Abstract
Genome-wide association study indicates that STAT4 is a plausible candidate for an association study with HBV-related liver diseases. We aimed to examine the roles of STAT4 polymorphisms on HBV-related liver diseases in a Chinese Han population. We selected 13 SNPs in STAT4 based on the HapMap database to investigate their associations in 3,033 participants. SNP rs7574865 was significantly associated with HBV infection [odds ratio (OR) 1.15; 95 % confidence interval (CI) 1.00, 1.31; P = 0.046] and clearance (OR 1.17; 95 % CI 1.02, 1.33; P = 0.028). Further, haplotype-based association analysis indicated that the haplotype CTCTT, formed by SNPs rs8179673, rs7574865, rs4274624, rs11889341, and rs10168266, was significantly associated with HBV infection (OR 0.87; 95 % CI 0.76, 0.99; P = 0.022) and clearance (OR 0.86; 95 % CI 0.75, 0.99; P = 0.018). Bioinformatics analysis of these SNPs predicted that they participate in transcriptional regulation. Taken together, our findings demonstrate that variants in STAT4 play a critical role in HBV infection and clearance in the Chinese Han population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Infection with HBV is a major public health issue throughout the world, especially in Asian countries (Schweitzer et al. 2015). Despite the effective use of the potent HBV vaccine and antiviral treatments, HBV infection maintains a high prevalence worldwide, with an estimated 240 million people affected chronically in 2015 (World Health Organization 2015; http://www.who.int/mediacentre/factsheets/fs204/en/), and is the world’s tenth leading cause of death (Trepo et al. 2014). Approximately 60 % of hepatocellular carcinoma (HCC) is associated with chronic hepatitis B (CHB), and the infection is responsible for many cases of liver cirrhosis (LC) (Lai et al. 2003; Perz et al. 2006).
Infection with HBV and its progression to LC and HCC are influenced by viral, environmental, and host factors (Thursz 2001). Genome-wide association studies (GWAS) have been recently used increasingly to identify genetic variants in HBV-related liver diseases, which revealed several novel common variants located in ubiquitin-conjugating enzyme E2L3 (Hu et al. 2013), solute carrier family 10 member 1 (Peng et al. 2015), glutamate receptor ionotropic kainate 1 (Li et al. 2012), kinesin family member 1B (Li et al. 2012), and signal transducer and activator of transcription 4 (STAT4) (Jiang et al. 2013). However, many of these newly identified susceptibility gene findings have not been replicated in independent studies.
STAT4 is located on chromosome 2q32.3 and encodes a crucial transcription factor (Zhong et al. 1994). Several cytokines, such as interleukin (IL)-2 (Wang et al. 1999), IL-12 (Bacon et al. 1995) and interferon (IFN)-α/β (Nguyen et al. 2002), are reported to activate STAT4 for transmitting signals to its downstream target genes. In natural killer (NK) and T cells, STAT4 is phosphorylated predominately by the binding of IL-12 and its receptor. Then the phosphorylated STAT4 stimulates NK and T cells to produce IFN-γ, an important cytokine for antiviral replication and antitumor activity (Wang et al. 2015). Moreover, it has been reported that STAT4 plays critical roles in other viral infections, such as respiratory syncytial virus (Dulek et al. 2014), herpes simplex virus (HSV) 2 (Svensson et al. 2012), and lymphocytic choriomeningitis virus (Holz et al. 1999). A recent GWAS found that SNP rs7574865 in STAT4 was associated with HBV infection and HBV-related HCC (Jiang et al. 2013), but this association has not been replicated yet in any other Chinese Han population.
To further investigate the association of STAT4 polymorphisms with HBV-related liver diseases, we performed an association study of single and multiple SNPs within STAT4 in a large Chinese Han sample.
Materials and methods
Subject recruitment and demographic characteristics
This case–control study included 3033 Chinese Han individuals comprising 1610 with chronic HBV infection (CHBVI) and 1423 uninfected control subjects (Table 1). A detailed description of this sample has been reported previously (Tao et al. 2015).
Based on the laboratory testing results and the “Guideline of Prevention and Treatment for Chronic Hepatitis B” defined by the Chinese Medical Association (Chinese Society of and Chinese Society of Infectious Diseases 2011), 1610 CHBVI patients could be identified as asymptomatic carriers (AsC; n = 388), CHB (n = 853), LC (n = 296), or HCC (n = 73). The diagnostic criteria for these HBV-related liver diseases were as follows: (1) AsC: seropositive for either HBsAg or HBV-DNA and persistently normal ALT; (2) CHB: seropositive for either HBsAg or HBV-DNA and recurrently abnormal or continuously elevated ALT for >6 months; (3) LC: presence of ascites, hypersplenism, elevated ALT, and appropriate imaging findings; (4) HCC: abnormal serum alpha-fetoprotein and imaging findings. The 1423 control subjects were classified as healthy (n = 749) or HBV clearance (n = 674). The definition of healthy subjects and clearance was: (1) healthy: negative for HBsAg, HBcAb, and HBsAb; and (2) clearance: negative for HBsAg but positive for HBcAb and HBsAb.
Subjects were excluded if they were: (1) co-infected with any other hepatitis virus; (2) suffered from any other type of cancer or autoimmune disease; (3) positive for human immunodeficiency virus; (4) effectively immunized against HBV; (5) not Chinese Han ethnicity; or (6) pregnant.
DNA extraction, SNP selection and genotyping, and quality control
Genomic DNA was extracted from 3 ml of peripheral blood of each subject using the Qiagen DNA purification kit. Selection of 13 SNPs in STAT4 was based on the HapMap database (Release #27) with a minor allele frequency (MAF) >0.1 in the Chinese population (Fig. 1; Table S1). Of the 13 selected variants, SNPs rs3821236, rs1517352, rs10168266, rs11889341, rs4274624, rs7574865, rs8179673, rs11685878, rs7572482, and rs897200 have been reported to be associated with autoimmune or viral disease, including systemic lupus erythematosus (SLE) (Kawasaki et al. 2008), inflammatory bowel disease (Jostins et al. 2012), primary biliary cirrhosis (PBC) (Joshita et al. 2014), autoimmune Addison’s disease (Mitchell et al. 2014), Behcet’s disease (Hou et al. 2012), HBV-related HCC (Jiang et al. 2013), or HSV-2 infection (Svensson et al. 2012).
All SNPs included in the study were genotyped in a 384-well microplate format using the TaqMan SNP Genotyping Assay (Applied Biosystems, Foster City, CA, USA). The genotyping rate for each SNP was required to be >95 %, and the P value for the Hardy–Weinberg equilibrium was set to >0.05.
Literature search for meta-analyses on the association of rs7574865 with HBV infection and clearance
Our meta-analysis was performed according to the criteria suggested in the PRISMA guidelines (Moher et al. 2009). We searched PubMed and Web of Science for all eligible studies that could be included in this study. The key words used in the search strategy were: “rs7574865” or “STAT4” or “signal transducer and activator of transcription 4” and “hepatitis B” or “hepatitis B virus” or “HBV”. The identified reports were checked individually for reference to studies that were not found in the abovementioned electronic databases. When different samples of the same population were included in a study, each one was treated as an independent one in this report. For each study, the following data were extracted: author, publication year, ethnicity, study design, genotyping method, sample size, mean age, and sex ratio.
The inclusion criteria for the meta-analysis were that: (1) the study was designed as a case–control study; (2) the study provided the number of CHBVI patients, healthy individuals, and subjects with HBV clearance, and the allele frequency of rs7574865; (3) the study presented sufficient information for calculating the odds ratio (OR) and the 95 % confidence interval (CI).
Statistical analysis
The population admixture of subjects was assessed by principal component (PC) analysis using 291 ancestry informative markers (AIMs) selected from the Illumina arrays. For individual SNP-based association analysis, the association of STAT4 polymorphisms with HBV-related liver diseases was performed under an additive genetic model using PLINK (v. 1.07) (Purcell et al. 2007). The ORs and 95 % CIs were calculated by logistic regression with age, sex, and the first seven PCs as covariates. After Bonferroni correction for multiple tests, the appropriate significance of individual SNP was set to P = 3.8 × 10−3 (0.05/13). Pairwise linkage disequilibrium (LD) was determined by the D′ value, and haplotype blocks were generated by the Haploview software (v. 4.2) (Barrett et al. 2005). The association of each haplotype with HBV-related liver diseases was analyzed using R package “haplo.stats” (http://cran.r-project.org/web/packages/haplo.stats; Yang and Li 2014). The P value was given by the score test under the same genetic model and covariates as used in the individual SNP-based association analysis (Schaid et al. 2002). The significance of each haplotype was adjusted by the number of major haplotypes (frequency >5 %) included in the analysis. For the SNPs within the significant haplotypes, variant effects on the regulation of transcription were analyzed using HaploReg (v. 2) (http://www.broadinstitute.org/mammals/haploreg/haploreg_v2.php). In this database, evolutionarily conserved regions are predicted by genome evolutionary rate profiling (GERP) (Ward and Kellis 2012), the enhancer histone mark prediction was based on the evidence of local H3K4Me1 modification, and variant effects on regulatory motifs were evaluated according to TRANSFAC, JASPAR and protein-binding microarray experiments (Ward and Kellis 2012).
Meta-analysis was performed to assess the association of rs7574865 with HBV infection and clearance using Comprehensive Meta-Analysis software (v. 2.0; Biostat Inc., Englewood, NJ, USA, as we described previously (Ma et al. 2015a, b)). Heterogeneity among the studies was assessed by Cochran’s Q and I 2 statistic tests (Higgins et al. 2003). If the P value of the Q test is <0.1, indicating heterogeneity among the studies, the random-effects model is more applicable. Otherwise, use of the fixed-effects model is encouraged. The I 2 test is used to analyze the effect of heterogeneity and the degree of inconsistency among the studies. If Cochran’s Q P value is >0.1 and the I 2 value is ≤50 %, we consider that there exists no strong evidence for heterogeneity (Higgins et al. 2003; Taylor et al. 2015). In this work, both the random-effects and fixed-effects models were used in the meta-analysis. The Z statistical test was used to calculate the significance of pooled ORs. A funnel plot was applied to assess potential publication bias. Sensitivity analysis was performed to detect the stability of results. The significance level was set to P < 0.05 (two-sided) for meta-analysis.
Results
Association analysis of STAT4 polymorphisms with HBV infection and clearance
To identify the association of STAT4 polymorphisms with HBV infection and clearance, we compared individual SNP-based association analysis of 13 SNPs in 1610 CHBVI patients vs. 749 healthy subjects and 1610 CHBVI patients vs. 674 clearance subjects under an additive genetic model after adjusting for age, sex, and the first seven PCs (Table 2).
We found that SNP rs7574865 was significantly associated with HBV infection risk (OR 1.15; 95 % CI 1.00, 1.31; P = 0.046), providing an independent replication of a previous report (Jiang et al. 2013). Furthermore, this SNP was negatively correlated with HBV clearance (OR 1.17; 95 % CI 1.02, 1.33; P = 0.028). That is, rs7574865 plays a role in HBV susceptibility, as well as a retarding role in HBV clearance.
An additional four SNPs showed a nominally significant correlation with HBV infection and clearance (Table 2). When comparing CHBVI with the healthy group, we obtained the OR and its corresponding P value for SNPs rs10168266 (OR 1.19; P = 0.013), rs11889341 (OR 1.18; P = 0.018), rs4274624 (OR 1.13; P = 0.056), and rs8179673 (OR 1.14; P = 0.047). As for the HBV clearance, the ORs and their corresponding P values were as follows: rs10168266 OR 1.15; P = 0.043; rs11889341 OR 1.15; P = 0.041; rs4274624 OR 1.15; P = 0.050; and rs8179673 OR 1.15; P = 0.040. Thus, all of the SNPs showed a risk effect on HBV infection and clearance.
Association analysis of STAT4 polymorphisms with disease progression
To reveal the relation of STAT4 variants to the progression of HBV-related liver diseases, we performed the following three comparisons: (1) AsC vs. CHB; (2) CHB vs. LC; and (3) LC vs. HCC. We found no significant association of the STAT4 variants with disease progression after adjusting for age, sex, and the first seven PCs (detailed results not shown).
Haplotype analysis of STAT4 polymorphisms with HBV infection and clearance
Haplotype-based association analysis was performed for all SNPs in STAT4 regardless of whether they were significant or non-significant according to the individual association analysis. One haplotype, CTCTT, formed by rs8179673, rs7574865, rs4274624, rs11889341, and rs10168266, showed a significant association with HBV infection and clearance (Table 3; Fig. 2). When comparing the CHBVI with the healthy group, the haplotype CTCTT was significantly associated with HBV infection (Hap Score = −2.290; P = 0.022). Similarly, the haplotype CTCTT was significantly associated with HBV clearance when comparing the CHBVI with the clearance group (Hap Score = −2.370; P = 0.018). Together, these results indicate that the haplotype formed by the minor alleles of the abovementioned SNPs is protective against HBV susceptibility and beneficial for HBV clearance.
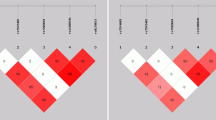
LD maps and haplotype blocks of STAT4 generated by Haploview software (v. 4.2). The LD map was determined by the D′ value (D′ >0.9), which was drawn from the genotypic data of the CHBVI and healthy subjects (a) and the genotypic data of the CHBVI and clearance subjects (b). LD linkage disequilibrium, CHBVI chronic HBV infection (includes AsC, CHB, LC, and HCC), AsC asymptomatic carrier, CHB chronic hepatitis B, LC liver cirrhosis, HCC hepatocellular carcinoma
Meta-analysis of association of rs7574865 with HBV infection and clearance
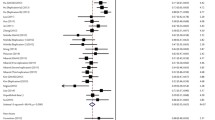
To determine the effects of rs7574865 on HBV infection and clearance with a large sample, we performed meta-analysis for those reported studies along with the current one. Based on this search strategy, we included 10 reported studies and one unpublished data set totaling 8944 CHBVI patients, 8451 healthy individuals, and 2081 subjects with HBV clearance (Table S2). Under the fixed-effects model, rs7574865 was significantly associated with HBV infection (pooled OR 1.14; 95 % CI 1.07, 1.21; pooled P = 3.8 × 10−5; Fig. 3a) and clearance (OR 1.20; 95 %CI 1.07, 1.35; P = 0.002; Fig. 4a). Similarly, we performed a meta-analysis under the random-effects model, which again revealed rs7574865 to be significantly associated with HBV infection (pooled OR 1.14; 95 % CI 1.07, 1.21; pooled P = 3.8 × 10−5; Fig. 3b) and clearance (pooled OR 1.25; 95 % CI 1.01, 1.55; pooled P = 0.044; Fig. 4b). Considering that we found no significant evidence for the presence of heterogeneity among the studies for the meta-analysis of HBV infection (P = 0.82; I 2 = 0) and clearance (P = 0.19; I 2 = 34 %), we believed the results from either model are acceptable, and both models provide further evidence for the significant association of rs7572865 with HBV infection and clearance. Next, we performed sensitivity analysis by sequentially omitting individual studies under the fixed-effects model and found that the P value remained significant for all cases (data not shown).
Forest plots of meta-analysis results for the association of rs7574865 with HBV infection in the CHBVI and healthy groups under the fixed-effects model (a) and the random-effects model (b). The Z and P values of each study are shown by rows. The central vertical solid line represents ORs that equal 1 under the null hypothesis. The ORs and 95 % CIs are represented by the square and horizontal bars, respectively. The pooled OR is indicated by the diamond
Forest plots of meta-analysis results for the association of rs7574865 with HBV clearance in the CHBVI and clearance groups under the fixed-effects model (a) and the random-effects model (b). The Z and P values of each study are shown by rows. The central vertical solid line represents ORs that equal 1 under the null hypothesis. The ORs and 95 % CIs are represented by the square and horizontal bars, respectively. The pooled OR is indicated by the diamond symbol
Function prediction analysis of SNPs in STAT4
We applied the HaploReg tool (Ward and Kellis 2012) to predict the function of the five SNPs in the significant haplotype CTCTT (Table S3). Of the five, SNP rs7574865 was predicted to be conserved based on the GERP; SNP rs11889341 was associated with histone modifications in B cells from peripheral blood that were suggestive of enhancer activity. In addition, four SNPs (rs10168266, rs11889341, rs7574865, and rs8179673) were predicted to affect specific regulatory motifs such as Foxp1, Foxo3, HNF1, and HNF4. These predictions indicated that the SNPs have important effects on transcription by influencing the binding of transcription factors or regulatory motifs.
Discussion
A recent GWAS study identified STAT4 as a susceptibility gene for HBV infection and HBV-related HCC in the Chinese Han population (Jiang et al. 2013). However, Liao et al. failed to validate such an association using 1312 Chinese Han individuals (Liao et al. 2014). To determine whether this gene indeed plays any role in HBV-related liver diseases, we carried out this study with a much larger sample.
In individual SNP-based association analysis under the additive genetic model, SNP rs7574865 in STAT4 not only increased the probability of HBV infection (OR = 1.15; P = 0.046) but also reduced the probability of viral clearance (OR = 1.17; P = 0.028). The meta-analysis of the data on this SNP, incorporating all the data from 8944 CHBVI patients, 8451 healthy and 2081 HBV clearance individuals, also indicated SNP rs7574865 to be a risk factor for HBV infection and an inhibitor of clearance. During the preparation of this manuscript, Lu et al. (2015) reported that rs7574865 is related to HBV clearance with 596 subjects, whose result is consistent with ours. It has also been reported that rs7574865/G was significantly associated with lower mRNA values for STAT4 (Jiang et al. 2013). Because STAT4 was able to bind to the first intron of interferon-gamma, higher STAT4 expression was expected to enhance the activation of IFN-γ, a soluble cytokine with important effects on modulating antiviral activity and immunoregulatory functions (Morinobu et al. 2002), leading to the increased ability for HBV clearance and resistance to viral infection. Also, we found SNPs rs10168266, rs11889341, rs4274624, and rs8179673 to be nominally associated with HBV infection and viral clearance. It has been reported that SNPs rs10168266, rs11889341, and rs8179673 are significantly correlated with PBC and SLE susceptibility (Joshita et al. 2014; Kawasaki et al. 2008); and SNP rs4274624 is related to susceptibility to autoimmune Addison’s disease (Mitchell et al. 2014). These results suggest that STAT4 plays dual roles in autoimmune disease and viral infection.
We also investigated the roles of STAT4 polymorphisms in each stage of liver disease progression. None of the SNPs showed a significant association, which indicates that STAT4 plays different roles at different stages of HBV infection. STAT4 protects against HBV infection and increases HBV clearance, but it may play no significant role in disease progression. However, in a recent comparison of 712 LC patients and 2601 HBV-positive controls, Jiang et al. (2015) found that rs7574865 was significantly associated with disease progression, which is inconsistent with our findings. This discrepancy might be caused by a relatively small sample in a few HBV-related liver disease categories, and a large sample is required for future study.
Further, we performed haplotype-based association analysis and found a high protective CTCTT haplotype formed by rs8179673-rs7574865-rs4274624-rs11889341-rs10168266, which showed significant activity against HBV infection (Hap Score = −2.290; P = 0.022) and increased the probability of viral clearance (Hap Score = −2.370; P = 0.018). This suggested that these five SNPs jointly contribute to HBV infection and clearance, although they were not significantly associated with HBV-related liver diseases after Bonferroni correction based on individual SNP-based association analysis.
We then evaluated the potential biological functions of these five SNPs located in noncoding regions using the HaploReg tool (Ward and Kellis 2012). It predicted that SNP rs11889341 influences the enhancer activity in B cells on the basis of its location with a histone modification. Additionally, regulatory motif prediction suggested that SNPs rs10168266, rs11889341, rs7574865, and rs8179673 affect the affinity for binding of transcription factors. Many transcription factors are associated with viral infection and liver diseases. For rs11889341, the alternate allele increases affinity for Foxo3, which regulates CD8 T cell responses during chronic viral infection (Sullivan et al. 2012). For rs7574865, the alternate allele is predicted to have a lower affinity for Foxp1, which is reported to be associated with the risk of LC progression to HBV-related HCC (Li et al. 2015). We suspect that rs7574865 influences oncogenesis by changing the affinity for Foxp1. For rs8179673, its alternate allele is predicted to have lower binding of transcription factors HNF1 and HNF4 to STAT4. HNF1α repression can lead to activation of JAK/STAT pathways and facilitate the progression of chronic liver diseases (Qian et al. 2015), and HNF4 also plays an important role in HBV replication (Shin et al. 2014). However, it remains to be determined how these five SNPs contribute to HBV infection and clearance by affecting STAT4 transcriptional regulation.
In conclusion, this study confirmed the association of STAT4 polymorphisms with HBV susceptibility and viral clearance in a much larger sample than previously studied. In addition, haplotype-based association analysis revealed a highly protective CTCTT haplotype, which is also significantly associated with HBV infection and clearance. Finally, functional prediction analysis using the HaploReg tool indicated that these five SNPs likely play critical roles in the regulation of transcription of the STAT4 gene. However, further functional study is required to validate these predictions.
Abbreviations
- HBV:
-
Hepatitis B virus
- HCC:
-
Hepatocellular carcinoma
- CHB:
-
Chronic hepatitis B
- LC:
-
Liver cirrhosis
- STAT4:
-
Signal transducer and activator of transcription 4
- HBsAg:
-
Hepatitis B surface antigen
- HBsAb:
-
Hepatitis B surface antibody
- HBeAg:
-
Hepatitis B e antigen
- HBeAb:
-
Hepatitis B e antibody
- HBcAb:
-
Hepatitis B core antibody
- HCsAb:
-
Hepatitis C surface antibody
- ALT:
-
Serum alanine aminotransferase
- AsC:
-
Asymptomatic carriers
References
Bacon CM, Petricoin EF 3rd, Ortaldo JR, Rees RC, Larner AC, Johnston JA, O’Shea JJ (1995) Interleukin 12 induces tyrosine phosphorylation and activation of STAT4 in human lymphocytes. Proc Natl Acad Sci USA 92(16):7307–7311
Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21(2):263–265. doi:10.1093/bioinformatics/bth457
Chinese Society of H, Chinese Society of Infectious Diseases CMA (2011) The guideline of prevention and treatment for chronic hepatitis B (2010 version). Zhonghua liu xing bing xue za zhi Zhonghua liuxingbingxue zazhi 32(4):405–415
Dulek DE, Newcomb DC, Toki S, Goliniewska K, Cephus J, Reiss S, Bates JT, Crowe JE Jr, Boyd KL, Moore ML, Zhou W, Peebles RS Jr (2014) STAT4 deficiency fails to induce lung Th2 or Th17 immunity following primary or secondary respiratory syncytial virus (RSV) challenge but enhances the lung RSV-specific CD8+ T cell immune response to secondary challenge. J Virol 88(17):9655–9672. doi:10.1128/JVI.03299-13
Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. Bmj 327(7414):557–560. doi:10.1136/bmj.327.7414.557
Holz A, Bot A, Coon B, Wolfe T, Grusby MJ, von Herrath MG (1999) Disruption of the STAT4 signaling pathway protects from autoimmune diabetes while retaining antiviral immune competence. J Immunol 163(10):5374–5382
Hou S, Yang Z, Du L, Jiang Z, Shu Q, Chen Y, Li F, Zhou Q, Ohno S, Chen R, Kijlstra A, Rosenbaum JT, Yang P (2012) Identification of a susceptibility locus in STAT4 for Behcet’s disease in Han Chinese in a genome-wide association study. Arthritis Rheum 64(12):4104–4113. doi:10.1002/art.37708
Hu Z, Liu Y, Zhai X, Dai J, Jin G, Wang L, Zhu L, Yang Y, Liu J, Chu M, Wen J, Xie K, Du G, Wang Q, Zhou Y, Cao M, Liu L, He Y, Wang Y, Zhou G, Jia W, Lu J, Li S, Liu J, Yang H, Shi Y, Zhou W, Shen H (2013) New loci associated with chronic hepatitis B virus infection in Han Chinese. Nat Genet 45(12):1499–1503. doi:10.1038/ng.2809
Jiang DK, Sun J, Cao G, Liu Y, Lin D, Gao YZ, Ren WH, Long XD, Zhang H, Ma XP, Wang Z, Jiang W, Chen TY, Gao Y, Sun LD, Long JR, Huang HX, Wang D, Yu H, Zhang P, Tang LS, Peng B, Cai H, Liu TT, Zhou P, Liu F, Lin X, Tao S, Wan B, Sai-Yin HX, Qin LX, Yin J, Liu L, Wu C, Pei Y, Zhou YF, Zhai Y, Lu PX, Tan A, Zuo XB, Fan J, Chang J, Gu X, Wang NJ, Li Y, Liu YK, Zhai K, Zhang H, Hu Z, Liu J, Yi Q, Xiang Y, Shi R, Ding Q, Zheng W, Shu XO, Mo Z, Shugart YY, Zhang XJ, Zhou G, Shen H, Zheng SL, Xu J, Yu L (2013) Genetic variants in STAT4 and HLA-DQ genes confer risk of hepatitis B virus-related hepatocellular carcinoma. Nat Genet 45(1):72–75. doi:10.1038/ng.2483
Jiang DK, Ma XP, Wu X, Peng L, Yin J, Dan Y, Huang HX, Ding DL, Zhang LY, Shi Z, Zhang P, Yu H, Sun J, Lilly Zheng S, Deng G, Xu J, Liu Y, Guo J, Cao G, Yu L (2015) Genetic variations in STAT4, C2, HLA-DRB1 and HLA-DQ associated with risk of hepatitis B virus-related liver cirrhosis. Sci Rep 5:16278. doi:10.1038/srep16278
Joshita S, Umemura T, Nakamura M, Katsuyama Y, Shibata S, Kimura T, Morita S, Komatsu M, Matsumoto A, Yoshizawa K, Ishibashi H, Tanaka E, Ota M (2014) STAT4 gene polymorphisms are associated with susceptibility and ANA status in primary biliary cirrhosis. Dis Markers 2014:727393. doi:10.1155/2014/727393
Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Buning C, Cohain A, Cichon S, D’Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, Hawkey C, Hedl M, Hu X, Karlsen TH, Kupcinskas L, Kugathasan S, Latiano A, Laukens D, Lawrance IC, Lees CW, Louis E, Mahy G, Mansfield J, Morgan AR, Mowat C, Newman W, Palmieri O, Ponsioen CY, Potocnik U, Prescott NJ, Regueiro M, Rotter JI, Russell RK, Sanderson JD, Sans M, Satsangi J, Schreiber S, Simms LA, Sventoraityte J, Targan SR, Taylor KD, Tremelling M, Verspaget HW, De Vos M, Wijmenga C, Wilson DC, Winkelmann J, Xavier RJ, Zeissig S, Zhang B, Zhang CK, Zhao H, International IBDGC, Silverberg MS, Annese V, Hakonarson H, Brant SR, Radford-Smith G, Mathew CG, Rioux JD, Schadt EE, Daly MJ, Franke A, Parkes M, Vermeire S, Barrett JC, Cho JH (2012) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491(7422):119–124. doi:10.1038/nature11582
Kawasaki A, Ito I, Hikami K, Ohashi J, Hayashi T, Goto D, Matsumoto I, Ito S, Tsutsumi A, Koga M, Arinami T, Graham RR, Hom G, Takasaki Y, Hashimoto H, Behrens TW, Sumida T, Tsuchiya N (2008) Role of STAT4 polymorphisms in systemic lupus erythematosus in a Japanese population: a case-control association study of the STAT1-STAT4 region. Arthritis Res Therapy 10(5):R113. doi:10.1186/ar2516
Lai CL, Ratziu V, Yuen MF, Poynard T (2003) Viral hepatitis B. Lancet 362(9401):2089–2094. doi:10.1016/S0140-6736(03)15108-2
Li S, Qian J, Yang Y, Zhao W, Dai J, Bei JX, Foo JN, McLaren PJ, Li Z, Yang J, Shen F, Liu L, Yang J, Li S, Pan S, Wang Y, Li W, Zhai X, Zhou B, Shi L, Chen X, Chu M, Yan Y, Wang J, Cheng S, Shen J, Jia W, Liu J, Yang J, Wen Z, Li A, Zhang Y, Zhang G, Luo X, Qin H, Chen M, Wang H, Jin L, Lin D, Shen H, He L, de Bakker PI, Wang H, Zeng YX, Wu M, Hu Z, Shi Y, Liu J, Zhou W (2012) GWAS identifies novel susceptibility loci on 6p21.32 and 21q21.3 for hepatocellular carcinoma in chronic hepatitis B virus carriers. PLoS Genet 8(7):1002791. doi:10.1371/journal.pgen.1002791
Li F, Liu T, Xiao CY, Yu JX, Lu LG, Xu MY (2015) FOXP1 and SPINK1 reflect the risk of cirrhosis progression to HCC with HBV infection. Biomed Pharmacother Biomedecine Pharmacotherapie 72:103–108. doi:10.1016/j.biopha.2015.04.006
Liao Y, Cai B, Li Y, Chen J, Tao C, Huang H, Wang L (2014) Association of HLA-DP/DQ and STAT4 polymorphisms with HBV infection outcomes and a mini meta-analysis. PLoS One 9(11):e111677. doi:10.1371/journal.pone.0111677
Lu Y, Zhu Y, Peng J, Wang X, Wang F, Sun Z (2015) STAT4 genetic polymorphisms association with spontaneous clearance of hepatitis B virus infection. Immunol Res 62(2):146–152. doi:10.1007/s12026-015-8645-1
Ma Y, Wang M, Yuan W, Su K, Li MD (2015a) The significant association of Taq1A genotypes in DRD2/ANKK1 with smoking cessation in a large-scale meta-analysis of Caucasian populations. Transl Psychiatry 5:e686. doi:10.1038/tp.2015.176
Ma Y, Yuan W, Cui W, Li MD (2015b) Meta-analysis reveals significant association of 3′-UTR VNTR in SLC6A3 with smoking cessation in Caucasian populations. Pharmacogenomics J. doi:10.1038/tpj.2015.44
Mitchell AL, Macarthur KD, Gan EH, Baggott LE, Wolff AS, Skinningsrud B, Platt H, Short A, Lobell A, Kampe O, Bensing S, Betterle C, Kasperlik-Zaluska A, Zurawek M, Fichna M, Kockum I, Nordling Eriksson G, Ekwall O, Wahlberg J, Dahlqvist P, Hulting AL, Penna-Martinez M, Meyer G, Kahles H, Badenhoop K, Hahner S, Quinkler M, Falorni A, Phipps-Green A, Merriman TR, Ollier W, Cordell HJ, Undlien D, Czarnocka B, Husebye E, Pearce SH (2014) Association of autoimmune Addison’s disease with alleles of STAT4 and GATA3 in European cohorts. PLoS One 9(3):e88991. doi:10.1371/journal.pone.0088991
Moher D, Liberati A, Tetzlaff J, Altman DG, Group P (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med 151(4):264–269 (W264)
Morinobu A, Gadina M, Strober W, Visconti R, Fornace A, Montagna C, Feldman GM, Nishikomori R, O’Shea JJ (2002) STAT4 serine phosphorylation is critical for IL-12-induced IFN-gamma production but not for cell proliferation. Proc Natl Acad Sci USA 99(19):12281–12286. doi:10.1073/pnas.182618999
Nguyen KB, Watford WT, Salomon R, Hofmann SR, Pien GC, Morinobu A, Gadina M, O’Shea JJ, Biron CA (2002) Critical role for STAT4 activation by type 1 interferons in the interferon-gamma response to viral infection. Science 297(5589):2063–2066. doi:10.1126/science.1074900
Peng L, Zhao Q, Li Q, Li M, Li C, Xu T, Jing X, Zhu X, Wang Y, Li F, Liu R, Zhong C, Pan Q, Zeng B, Liao Q, Hu B, Hu ZX, Huang YS, Sham P, Liu J, Xu S, Wang J, Gao ZL, Wang Y (2015) The p.Ser267Phe variant in SLC10A1 is associated with resistance to chronic hepatitis B. Hepatology 61(4):1251–1260. doi:10.1002/hep.27608
Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP (2006) The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol 45(4):529–538. doi:10.1016/j.jhep.2006.05.013
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81(3):559–575. doi:10.1086/519795
Qian H, Deng X, Huang ZW, Wei J, Ding CH, Feng RX, Zeng X, Chen YX, Ding J, Qiu L, Hu ZL, Zhang X, Wang HY, Zhang JP, Xie WF (2015) An HNF1alpha-regulated feedback circuit modulates hepatic fibrogenesis via the crosstalk between hepatocytes and hepatic stellate cells. Cell Res 25(8):930–945. doi:10.1038/cr.2015.84
Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA (2002) Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet 70(2):425–434. doi:10.1086/338688
Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ (2015) Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet 386(10003):1546–1555. doi:10.1016/S0140-6736(15)61412-X
Shin GC, Ahn SH, Choi HS, Kim J, Park ES, Kim DH, Kim KH (2014) Hepatocystin contributes to interferon-mediated antiviral response to hepatitis B virus by regulating hepatocyte nuclear factor 4alpha. Biochim Biophys Acta 1842(9):1648–1657. doi:10.1016/j.bbadis.2014.04.016
Sullivan JA, Kim EH, Plisch EH, Suresh M (2012) FOXO3 regulates the CD8 T cell response to a chronic viral infection. J Virol 86(17):9025–9034. doi:10.1128/JVI.00942-12
Svensson A, Tunback P, Nordstrom I, Shestakov A, Padyukov L, Eriksson K (2012) STAT4 regulates antiviral gamma interferon responses and recurrent disease during herpes simplex virus 2 infection. J Virol 86(17):9409–9415. doi:10.1128/JVI.00947-12
Tao J, Su K, Yu C, Liu X, Wu W, Xu W, Jiang B, Luo R, Yao J, Zhou J, Zhan Y, Ye C, Yuan W, Jiang X, Cui W, Li MD, Li L (2015) Fine mapping analysis of HLA-DP/DQ gene clusters on chromosome 6 reveals multiple susceptibility loci for HBV infection. Amino Acids. 47(12):2623–2634. doi:10.1007/s00726-015-2054-6
Taylor PN, Porcu E, Chew S, Campbell PJ, Traglia M, Brown SJ, Mullin BH, Shihab HA, Min J, Walter K, Memari Y, Huang J, Barnes MR, Beilby JP, Charoen P, Danecek P, Dudbridge F, Forgetta V, Greenwood C, Grundberg E, Johnson AD, Hui J, Lim EM, McCarthy S, Muddyman D, Panicker V, Perry JR, Bell JT, Yuan W, Relton C, Gaunt T, Schlessinger D, Abecasis G, Cucca F, Surdulescu GL, Woltersdorf W, Zeggini E, Zheng HF, Toniolo D, Dayan CM, Naitza S, Walsh JP, Spector T, Davey-Smith G, Durbin R, Richards JB, Sanna S, Soranzo N, Timpson NJ, Wilson SG, Consortium UK (2015) Whole-genome sequence-based analysis of thyroid function. Nat Commun 6:5681. doi:10.1038/ncomms6681
Thursz M (2001) Genetic susceptibility in chronic viral hepatitis. Antiviral Res 52(2):113–116
Trepo C, Chan HL, Lok A (2014) Hepatitis B virus infection. Lancet 384(9959):2053–2063. doi:10.1016/S0140-6736(14)60220-8
Wang KS, Ritz J, Frank DA (1999) IL-2 induces STAT4 activation in primary NK cells and NK cell lines, but not in T cells. J Immunol 162(1):299–304
Wang Y, Qu A, Wang H (2015) Signal transducer and activator of transcription 4 in liver diseases. Int J Biol Sci 11(4):448–455. doi:10.7150/ijbs.11164
Ward LD, Kellis M (2012) HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 40(Database issue):D930–D934. doi:10.1093/nar/gkr917
Yang J, Li MD (2014) Association and interaction analyses of 5-HT3 receptor and serotonin transporter genes with alcohol, cocaine, and nicotine dependence using the SAGE data. Hum Genet 133(7):905–918. doi:10.1007/s00439-014-1431-7
Zhong Z, Wen Z, Darnell JE Jr (1994) Stat3 and Stat4: members of the family of signal transducers and activators of transcription. Proc Natl Acad Sci USA 91(11):4806–4810
Acknowledgments
We thank Dr. David L. Bronson for his excellent editing of this manuscript. This study was in part supported by the National Science and Technology Major Project (No. 2012ZX10002004) and the Chinese High Technology Research and Development program (No. 2012AA020204), Research Center for Air Pollution and Health of Zhejiang University, and Ministry of Science and Technology of China (2012AA020405).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflicts of interest regarding this report.
Research involving human participants and/or animals
Statement of human rights All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Statement on the welfare of animals Not applicable.
Informed consent
Informed consent was obtained from all individual participants included in the study and the Ethics Committee of the First Affiliated Hospital of Zhejiang University School of Medicine approved the project.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jiang, X., Su, K., Tao, J. et al. Association of STAT4 polymorphisms with hepatitis B virus infection and clearance in Chinese Han population. Amino Acids 48, 2589–2598 (2016). https://doi.org/10.1007/s00726-016-2283-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-016-2283-3