from Desulfovibrio gigas (DgAOR) does not have a sulfido ligand [10]. This observation was controversial, as it was demonstrated that in XO the sulfido ligand has an essential role in the reaction mechanism.
In the present work, steady-state kinetics in solution, X-ray crystallography, and NMR spectroscopy were applied to understand the reactivity of different substrates according to their stereochemistry to study the enzyme–substrate interaction, and to analyze the substrate and product of the hydroxylation reaction catalyzed by DgAOR. High-resolution diffracting crystals of native DgAOR soaked with benzaldehyde, trans-cinnamaldehyde and 3-phenyl propionaldehyde showed structural features that explain the interaction mode of the substrate and product in the active site, as well as the mechanism of enzyme inhibition and/or inactivation by the substrate.
Materials and methods
DgAOR purification and protein quantification
Desulfovibrio gigas was purified as previously reported [11]. Protein quantification was performed using either the bicinchoninic acid assay (Sigma-Aldrich) or using the enzyme molar extinction coefficient at 462 nm (ε = 24.6 mM−1 cm−1). The enzyme was stored in 50 mM tris(hydroxymethyl)aminomethane (Tris–HCl) buffer pH 7.6 at −20 °C until use.
Enzyme kinetic assays in solution
Steady-state kinetic studies of DgAOR were performed aerobically at 308 K by measuring the rate of 2,6-dichlorophenol-indophenol (DCPIP) reduction at 600 nm (ε = 21 mM−1 cm−1) in a 1 cm optical path length cell containing 50 mM Tris–HCl buffer pH 7.6, 35 μM DCPIP, 40 or 400 nM enzyme depending on the substrate, and variable concentration of the substrate (benzaldehyde, 2-phenyl acetaldehyde, 3-phenyl propionaldehyde, trans-cinnamaldehyde, 2-hydroxybenzaldehyde, 3-hydroxybenzaldehyde, 2,3-dihydroxybenzaldehyde, 2,5-dihydroxybenzaldehyde, hypoxanthine, xanthine, 1-methyl xanthine, 3-methyl xanthine, 1,3,7-trimethyl xanthine, allopurinol, oxypurinol, isoquinoline, nicotinic acid, formamide, dimethyl formamide). A broad range of substrate concentrations was tested and the initial reaction rates values were plotted and fitted to appropriate models (Michaelis–Menten or Haldane) to determine the respective kinetics parameters. In all the experiments, the enzyme was equilibrated for a few minutes with the reaction mixture until the absorbance of the DCPIP was stable. The reaction was started by substrate addition. Under these experimental conditions, one enzymatic unit (U) corresponds to 1 μmol of aldehyde oxidized per min and the specific activity is U/mg of enzyme.
Detection of substrates and products by NMR spectroscopy
The initial and final stages of the enzymatic conversion of aldehydes to their respective products were monitored by NMR spectroscopy. 13C-labeled benzaldehyde (α-13C, Sigma-Aldrich) was used as representative substrate. The reactions were performed in NMR tubes containing 500 μL of 1 mM DCPIP, 200 μM benzaldehyde and 50 mM Tris–HCl buffer pH 7.8. To establish the spectrometer lock, a sealed capillary containing pure D2O was inserted into the NMR tube. The 13C-NMR spectra were obtained at 298 K on a Bruker AMX-400 NMR spectrometer equipped with a QNPz probe and a temperature control unit. Spectra were processed using TOPSPIN 3.0 software (Bruker).
Crystallization, soaking procedures, data collection, and refinement
Single crystals of DgAOR were obtained at 4 °C using the sitting-drop vapor-diffusion method. The precipitating solution contained 30 % isopropanol, 0.2 M magnesium chloride in 0.2 M 4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid (HEPES) buffer pH 7.6 as described before [12, 13]. The crystallization drops were prepared by adding 4 μL of DgAOR (10 mg/mL) in 10 mM Tris-HCl buffer pH 7.6–2 μL of precipitating solution. Crystals appeared in 2 weeks and were then stabilized for at least 2 days with a harvesting buffer 1 (HB1 composition: 30 % isopropanol, 30 % polyethylene glycol 3,350, 0.2 M magnesium chloride and 0.2 M HEPES buffer). To remove isopropanol from the active site, harvesting buffer 2 (HB2 composition: 30 % polyethylene glycol 3,350, 0.2 M magnesium chloride and 0.2 M HEPES buffer) was slowly added to the drop for 2 more days. The crystals were then transferred to new drops containing only HB2. After 3 days of stabilization, the single crystals were transferred to new drops containing HB2 solution plus the substrates at 1.4 mM benzaldehyde, 1.1 mM trans-cinnamaldehyde, and ~1 mM 3-phenyl propionaldehyde. After 1 h of soaking for benzaldehyde and 3-phenyl propionaldehyde and 20 h for trans-cinnamaldehyde, the crystals were flash frozen in liquid nitrogen.
Complete datasets were collected for each soaking experiment at two different beamlines: for the benzaldehyde soak, the dataset was collected at ID29 of the ESRF (Grenoble, France) at wavelength 0.976 Å while for the trans-cinnamaldehyde, and the 3-phenyl propionaldehyde soaks, the data were collected at Proxima I of SOLEIL (Paris, France) at wavelengths, 0.979 and 0.918 Å, respectively. The crystals diffracted beyond 1.49 Å and belong to the P6122 space group with cell constants similar to those of the native protein (PDB code: 1VLB). Data were processed using MOSFLM and SCALA from the CCP4 suite [14, 15]. Data collection statistics are presented in Table 1. Phases were obtained by molecular replacement using PHASER and the 1.28 Å resolution molecular model of the native DgAOR (PDB code: 1VLB) [16, 17]. REFMAC 5.5 was used to perform restrained refinement and COOT was used to inspect the electron density maps and manual model building [18, 19]. Geometrical restraints were not used to refine the PCD cofactor. Constructive validation and structure re-refinement were performed using PDB_REDO [20]. Geometrical validation was carried out by several programs such as PROCHECK, MOLPROBITY and STAN [21–23]. A summary of the refinement statistics is presented in Table 2.
Results
Kinetic studies of DgAOR with aromatic aldehydes and heterocyclic compounds
Mammalian aldehyde oxidase (AO) and xanthine oxidase (XO) show a high level of analogy in both molecular and structural properties. Earlier studies, aimed to determine their substrate specificities, found out that both AO and XO catalyze the hydroxylation of aldehydes and purines, though AO catalyzes aldehydes hydroxylation more efficiently than XO [24]. Preliminary kinetic studies showed that in contrast to mammalian AO, DgAOR catalyzes the hydroxylation of aldehydes but not of heterocyclic compounds [25]. In the present work, we analyzed the DgAOR substrate specificity using a wide range of aromatic aldehydes and heterocyclic compounds, which are the natural substrates of different XO family members (Table 3).
The enzymatic activity of DgAOR was firstly tested using aromatic aldehydes such as benzaldehyde, 2-phenyl acetaldehyde and 3-phenyl propionaldehyde to determine how the distance of the aldehyde function to the aromatic ring affects the reaction rate and substrate affinity. Benzaldehyde yielded kinetic parameters similar to those already published [25], confirming the high reactivity and affinity of DgAOR for this compound. The reaction rates of 2-phenyl acetaldehyde and 3-phenyl proprionaldehyde were comparable to that of benzaldehyde, though the k cat/K M ratios indicated a lower affinity for these compounds (Table 3).
High concentrations of benzaldehyde, 2-phenyl acetaldehyde and 3-phenyl propionaldehyde affect negatively the reaction rates, most likely binding to the ES complex yielding the ternary complex SES according to Scheme 1. Least-square fitting of the experimental data obtained with these three aromatic aldehydes to the Haldane equation [26] shown in the Scheme 1 yielded satisfactory fittings and generated the kinetic parameters that are summarized in Table 3.
Unexpectedly, besides the reversible substrate inhibition observed at high substrate concentrations, 3-phenyl propionaldehyde also produced the irreversible inactivation of DgAOR through a suicide substrate behavior. This phenomenon was evidenced by the fact that the DCPIP absorbance does not reach zero despite the large molar excess of 3-phenyl propionaldehyde which should be enough to reduce all the electron acceptor available in the reaction mixture. This result suggests that the enzyme gets inactive during the course of the reaction. This phenomenon was confirmed and explained at molecular level after solving the X-ray crystallographic structure of single crystals of DgAOR soaked with 3-phenyl propionaldehyde (see X-ray crystallography results in “Molecular picture of the suicide behavior of 3-phenyl propionaldehyde”).
The presence of a sterical restrain in the substituent group of the aldehyde also affects the kinetic parameters. As it is clear from the obtained data, the unsaturation present in the aliphatic portion of trans-cinnamaldehyde affected negatively the reaction rate (k cat) and yielded lower K M values, resulting in k cat/K M ratio similar to that obtained for 3-phenyl propionaldehyde. The lower reaction rate observed for cinnamaldehyde might be produced by the presence of the carbonyl group conjugated with an alkene, which lowers the electrophilic character of the aldehyde carbon atom and hence increasing the activation energy of the hydroxylation reaction. As observed for the other aldehydes with aromatic substituents, substrate inhibition was also observed at high cinnamaldehyde concentrations and least-square fitting of the experimental data to the Haldane equation yielded a K iS value similar to that of 3-phenyl propionaldehyde (Table 3).
Previous studies described that the kinetic parameters of DgAOR are deeply affected by the presence of substituent groups in the aromatic ring of benzaldehyde [25]. We found that 2-OH benzaldehyde (salicylaldehyde) yield the lowest k cat and K M values of all substrates tested in this work, suggesting that the OH group at the ortho position participates in a strong interaction with the active site pocket, which in turn affects negatively the reaction rate. Crystal soaks of DgAOR with this substrate were also carried out but, unfortunately, an X-ray structure of DgAOR with salicylaldehyde in the active site of could not be obtained. The position of the hydroxyl group appears to be highly relevant as the kinetic parameters obtained with 3-OH benzaldehyde were considerably different to those of 2-OH benzaldehyde. In fact, the k cat value obtained is similar to that of benzaldehyde but the K M is much higher (Table 3), indicating that the interaction of amino acids near the active site with the 3-OH substituent destabilized the enzyme–substrate complex but does not interfere with the rate-limiting step of the reaction. To analyze the combined effect of both 2-OH and 3-OH substituents in the enzyme activity, 2,3-diOH benzaldehyde was used as substrate. It was observed that the ortho-OH overrides the effect of the meta-OH interfering with the rate-limiting step of the global reaction, as the k cat value decreased in the order of magnitude found for the single substituted 2-OH benzaldehyde, whereas the K M also decreased relative to the 3-OH benzaldehyde though the values were still considerably higher than that of 2-OH benzaldehyde. This result proves the positive and negative contributions of the ortho-OH and meta-OH, respectively, on the formation of the ES complex. This negative effect was more evident when 2,5-diOH benzaldehyde was used as substrate, as the presence of the meta’-OH substituent outweighed the effect of the ortho-OH, as the k cat/K M ratio decreased one and two orders of magnitude compared to the 2,3-diOH benzaldehyde and 2-OH benzaldehyde, respectively (Table 3). It is important to note that all the OH-substituted benzaldehydes did not show substrate inhibition as observed for the other aromatic aldehydes. This might indicate that the presence of the hydroxyl group hinders the formation of the SES ternary complex (Scheme 1). This can be explained by the hydrophobic character of the substrates, which is lower for the hydroxyl-substituted aromatic groups of 2-OH, 3-OH, 2,3-diOH, and 2,5-diOH benzaldehydes, when compared to benzaldehyde, 2-phenyl acetaldehyde, 3-phenyl propionaldehyde and trans-cinnamaldehyde. In other words, for substrate inhibition to occur, the second aldehyde molecule responsible for the inhibition should be stabilized by hydrophobic interactions in the substrate entrance channel, which is probably the case for the benzaldehyde, 2-phenyl acetaldehyde, 3-phenyl propionaldehyde and trans-cinnamaldehyde. This hypothesis is in line with our crystallographic results presented below in “Molecular picture of the enzyme–substrate interaction: crystal structure of DgAOR with benzaldehyde”, in which a second benzaldehyde molecule can be observed in the catalytic pocket stabilized by hydrophobic interactions with amino acids side chains, blocking substrate entrance and/or product exit. However, in the case of 2-OH, 3-OH, 2,3-diOH, and 2,5-diOH benzaldehydes further interactions, namely H-bonds, must be required for substrate and/or product stabilization at the catalytic pocket and, probably, due to the hydrophobic character of DgAOR catalytic pocket stable enzyme–substrate or enzyme–product complexes with OH-substituted benzaldehydes were not obtained.
The substrates of other enzymes of the XO family were also tested, as the active sites of the members of this enzyme family share high similarity. DgAOR was unable to catalyze the oxidative hydroxylation of hypoxanthine, xanthine, 1-methyl xanthine, 3-methyl xanthine, 1,3,7-trimethyl xanthine (caffeine), allopurinol and oxypurinol, isoquinoline, nicotinate, formamide and dimethylformamide. In contrast to aldehydes, in which a carbonyl carbon atom is hydroxylated, in the heterocyclic compounds the target carbon atom (highlighted in red in Table 3) is not from a carbonyl but instead, is adjacent to nitrogen atoms. This indicates that, in contrast to XO and AO, DgAOR is only capable of hydroxylating carbonyl functions. A possible explanation is that the lack of sulfido ligand in the catalytically competent DgAOR modifies the electronic properties of the Mo site turning this enzyme incompetent to react with carbon atoms of N-heterocycles.
Molecular picture of the enzyme–substrate interaction: crystal structure of DgAOR with benzaldehyde
To study at the molecular level, the enzyme–substrate interaction and determine which amino acids are involved in substrate orientation and specificity, single crystals of DgAOR obtained from an enzyme batch showing high specific activity were soaked with the compounds as summarized in Table 3. The soaking experiments with benzaldehyde, trans-cinnamaldehyde and 3-phenyl propionaldehyde yielded high-resolution diffracting crystals, which served for the purpose of this work. As expected, the final overall structures obtained from soaked crystals of DgAOR were very similar to the native DgAOR (PDB code: 1VLB), showing rmsd values in the range 0.15–0.20 Å for all backbone atoms.
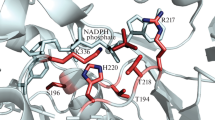
The single-crystal of DgAOR soaked with benzaldehyde diffracted up to 1.49 Å resolution (PDB code: 4US8). The protein structure was refined using anisotropic B factors with final R and R-free values of 9.11 and 12.22 %, respectively. After a detailed analysis of the electron density maps in the Mo site region, it was possible to identify the aromatic rings of two benzaldehyde molecules with partial occupancy. The first one (B1) is in close contact with the Mo atom while the other one (B2) is at the substrate entrance channel (Fig. 2).
1.49 Å resolution crystallographic structure obtained from DgAOR crystals soaked with 1.4 mM benzaldehyde, after isopropanol removal. Atom distances are in Å. Atoms color code: Mo in light teal, S in yellow, O in red, C in cyan. The 2mFo–DFc maps (blue mesh) are contoured at 1σ. The benzaldehyde molecules B1 and B2 were modeled in two alternative conformations (orange/green and blue/violet, respectively) with occupancy factors of 0.3 for each conformation
Benzaldehyde molecule B1 is approximately in the same position as the isopropanol molecule found in the native structure of DgAOR [12]. The aromatic ring of B1 establishes π-π interactions (π-stacking) with the side chains of Phe425 and Tyr535 in a parallel-dislocated conformation. This type of interaction stabilizes the substrate in the active site and is probably responsible for the high affinity of DgAOR towards benzaldehyde. The aldehyde function of the benzaldehyde molecule is not unambiguously observed in the electron density map with a 1σ contour. After lowering the contour, the position of the aldehyde function toward the Mo atom becomes clearer, though electron density is not continuous. In this case, the oxygen atom of the aldehyde is at 2.42 Å from the metal with an orientation suitable for catalysis (orange conformation of molecule B1 in Fig. 2). An alternative conformation for B1 has also been considered where the aldehyde function is pointing away from the active site, suggesting that the B1 molecule is disordered. The two conformations of B1 have been modeled with occupancy of 0.3 and in both cases the temperature factors are in the same order of magnitude as the average B factors of the protein. Additionally, inspection of the Fo-Fc map indicates the presence of a second benzaldehyde molecule B2, in the substrate entrance channel next to B1 (Fig. 2). Even though the electron density is very poor, probably due to low occupancy and disorder, B2 was also modeled with two distinct conformations, each with 0.3 occupancy.
The molecular disorder observed in the X-ray data suggests that benzaldehyde molecule B1 can adopt different conformations when is close to the Mo atom. This evidence and the discontinuous electron density of the B1 molecule (Fig. 2) prompted us to evaluate the possibility of an unexpected catalytic reaction (e.g., decarboxylation upon hydroxylation with production of benzene, hydroxylation of the aromatic ring, within others). NMR spectroscopy can be successfully applied to understand the chemical and biological behavior of aldehydes in aqueous solution. For this reason, it can be used to determine precisely which compounds are formed upon enzymatic oxidation. In the present work, the enzymatic oxidation of benzaldehyde (enriched with 13C on the α-carbon atom) was analyzed by means of 13C-NMR spectroscopy. Considering the aromatic nature of the artificial electron acceptor (DCPIP) used in the enzymatic reaction, 1H-NMR was ruled out. The 13C-NMR spectrum of benzaldehyde in buffered aqueous solution yielded a peak at ca. 196 ppm (Fig. 3). No evidence of the hydrated form or any other compound was observed. This result was expected since the hydration equilibrium constant of aromatic aldehydes is smaller than those for aliphatic aldehydes, as the positive charge on the carbonyl carbon atom is partially distributed on the phenyl group, decreasing its electrophilicity. This behavior is explained by the slow isotropic exchange of oxygen between the carbonyl group and water [27]. Upon addition of 1 μM DgAOR and 3 min incubation at 298 K, a new peak at 175 ppm was clearly observed, typical of the carboxylic carbon atom of benzoate/benzoic acid (Fig. 3). This result ruled out the possibility of an unexpected enzymatic reaction.
13C-NMR spectra of 200 μM benzaldehyde-α-13C plus 1 mM DCPIP in 50 mM TrisHCl pH 7.8 before (a) and after (b) addition of 1 μM DgAOR and 3 min incubation at 298 K. The peaks corresponding to the carbon of benzaldehyde and benzoic acid are indicated with red arrows. The peak at ca. 60 ppm corresponds to the natural abundance of 13C present in the buffer Tris–HCl
Molecular picture of the enzyme–product interaction: crystal structure of DgAOR with trans-cinnamic acid
The single-crystal of DgAOR soaked with trans-cinnamaldehyde diffracted up to 1.13 Å (PDB code: 4USA). The crystal structure was refined using anisotropic B factors with final R and R-free values of 10.17 and 12.17 %, respectively. By inspecting the electron density maps in the substrate entrance channel, it was possible to identify the aromatic ring as well as the aliphatic portion of what could be the soaked substrate (Fig. 4). However, careful analysis of the mFo-DFc difference map, occupancy and B factors, suggested that instead of trans-cinnamaldehyde, the crystal structure trapped the catalytic reaction product trans-cinnamic acid with 0.5 occupancy.
1.13 Å resolution crystallographic structure obtained from DgAOR crystals soaked with 1.1 mM trans-cinnamaldehyde, after isopropanol removal. Atom distances are in Å. Atoms color code: Mo in light teal, S in yellow, O in red, C in cyan. The 2mFo–DFc maps (blue mesh) are contoured at 1σ. In orange an alternate conformation of the side chain of Phe425 and a molecule of trans-cinnamaldehyde, both modeled with occupancies of 0.5, and interacting through a parallel-dislocated π-π interaction is represented
Interestingly, the aromatic ring of the trans-cinnamic acid is slightly farther from the Mo ion and its aromatic ring is tilted ca. 50º when compared to the benzaldehyde B1 molecule (Fig. 2). Furthermore, the side chain of Phe425 could also been modeled with alternate conformations, with 60º differences between the planes of the aromatic rings. The unusual rotamer of the residue establishes a parallel-dislocated π–π interaction with the trans-cinnamic acid ring. Furthermore, the carboxylate function is pointing away from the Mo ion, suggesting that after the hydroxylation of the aldehyde, the trans-cinnamic acid has to rearrange itself and that the migration of the product is aided with the conformational re-arrangement of the Phe425 side chain, promoting the diffusion of the carboxylic acid to the enzyme surface.
Molecular picture of the suicide behavior of 3-phenyl propionaldehyde
Similar to the other soaking experiments presented in this work, DgAOR single crystals were soaked with 3-phenyl propionaldehyde (PDB code: 4US9). The crystals diffracted up to a resolution of 1.40 Å and the protein structure was refined using anisotropic B factors up to 9.34 % R and 11.70 % R-free values. In contrast to the other complexes analyzed in this work, no electron density corresponding to the substrate or product molecules could be observed close to the Mo ion or in the catalytic pocket. Nevertheless, clear positive electron density corresponding to the substrate molecule was found at the protein surface at the beginning of the substrate entrance channel. The 3-phenyl propionaldehyde molecule could be modeled with full occupancy and it was possible to observe that the Nε 2 of His752 was covalently bound to the carbonyl carbon atom of the aldehyde at 1.34 Å (Fig. 5). The carbonyl of the aldehyde and the side chain of the histidine are not coplanar, with angles of ca. 114º. Such geometry suggests that the imidazole ring attacked the aldehyde due to its high concentration during the soaking experiment. This kind of interaction has been reported before, where formaldehyde was found to react with imidazole producing 1-hydroxymethyl imidazole [28]. This covalent bond probably arises from an unspecific reaction that occurred during the soaking experiment which was performed using a high concentration of 3-phenyl propionaldehyde. The obtained structure shows the result of this unspecific reaction where the bulky substituent group of 3-phenyl propionaldehyde emerges at the entrance of the substrate channel, blocking the access to new substrate molecules. This structural evidence might be correlated with the suicide substrate behavior observed in the kinetic studies, where inactivation of the enzyme is observed after a few catalytic cycles, when high concentrations of the substrate are used.
1.40 Å resolution crystallographic structure obtained from DgAOR crystals soaked with 1 mM 3-phenyl propionaldehyde, after isopropanol removal. a Enzyme surface close to the entrance channel showing the 3-phenyl propionaldehyde bound to His752 side chain. b 2mFo-DFc map (blue mesh, contoured at 1σ) showing the adduct between the His752 side chain (in cyan) and the 3-phenyl propionaldehyde molecule (in orange, modeled with occupancy of 1)
Discussion
Aldehyde oxidoreductase from Desulfovibrio gigas was studied using steady-state enzyme kinetics and X-ray crystallography to evaluate the ability of this enzyme to hydroxylate aromatic aldehydes and heterocyclic compounds and elucidate how the interaction of such compounds occurs within the catalytic pocket.
NMR spectroscopy studies using 13C-labeled benzaldehyde confirmed that the substrate of DgAOR is converted into the respective carboxylic acid (Fig. 3). The X-ray crystallographic data showed that the benzaldehyde (B1) binds at the active site being stabilized mainly by hydrophobic interactions. The side chains of Phe425 and Tyr535 have a key role in substrate stabilization, establishing π-π stacking interactions with the aromatic ring of the substrate when this enters into the active site. These interactions seem to be of utmost importance to correctly orientate the substrate to react with the Mo ion (Fig. 2). Yet, the disorder observed in the benzaldehyde molecule suggests that it could enter into the catalytic channel with a wrong orientation leading to non-productive binding. Kinetic data were obtained using benzaldehyde as substrate and the decrease in the reaction rates observed at high substrate concentrations was modeled using the Haldane equation (see Scheme 1). This model states that enzyme inhibition occurs by the formation of an SES ternary complex. We believe that this complex is evident in the crystal structure here presented where a second, partially occupied benzaldehyde molecule (B2) was modeled in the substrate entrance channel. This SES ternary complex observed in the X-ray data explains the substrate inhibition observed at high substrate concentrations in the kinetic studies (Table 3). Nevertheless, it also shows the pathway and the interaction that guide the benzaldehyde molecule through the entrance channel to the catalytic site.
The kinetic data obtained with the two similar molecules 3-phenyl propionaldehyde (Ar–CH2–CH2–COH) and trans-cinnamaldehyde (Ar–CH=CH–COH) as substrates indicate that when a structural restrain is present in the substituent group, both the maximum reaction rate and K M value decrease. The latter might be produced not only by the important decrease in k cat [K M = (k cat + k Off)/k On], but also due to considerable changes in the k On and k Off values attributed to the different stereochemistry of the compounds. On the other hand, the lower reaction rate observed could also be due to the lower electrophilic character of the aldehyde carbon atom of cinnamaldehyde.
The X-ray data obtained from DgAOR soaked with trans-cinnamaldehyde (Fig. 4) could not show how this substrate interacts with the Mo ion. Nevertheless, after careful inspection of the 2mFo-DFc map, it was possible to determine that the extra electron density observed in the 10 Å deep channel of the enzyme corresponds to a trans-cinnamic acid molecule. The X-ray structure in Fig. 4 suggests that the reaction product has been trapped while exiting from the catalytic pocket. If this is the case, the Phe425 role is not only to stabilize and orientate the substrate molecule, but also to promote the product release from the active site. We are confident that the X-ray structure depicted in Fig. 4 does not represent an event associated with inhibition by product of the reaction since further kinetic studies showed that high concentrations (up to 20 mM) of carboxylic acids (e.g., benzoic acid, acetic acid) do not affect neither the reaction rates nor the K M values when benzaldehyde is used as substrate. This can be explained by the highly hydrophobic character of the DgAOR substrate entrance channel and the low pKa value of carboxylic acids (pKa ~4). In this sense, once the carboxylic acid molecule diffuses from the enzyme channel into the bulk solution, it is deprotonated to produce the carboxylate form, and the negatively charged molecule cannot re-enter the channel.
The steady-state kinetics in solution showed that 3-phenyl propionaldehyde yields maximum reaction rates and K M values comparable to those of benzaldehyde. Nevertheless, at high substrate concentrations, this compound leads to enzyme inactivation through a process known as suicide substrate behavior. The X-ray data of DgAOR soaked with this aldehyde suggested that the enzyme inactivation might occur by the fact that the aromatic substituent of this bulky aldehyde blocks the entrance of new substrate molecules into the active site (Fig. 5). The formation of a covalent bond between the C-atom of the aldehyde with the side chain of His752 at the enzyme surface is evident from the short C–N bond distance (1.34 Å). In agreement with this observation, earlier reports showed that the reaction of imidazole with formaldehyde can produce 1-hydroxymethyl imidazole [28]. In this compound, the N-atom of the imidazole is added to the alpha carbon of the aldehyde, producing a C-atom with a sp3 hybridization. This explains the non-planar geometry of the alpha C-atom of 3-phenyl propionaldehyde observed in the X-ray structure depicted in Fig. 5.
The ability of DgAOR to catalyze the oxidation of aromatic aldehydes like benzaldehyde 2-phenyl acetaldehyde, 3-phenyl propionaldehyde and trans-cinnamaldehyde indicated that the catalytic pocket of the enzyme can accommodate both small and bulky substrates. Moreover, the fact that the aldehydes substituted with –OH groups in the aromatic ring are also substrates of DgAOR indicated that substrates with polar substituents (without net charge) which need the formation of H-bonds to be stabilized in the catalytic pocket can also enter and react at the Mo site. However, X-ray data could not be obtained from the soaking experiments with these compounds. This suggests that these substrates and their reaction products do not form a stable complex with DgAOR in the catalytic pocket as in the case of benzaldehyde (substrate) or trans-cinnamic acid (reaction product). Furthermore, when N-heterocyclic molecules known as good substrates for other members of the XO family (see Table 3) have been tested as substrates of DgAOR, no activity was detected. The fact that these compounds cannot be hydroxylated by DgAOR might be due to the different ligand composition of the Mo active site. DgAOR is a unique enzyme within the XO family of mononuclear Mo-enzymes since it does not need the essential sulfido ligand to catalyze the hydroxylation of aldehydes [10]. However, the lack of the third equatorial sulfur ligand at the Mo ion might be responsible for the incapability of DgAOR to hydroxylate N-heterocyclic compounds as for example xanthine.
Abbreviations
- AOR:
-
Aldehyde oxidoreductase
- AMP:
-
Adenosine monophosphate
- AO:
-
Aldehyde oxidase
- CMP:
-
Cytidine monophosphate
- DCPIP:
-
2,6-Dichlorophenol-indophenol
- Dg :
-
Desulfovibrio gigas
- DMSOr:
-
Dimethyl sulfoxide reductase
- FAD:
-
Flavin adenine dinucleotide
- HEPES:
-
4-(2-Hydroxyethyl)-1-piperazine ethanesulfonic acid
- PCD:
-
Pyranopterin cytidine dinucleotide
- PMP:
-
Pyranopterin monophosphate
- rmsd:
-
Root-mean-square deviation
- SO:
-
Sulfite oxidase
- Tris–HCl:
-
Tris(hydroxymethyl)aminomethane
- U:
-
Enzymatic unit
- W-AOR:
-
W-aldehyde oxidoreductase
- XO:
-
Xanthine oxidase
References
Gonzalez PJ, Rivas MG, Mota CS, Brondino CD, Moura I, Moura JJG (2013) Coord Chem Rev 257:315–331
Basu P, Burgmayer SJ (2011) Coord Chem Rev 255:1016–1038
Hille R (1996) Chem Rev 96:2757–2816
Hille R (2002) Trends Biochem Sci 27:360–367
Hille R (2002) Met Ions Biol Syst 39:187–226
Johnson MK, Rees DC, Adams MW (1996) Chem Rev 96:2817–2840
Brondino CD, Rivas MG, Romao MJ, Moura JJ, Moura I (2006) Acc Chem Res 39:788–796
Brondino CD, Romao MJ, Moura I, Moura JJ (2006) Curr Opin Chem Biol 10:109–114
Wahl RC, Rajagopalan KV (1982) J Biol Chem 257:1354–1359
Santos-Silva T, Ferroni F, Thapper A, Marangon J, Gonzalez PJ, Rizzi AC, Moura I, Moura JJG, Romao MJ, Brondino CD (2009) J Am Chem Soc 131:7990–7998
Marangon J, Correia HD, Brondino CD, Moura JJ, Romao MJ, Gonzalez PJ, Santos-Silva T (2013) PLoS One 8:e83234
Rebelo JM, Dias JM, Huber R, Moura JJ, Romao MJ (2001) J Biol Inorg Chem 6:791–800
Romao MJ, Archer M, Moura I, Moura JJ, LeGall J, Engh R, Schneider M, Hof P, Huber R (1995) Science 270:1170–1176
Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG (2011) Acta Crystallogr D Biol Crystallogr 67:271–281
Evans P (2006) Acta Crystallogr D Biol Crystallogr 62:72–82
McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) J Appl Crystallogr 40:658–674
McCoy AJ (2007) Acta Crystallogr D Biol Crystallogr 63:32–41
Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Acta Crystallogr D Biol Crystallogr 66:486–501
Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA (2011) Acta Crystallogr D Biol Crystallogr 67:355–367
Joosten RP, Joosten K, Murshudov GN, Perrakis A (2012) Acta Crystallogr D Biol Crystallogr 68:484–496
Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) Acta Crystallogr D Biol Crystallogr 66:12–21
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) J Appl Cryst 26:283–291
Nayal M, Di Cera E (1996) J Mol Biol 256:228–234
Krenitsky TA, Neil SM, Elion GB, Hitchings GH (1972) Arch Biochem Biophys 150:585–599
Barata BA, LeGall J, Moura JJ (1993) Biochemistry 32:11559–11568
Briggs GE, Haldane JB (1925) Biochem J 19:338–339
McClelland RA, Coe M (1983) J Am Chem Soc 105:2718–2725
Alley PW (1975) J Org Chem 40:1837–1838
Acknowledgments
H.C. and J.M. thank Fundação para a Ciência e a Tecnologia (FCT) for the fellowships SFRH/BD/86796/2012 and SFRH/BD/60725/2009. P.J.G. and T.S.S. thank program Ciência 2008 of FCT. C.D.B. and P.J.G. are members of CONICET (Argentina). We thank the beamline scientists of ID29 from the ESRF and of Proxima I from SOLEIL for their assistance in X-ray data collection. This work was supported by FCT through the project PTDC/BIA-PRO/118377/2010 and Grant No. PEst-C/EQB/LA0006/2011.1.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Responsible Editors: José Moura and Paul Bernhardt.
An Interactive 3D Complement page in Proteopedia is available at: http://proteopedia.org/wiki/index.php/Journal:JBIC:27.
Rights and permissions
About this article
Cite this article
Correia, H.D., Marangon, J., Brondino, C.D. et al. Aromatic aldehydes at the active site of aldehyde oxidoreductase from Desulfovibrio gigas: reactivity and molecular details of the enzyme–substrate and enzyme–product interaction. J Biol Inorg Chem 20, 219–229 (2015). https://doi.org/10.1007/s00775-014-1196-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00775-014-1196-4