Abstract
We analyzed the structure of horseradish peroxidase (HRP) under denaturing conditions of 9 M urea or 6 M guanidine hydrochloride (GdnHCl). Far-UV circular dichroism (CD) spectra indicated the existence of native-like secondary structure of holo-HRP in 9 M urea. In addition, slight changes in near-UV and Soret region CD spectra of holo-HRP in 9 M urea suggest that the tertiary structure of holo-HRP and the binding of heme remain partially intact in this condition. A transition in the thermal unfolding transition curve of holo-HRP in 9 M urea indicated the existence of a considerable amount of secondary structure. However, no secondary structure, tertiary structure, or interaction between heme and HRP were observed in holo-HRP in 6 M GdnHCl. Small-angle X-ray scattering indicated that although distal and proximal domains of holo-HRP in 9 M urea might be partially unfolded, the central region that contains the heme might maintain its tertiary structure. Our results suggest that retention of the heme is essential for maintenance of the structure of HRP under highly denaturing conditions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Peroxidases (EC 1.11.1.7) are heme-containing enzymes that catalyze oxidation of various organic and inorganic substrates by H2O2 (Poulos 1993). These enzymes are found widely in almost all organisms (Koua et al. 2009). Horseradish peroxidase (HRP) is an important monomeric enzyme (Veitch 2004). HRP consists of thirteen α-helices and two short antiparallel β-strands (Gajhede et al. 1997), and includes a heme prosthetic group, two calcium binding sites, four disulfide bridges, and eight carbohydrate chains (Gajhede et al. 1997).
HRP has numerous medical and industrial applications (Azevedo et al. 2003; Krainer and Glieder 2015). It has been used in medical diagnosis, in histochemical staining, and in biosensors (Veitch 2004). Moreover, it has been used to synthesize organic polymers and to remove phenols and aromatic amines from wastewaters (Won et al. 2004; Wagner and Nicell 2002). However, its industrial applications have been limited by its instability under various conditions such as elevated temperatures and excess H2O2. Although many experiments have been performed to increase the stability and activity of HRP by protein engineering, enzyme immobilization, and chemical modification (Ryan and O’Fagain 2007; Zakharova et al. 2011), more information concerning its structural stability is needed.
The stability of HRP is affected by several variables such as temperature, pH, glycosylation, and concentration of calcium ions (Chattopadhyay and Mazumdar 2000; Howes et al. 2001). Given that the catalytic activities of enzymes are dependent on their proper folding to generate three-dimensional structures, increased knowledge of the denaturation mechanism of HRP under denaturing conditions may guide design strategies to increase its conformational stability and to reduce loss of its activity.
HRP folds into an N-terminal domain and a C-terminal domain. The heme is located at the active-site cleft between a distal N-terminal domain and a proximal C-terminal domain (Fig. 1a), and is important to stabilize the conformation of HRP (Veitch 2004). Destruction of heme inactivates HRP; this result indicates that the heme molecule is important for HRP activity (Mao et al. 2013). However, the mechanism by which heme contributes to the stability of HRP and renders it resistant to denaturation remains largely unknown despite many investigations (Carvalho et al. 2003; Feng et al. 2008).
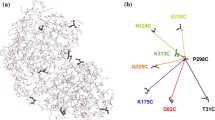
Structure of HRP. a Ribbon representation of HRP structure (PDB code 1W4W). Orange stick heme group; yellow distal N-terminal domain; purple: proximal C-terminal domain. b B-factor-labeled structure of HRP. Low B-factors (blue) indicate that the region is rigid. Figure was generated using PyMol (http://www.pymol.org) (color figure online)
In this study, we analyzed the role of heme in HRP under highly denaturing conditions (9 M urea and 6 M guanidine hydrochloride (GdnHCl) because these conditions are commonly used to completely unfold proteins (Tanford 1968). To monitor the unfolding process, we used circular dichroism (CD) and small-angle X-ray scattering (SAXS) methods. Holo-HRP retained a considerable amount of its secondary and tertiary structure, and maintained heme-binding ability in 9 M urea. Binding of heme at the interface between two domains contributed to the rigidity around the heme-binding site and thereby conferred this resistance to 9 M urea. Our results suggest that the heme contributes to the resistance to denaturation by providing a rigid binding site.
Materials and methods
Chemicals and sample preparation
All chemicals used in buffer were obtained from sigma. HRP was purchased from Roche as a lyophilized powder. Sample purity was assessed using SDS-PAGE analysis. The concentration of HRP was measured spectrophotometrically considering a molar extinction coefficient of 102 mM−1 cm−1 at 403 nm (Strickland et al. 1968). Apo-HRP was prepared from the holo-HRP by the acid butanone procedure (Yonetani 1967). The concentration of apo-HRP was estimated using an extinction coefficient of 20,000 mM−1 cm−1 at 280 nm (Tamura et al. 1972).
Circular dichroism
CD experiments were conducted using a Jasco J-810 spectropolarimeter at 25 °C. HRP was dissolved in a solution of 20 mM sodium phosphate (pH 7.4) and 1 mM CaCl2, in the absence and presence of 9 M urea or 6 M GdnHCl. Far-UV CD spectra were measured in a quartz cell of 2-mm path length. The spectra of near-UV and Soret regions were obtained using a rectangular quartz cell of 1-cm path length. Each spectrum was an average of at least three scans, corrected by subtracting the spectrum of the buffer solution and smoothed using Jasco software. Results were expressed as mean residue ellipticity (Greenfield 2006). In all measurements, HRP concentration [HRP] was 5 μM.
Thermal transitions
Thermal denaturation experiments were performed using CD spectroscopy as described previously (Cha et al. 2013). CD spectra were recorded at protein concentration of 3 μM in a 2 mm-quartz cell. Protein samples were prepared in the solution containing 20 mM sodium phosphate (pH 7.4) and 1 mM CaCl2, in the absence and presence of 9 M urea or 6 M GdnHCl. The samples were heated from 30 to 90 °C at 1 °C min−1 using a Peltier temperature control system. CD signals at 222 nm were collected every 1 °C.
Solution SAXS measurements
SAXS measurements were performed at the 4C SAXS II beamline of the Pohang Light Source II (PLS II) with 3 GeV power, Korea. A light source from an In-vacuum Undulator 20 (IVU20: 1.4-m length, 20-mm period) of the PLS II storage ring was focused using a vertical focusing toroidal mirror coated with rhodium, and monochromatized by a Si (111) double crystal monochromator to yield an X-ray beam wavelength λ = 0.734 Å. The X-ray beam size at the sample stage was 0.1 × 0.3 mm (V × H). A two-dimensional (2D) charge-coupled detector (Mar USA, Inc.) was used. For SAXS, sample-to-detector distances were 4.00 and 2.00 m. The scattering angle 2θ was calibrated against a polystyrene-b-polyethylene-b-polybutadiene-b-polystyrene block copolymer standard. The magnitude of scattering vector q = (4π/λ)sinθ was 0.15 nm−1 < q < 3.00 nm−1. We used quartz capillary with outside diameter of 1.5 mm and wall thickness of 0.01 mm as solution sample cells. Measurements of HRP protein solutions were performed over the range 2.0 ≤ [HRP] ≤ 7.0 g L−1. Each 2D SAXS pattern was averaged radially from the beam center and normalized to the transmitted X-ray beam intensity, which was monitored using a scintillation counter placed behind the sample. All scattering measurements were conducted at room temperature. The SAXS data were collected in six successive frames of 0.1 min each to monitor radiation damage, then processed as described previously (Cha et al. 2014; Soh et al. 2015; Kim et al. 2013; Jang et al. 2006). The scattering of specific buffer solutions was used as the experimental background. The radii of gyration R g,G were estimated by Guinier analysis of the scattering data (Glatter and Kratky 1982). The molecular mass (MM) was calculated from a BSA standard protein. The pair distance distribution p(r) function was obtained using indirect Fourier transform using the program GNOM (Semenyuk and Svergun 1991).
Construction of 3D structural models
To reconstruct the molecular shapes, the ab initio shape determination program DAMMIF (Franke and Svergun 2009) was used. For each reconstruction, 15 independent models were selected, then the program DAMAVER (Volkov and Svergun 2003) was used to filter the averaged aligned model at a given cutoff volume. The program CRYSOL (Svergun et al. 1995) was used to calculate the SAXS curves from the atomic models. For comparison of the overall shapes and dimensions, the program SUPCOMB (Kozin and Svergun 2001) was used to superimpose the ribbon diagrams of the atomic crystal models onto the reconstructed dummy atom models.
Results and discussion
Comparison of far-UV CD spectra between holo-HRP and apo-HRP
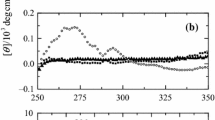
The far-UV CD spectrum of a protein can be used to evaluate its secondary structure (Greenfield 2006), so we first measured the far-UV CD spectra of holo-HRP (Fig. 2a) and apo-HRP (Fig. 2b) in native condition, 9 M urea, or 6 M GdnHCl. The far-UV CD spectrum of native holo-HRP showed characteristic bands of α-helix structure, consistent with previous reports that spectra of this enzyme had negative bands at 207 and 222 nm (Strickland 1968). CD spectrum of holo-HRP at 6 M GdnHCl showed the general pattern of unfolded polypeptide; previous studies have also showed that HRP was completely denatured in 6 M GdnHCl (Pappa and Cass 1993). However, the spectrum at 9 M urea showed an unexpected minor decrease in the molar ellipticity in the far-UV CD spectrum of holo-HRP; i.e., the enzyme retained most of its secondary structure. A previous paper reported that HRP retained ~50% of its secondary structure even in 8 M urea (Pappa and Cass 1993). By contrast, the far-UV CD spectra of apo-HRP under 9 M urea and 6 M GdnHCl denaturing conditions showed that apo-HRP was completely unfolded in both conditions. This comparison of the far-UV CD spectra of holo-HRP and apo-HRP suggests that the heme may be important in maintaining the secondary structure of holo-HRP in 9 M urea.
Analysis of tertiary structure and heme pocket of holo-HRP by CD spectroscopy
CD spectroscopy also provides information about the tertiary structure and the binding of non-protein cofactors, such as heme of protein (Kelly et al. 2005). The tertiary structure of protein and the binding of heme to protein can be estimated by CD spectroscopy in the near-UV and the Soret regions. Therefore, we further measured the near-UV and Soret CD spectra of holo-HRP under different denaturing conditions (Fig. 3). The near-UV CD spectrum of native holo-HRP had a negative peak near 284 nm, which is attributed to overlapping contribution from tyrosine and tryptophan residues. Although the CD signal of holo-HRP at 284 nm in 9 M urea decreased, the change was not significant; i.e., HRP maintained the majority of its tertiary structure in 9 M urea.
The Soret region CD spectra are sensitive to changes in heme surroundings, and, therefore, give information of the integrity of the heme pocket (Myer 1968). The CD bands in this region are due to the coupled interaction between the heme transitions and π–π* transitions in surrounding aromatic residues (Hsu and Woody 1971). The Soret region CD spectra of native holo-HRP showed a strong positive band at 407 nm, which is consistent with the previous report (Mogharrab et al. 2007). A slight decrease in the 406-nm CD band indicates that the holo-HRP in 9 M urea retains the integrity of the heme pocket. The intensity of the Soret CD signal is related to the catalytic activity of HRP (Akita et al. 2001); this may explain why holo-HRP retains its catalytic activity in 8 M urea (Haque et al. 1999). Our data together with previous published data suggest that 9 M urea could not significantly affect the secondary and tertiary structures or the heme pocket of holo-HRP, so that some catalytic activity was retained under highly denaturing condition.
Thermal unfolding of holo-HRP
Thermal denaturation of holo-HRP under different denaturing conditions was investigated by monitoring the molar ellipticity at 222 nm (Fig. 4). Native holo-HRP showed a cooperative unfolding transition, which indicates that the thermal unfolding process follows a simple two-state behavior. In 9 M urea the transition temperature of holo-HRP was decreased, but the enzyme also exhibited cooperative thermal unfolding. In contrast, the thermally denatured state of holo-HRP in 6 M GdnHCl had no signal in the far-UV CD spectrum. These results indicated that the holo-HRP at 9 M urea retained a considerable amount of secondary structure and, therefore, can undergo further cooperative thermal unfolding.
SAXS data analysis of holo-HRP
To investigate the structure of holo-HRP under different conditions, synchrotron SAXS measurements were performed at room temperature in the range 2.0 ≤ [holo-HRP] ≤ 7.0 g L−1. Guinier analysis (Glatter and Kratky 1982) of the SAXS data revealed linearity in a small q region (Fig. 5). Straight lines were obtained from the linear regression of the scattering data in the q 2-region. These analysis results suggest that holo-HRP in the respective conditions was present in a monodispersed state without aggregation. R g,G (Table 1) was estimated from the slope of the plot, and gave information about the enzyme’s size in solution. R g,G of holo-HRP in the native condition was 21.85 ± 0.20 Å, which is very similar to that (22.11 ± 0.01 Å) of the crystal structure of holo-HRP (PDB code 1W4W). R g,G increased in the order holo-HRP in native condition (21.85 ± 0.20 Å) < holo-HRP in 9 M urea (26.31 ± 0.36 Å) < holo-HRP in 6 M GdnHCl (40.50 ± 1.27 Å); i.e., holo-HRP molecules were partially unfolded in 9 M urea, but almost completely unfolded in 6 M GdnHCl.
Guinier plots of X-ray scattering profiles of holo-HRP under various conditions. a Holo-HRP in native condition, b holo-HRP in 9 M urea, c holo-HRP in 6 M GdnHCl. For clarity, each plot is shifted along the ln I(q) axis. The straight lines were obtained from the linear regression of the scattering data in the q 2-region
X-ray scattering profiles (Fig. 6) of holo-HRP were measured under different conditions, and compared with the theoretical SAXS curve calculated from the monomeric crystal structure of holo-HRP (PDB code 1W4W) using CRYSOL. The X-ray scattering pattern of holo-HRP in native condition (Fig. 6a) was very close to that of the holo-HRP crystal structure. The solution scattering pattern of holo-HRP in 9 M urea (Fig. 6b) was slightly different from that of the holo-HRP in native condition (Fig. 6a). The SAXS curve of holo-HRP in 6 M GdnHCl (Fig. 6c) differed completely from those of holo-HRP in native condition or in 9 M urea; this result suggests that the holo-HRP molecules in 6 M GdnHCl had different size and shape than those of holo-HRP in native condition.
X-ray scattering profiles of holo-HRP under different conditions, at room temperature. a Holo-HRP in native condition, b holo-HRP in 9 M urea, c holo-HRP in 6 M GdnHCl. Open symbols experimental data; solid lines X-ray scattering profiles obtained from dummy atoms models by the ab initio molecular shape determination program DAMMIF [χ 2 = 0.714 for (a), 0.675 for (b), 1.631 for (c)]; dashed line theoretical SAXS curve calculated from the crystal structure of holo-HRP (PDB code 1W4W) using the program CRYSOL (χ 2 = 2.355). For clarity, plots are shifted along the log I(q) axis
A Porod analysis (Rambo and Tainer 2011) was applied to the measured scattering data (Fig. 7). The X-ray scattering curve of holo-HRP in native condition (Fig. 7a) revealed a Porod plateau at q ≈ 0.1 Å−1. A similar plateau was observed in the X-ray scattering curve of holo-HRP even in 9 M urea (Fig. 7b). The presence of plateaus usually implies that the protein exists with a sharp homogenous electron density contrast between the particle and solvent (Rambo and Tainer 2011). We, therefore, concluded that holo-HRP molecules even in 9 M urea are in a partially unfolded state that is different slightly from the state of the holo-HRP in native condition. However, in the case of the holo-HRP in 6 M GdnHCl, the analysis did not show this plateau in the X-ray scattering curve (Fig. 7c); this absence indicates that the secondary and tertiary structures were almost completely destroyed in this strong denaturing condition.
Porod plots of X-ray scattering profiles of holo-HRP in Fig. 6. a Holo-HRP in native condition, b holo-HRP in 9 M urea, c holo-HRP in 6 M GdnHCl
We used GNOM to estimate the pair distance distribution p(r) function (Table 1) of holo-HRP under different conditions from the indirect Fourier transform of the scattering data (Fig. 8). This function provides the radius of gyration R g,p(r), which is based on the full scattering curve, and gives the maximum dimension D max (Table 1) of the protein molecule as the distance at which p(r) approaches 0. The resulting R g,p(r) values are in reasonable agreement with those from the Guinier analysis (Table 1). We computed a theoretical p(r) function for the atomic coordinates of the holo-HRP (PDB code 1W4W) monomeric crystal structure and compared it with those measured for holo-HRP under different conditions. The p(r) function for the holo-HRP in native condition (Fig. 8a) exhibited a symmetrical peak pattern with R g,p(r) = 22.70 ± 0.06 Å and D max = 68 Å, which is characteristic of a compact globular conformation, and is fairly similar to the peak (R g,p(r) = 21.93 ± 0.01 Å, D max = 70 Å) of the crystal structure of holo-HRP. The p(r) functions for the holo-HRP in both 9 M urea (Fig. 8b) and 6 M GdnHCl (Fig. 8c) appear to be quite different from that of the holo-HRP in native condition (Fig. 8a), because D max increased to 109 Å in 9 M urea and to 179 Å in 6 M GdnHCl.
We also used the program PRIMUS (Konarev et al. 2003) to estimate the Porod volume V p of holo-HRP under different conditions. The V p value increased in the order holo-HRP in native condition (66403 Å3) < holo-HRP in 9 M urea (70477 Å3) < holo-HRP in 6 M GdnHCl (152151 Å3). These V p values were directly correlated with the p(r) analysis results (Table 1).
The correlation between the obtained p(r) functions and the molecular structure was further examined using DAMMIF to reconstruct structural models ab initio from the X-ray scattering data. To improve the reliability of the final solution models, multiple runs of DAMMIF were used. For the protein, 15 independent models were selected, then DAMAVER was used to filter the averaged aligned model at given cutoff volume. The structural models reconstructed for holo-HRP were affected by the conditions (Fig. 9). The p(r) function (Fig. 8a) suggests that the conformation in the holo-HRP in native condition appeared to be close to that of the monomeric crystal structure in overall shape and dimension. For the holo-HRP in 6 M GdnHCl (Fig. 9c), the reconstructed domain appeared to be enormously extended along the vertical axis compared with that of the holo-HRP in native condition (Fig. 9a). In consideration of the Guinier, p(r), and Porod analyses results, we concluded the holo-HRP molecules were fully unfolded and linear in 6 M GdnHCl. The structural models of the holo-HRP in 9 M urea (Fig. 9b) differed from those of holo-HRP in native condition (Fig. 9a) and in 6 M GdnHCl (Fig. 9c). Its reconstructed 3D model seems to be somewhat extended in the upper and lower directions from the centre of the holo-HRP molecule (front view, Fig. 9b), when compared with the holo-HRP in native condition (Fig. 9a). This change suggests that distal and proximal domains of HRP may partially unfold, whereas the heme-binding site between both domains could maintain its tertiary structure. The B-factor was lower in the central region than in the outer region; this result means that the central region was more rigid than the outer region (Fig. 1b). The rigidity could be due to the interactions between heme and residues near the heme-binding site. Given that rigidity contributes to stability (Yu and Huang 2014), the rigid central region around the heme-binding site may confer resistance to denaturation.
Structural models of holo-HRP in solution as reconstructed using the ab initio shape determination program DAMMIF. a Holo-HRP in native condition, b holo-HRP in 9 M urea, c holo-HRP in 6 M GdnHCl. For protein, 15 independent models were generated, compared, and averaged, and then the filtered model was calculated using the program DAMAVER (mean value of NSD = 0.947 for holo-HRP in native condition, 0.837 for holo-HRP in 9 M urea, 0.974 for holo-HRP in 6 M GdnHCl). Surface rendering in the structural model was achieved using the program Discovery Studio 3.0 (Accelrys Inc.). For comparison of overall shapes and dimensions, the ribbon diagram of the atomic crystal structure of holo-HRP were superimposed on the reconstructed dummy atoms models using SUPCOMB (NSD = 1.116)
Implication of heme retention ability under denaturing conditions
Many studies have shown that some proteins can retain stable under highly denaturation conditions, such as high concentrations of urea (Cockle et al. 1978; Monhemi et al. 2014). However, the factors affecting the resistance to denaturation are largely unknown. A recent study showed that the maintenance of hydrogen bonds could be one of factors contributing to resistance to unfolding induced by urea (Wang et al. 2014).
In some heme-binding proteins, the heme cofactor also makes the protein resistant to denaturation. Previous findings showed that the presence of heme in globins conferred increased resistance to denaturation induced by chemical perturbations (Hargrove and Olson 1996). In this study, we also observed that the heme retention contributed to maintain the conformational stability of HRP under denaturing condition. This result suggests that evolution has yielded a heme-binding site in HRP that is rigid in such a way that it retains the heme, because this state confers stability in harsh conditions such as 9 M urea. The intrinsic stability of heme-binding site in HRP can give basic information that can guide search and design of peroxidases that are more stable than existing ones.
The retention of heme under denaturing conditions can be also enhanced by other factors. Heme-myoglobin affinity might be the primary determinant of the resistance of holo-myoglobin to unfolding (Hargrove and Olson 1996). Furthermore, some crowding agents, such as sucrose increase the heme stability by increasing heme retention (Kundu et al. 2015).
Enhanced heme retention ability can be useful in medicine and industry. For example, increasing heme retention ability would be useful in recombinant hemoglobin-based oxygen carriers for use as blood substitutes (Uppal et al. 2015). Therefore, combined with our data, studies on heme retention under harsh conditions would expand knowledge of how to increase heme retention for medical applications.
In conclusion, a combination of SAXS, CD, and thermal unfolding experiments indicated that under 9 M urea denaturing condition, the structure of holo-HRP unfolds only partially, and that holo-HRP maintains a considerable amount of secondary and tertiary structure; in particular, that it retains most of the tertiary structure of the central region that hosts the heme and, therefore, retains the heme. The heme group might provide the rigidity of the structure of HRP, especially the secondary helical structure around heme-binding site, thereby contributing to the conformational stability under denaturing conditions.
References
Akita M, Tsutsumi D, Kobayashi M, Kise H (2001) Structural change and catalytic activity of horseradish peroxidase in oxidative polymerization of phenol. Biosci Biotechnol Biochem 65(7):1581–1588. doi:10.1271/bbb.65.1581
Azevedo AM, Martins VC, Prazeres DM, Vojinovic V, Cabral JM, Fonseca LP (2003) Horseradish peroxidase: a valuable tool in biotechnology. Biotechnol Annu Rev 9:199–247
Carvalho AS, Melo EP, Ferreira BS, Neves-Petersen MT, Petersen SB, Aires-Barros MR (2003) Heme and pH-dependent stability of an anionic horseradish peroxidase. Arch Biochem Biophys 415(2):257–267
Cha HJ, Jang DS, Kim YG, Hong BH, Woo JS, Kim KT, Choi KY (2013) Rescue of deleterious mutations by the compensatory Y30F mutation in ketosteroid isomerase. Mol Cells 36(1):39–46. doi:10.1007/s10059-013-0013-1
Cha HJ, Jang DS, Jin KS, Lee HJ, Hong BH, Kim ES, Kim J, Lee HC, Choi KY, Lee M (2014) Three-dimensional structures of a wild-type ketosteroid isomerase and its mutant in solution. Sci Adv Mater 6(11):2325–2333
Chattopadhyay K, Mazumdar S (2000) Structural and conformational stability of horseradish peroxidase: effect of temperature and pH. Biochemistry 39(1):263–270
Cockle SA, Epand RM, Moscarello MA (1978) Resistance of lipophilin, a hydrophobic myelin protein, to denaturation by urea and guanidinium salts. J Biol Chem 253(22):8019–8026
Feng JY, Liu JZ, Ji LN (2008) Thermostability, solvent tolerance, catalytic activity and conformation of cofactor modified horseradish peroxidase. Biochimie 90(9):1337–1346. doi:10.1016/j.biochi.2008.03.010
Franke D, Svergun DI (2009) DAMMIF, a program for rapid ab initio shape determination in small-angle scattering. J Appl Crystallogr 42:342–346
Gajhede M, Schuller DJ, Henriksen A, Smith AT, Poulos TL (1997) Crystal structure of horseradish peroxidase C at 2.15 A resolution. Nat Struct Biol 4(12):1032–1038
Glatter O, Kratky O (1982) Small angle X-ray scattering. Academic Press, London
Greenfield NJ (2006) Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat Protoc 1(6):2527–2535. doi:10.1038/nprot.2006.204
Haque E, Debnath D, Basak S, Chakrabarti A (1999) Structural changes of horseradish peroxidase in presence of low concentrations of urea. Eur J Biochem 259(1–2):269–274
Hargrove MS, Olson JS (1996) The stability of holomyoglobin is determined by heme affinity. Biochemistry 35(35):11310–11318. doi:10.1021/bi9603736
Howes BD, Feis A, Raimondi L, Indiani C, Smulevich G (2001) The critical role of the proximal calcium ion in the structural properties of horseradish peroxidase. J Biol Chem 276(44):40704–40711. doi:10.1074/jbc.M107489200
Hsu MC, Woody RW (1971) The origin of the heme Cotton effects in myoglobin and hemoglobin. J Am Chem Soc 93(14):3515–3525
Jang DS, Lee HJ, Lee B, Hong BH, Cha HJ, Yoon J, Lim K, Yoon YJ, Kim J, Ree M, Lee HC, Choi KY (2006) Detection of an intermediate during the unfolding process of the dimeric ketosteroid isomerase. FEBS Lett 580(17):4166–4171. doi:10.1016/j.febslet.2006.06.069
Kelly SM, Jess TJ, Price NC (2005) How to study proteins by circular dichroism. Biochim Biophys Acta 1751(2):119–139. doi:10.1016/j.bbapap.2005.06.005
Kim ES, Jang DS, Yang SY, Lee MN, Jin KS, Cha HJ, Kim JK, Sung YC, Choi KY (2013) Controlled release of human growth hormone fused with a human hybrid Fc fragment through a nanoporous polymer membrane. Nanoscale 5(10):4262–4269. doi:10.1039/c3nr00474k
Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr 36:1277–1282
Koua D, Cerutti L, Falquet L, Sigrist CJ, Theiler G, Hulo N, Dunand C (2009) PeroxiBase: a database with new tools for peroxidase family classification. Nucleic Acids Res 37(Database issue):D261–D266. doi:10.1093/nar/gkn680
Kozin MB, Svergun DI (2001) Automated matching of high- and low-resolution structural models. J Appl Crystallogr 34:33–41
Krainer FW, Glieder A (2015) An updated view on horseradish peroxidases: recombinant production and biotechnological applications. Appl Microbiol Biotechnol 99(4):1611–1625. doi:10.1007/s00253-014-6346-7
Kundu J, Kar U, Gautam S, Karmakar S, Chowdhury PK (2015) Unusual effects of crowders on heme retention in myoglobin. FEBS Lett 589(24 Pt B):3807–3815. doi:10.1016/j.febslet.2015.11.015
Mao L, Luo S, Huang Q, Lu J (2013) Horseradish peroxidase inactivation: heme destruction and influence of polyethylene glycol. Sci Rep 3:3126. doi:10.1038/srep03126
Mogharrab N, Ghourchian H, Amininasab M (2007) Structural stabilization and functional improvement of horseradish peroxidase upon modification of accessible lysines: experiments and simulation. Biophys J 92(4):1192–1203. doi:10.1529/biophysj.106.092858
Monhemi H, Housaindokht MR, Moosavi-Movahedi AA, Bozorgmehr MR (2014) How a protein can remain stable in a solvent with high content of urea: insights from molecular dynamics simulation of Candida antarctica lipase B in urea: choline chloride deep eutectic solvent. Phys Chem Chem Phys 16(28):14882–14893. doi:10.1039/c4cp00503a
Myer YP (1968) Conformation of cytochromes. III. Effect of urea, temperature, extrinsic ligands, and pH variation on the conformation of horse heart ferricytochrome c. Biochemistry 7(2):765–776
Pappa HS, Cass AE (1993) A step towards understanding the folding mechanism of horseradish peroxidase. Tryptophan fluorescence and circular dichroism equilibrium studies. Eur J Biochem 212(1):227–235
Poulos TL (1993) Peroxidases. Curr Opin Biotechnol 4(4):484–489
Rambo RP, Tainer JA (2011) Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers 95(8):559–571. doi:10.1002/bip.21638
Ryan BJ, O’Fagain C (2007) Arginine-to-lysine substitutions influence recombinant horseradish peroxidase stability and immobilisation effectiveness. BMC Biotechnol 7:86. doi:10.1186/1472-6750-7-86
Semenyuk AV, Svergun DI (1991) GNOM—a program package for small-angle scattering data processing. J Appl Crystallogr 24:537–540
Soh YM, Burmann F, Shin HC, Oda T, Jin KS, Toseland CP, Kim C, Lee H, Kim SJ, Kong MS, Durand-Diebold ML, Kim YG, Kim HM, Lee NK, Sato M, Oh BH, Gruber S (2015) Molecular basis for SMC rod formation and its dissolution upon DNA binding. Mol Cell 57(2):290–303. doi:10.1016/j.molcel.2014.11.023
Strickland EH (1968) Circular dichroism of horseradish peroxidase and its enzyme-substrate compounds. Biochim Biophys Acta 151(1):70–75
Strickland EH, Kay E, Shannon LM, Horwitz J (1968) Peroxidase isoenzymes from horseradish roots. 3. Circular dichroism of isoenzymes and apoisoenzymes. J Biol Chem 243(13):3560–3565
Svergun D, Barberato C, Koch MHJ (1995) CRYSOL—a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr 28:768–773
Tamura M, Asakura T, Yonetani T (1972) Heme-modification studies on horseradish peroxidase. Biochim Biophys Acta 268(2):292–304
Tanford C (1968) Protein denaturation. Adv Protein Chem 23:121–282
Uppal S, Salhotra S, Mukhi N, Zaidi FK, Seal M, Dey SG, Bhat R, Kundu S (2015) Significantly enhanced heme retention ability of myoglobin engineered to mimic the third covalent linkage by nonaxial histidine to heme (vinyl) in synechocystis hemoglobin. J Biol Chem 290(4):1979–1993. doi:10.1074/jbc.M114.603225
Veitch NC (2004) Horseradish peroxidase: a modern view of a classic enzyme. Phytochemistry 65(3):249–259
Volkov VV, Svergun DI (2003) Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr 36:860–864
Wagner M, Nicell JA (2002) Detoxification of phenolic solutions with horseradish peroxidase and hydrogen peroxide. Water Res 36(16):4041–4052
Wang C, Chen Z, Hong X, Ning F, Liu H, Zang J, Yan X, Kemp J, Musselman CA, Kutateladze TG, Zhao R, Jiang C, Zhang G (2014) The structural basis of urea-induced protein unfolding in beta-catenin. Acta Crystallogr D Biol Crystallogr 70(Pt 11):2840–2847. doi:10.1107/S1399004714018094
Won K, Kim YH, An ES, Lee YS, Song BK (2004) Horseradish peroxidase-catalyzed polymerization of cardanol in the presence of redox mediators. Biomacromolecules 5(1):1–4. doi:10.1021/bm034325u
Yonetani T (1967) Studies on cytochrome c peroxidase. X. Crystalline apo-and reconstituted holoenzymes. J Biol Chem 242(21):5008–5013
Yu H, Huang H (2014) Engineering proteins for thermostability through rigidifying flexible sites. Biotechnol Adv 32(2):308–315. doi:10.1016/j.biotechadv.2013.10.012
Zakharova GS, Uporov IV, Tishkov VI (2011) Horseradish peroxidase: modulation of properties by chemical modification of protein and heme. Biochemistry (Mosc) 76(13):1391–1401. doi:10.1134/S0006297911130037
Acknowledgements
This research was supported by a Grant from the National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) (2014R1A2A2A01002931) and the Next-Generation BioGreen 21 Program, Rural Development Administration, Republic of Korea (PJ01121601). This was also funded by the industrial research project from LG Chem.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: S. Dai.
H. J. Cha and D. S. Jang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Cha, H.J., Jang, D.S., Jin, K.S. et al. Structural analyses combined with small-angle X-ray scattering reveals that the retention of heme is critical for maintaining the structure of horseradish peroxidase under denaturing conditions. Amino Acids 49, 715–723 (2017). https://doi.org/10.1007/s00726-016-2372-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-016-2372-3