Abstract
Respiratory motion in abdomen generates motion artifacts during Dynamic Contrast Enhanced MRI (DCE-MRI) data acquisition and it is clinically challenging to minimize the motion artifacts. Extraction of self-gated respiratory signal from the acquired k-space data is one of the methods to deal with the respiratory motion artifacts. The literature shows that non-Cartesian trajectories are less sensitive to motion artifacts than Cartesian trajectories. Golden-angle data acquisition in radial trajectory is preferred to extract the self-gated signal that splits the free-breathing data into different respiratory phases; also called motion states or bins. Conventionally, XD-GRASP (eXtra-Dimension golden-angle-radial Sparse Parallel MRI) reconstructs the binned data, but this method has limitations such as it does not preserve noise like texture (MR images have noise like artifacts) and it is a computationally intensive method. This research work proposes the use of a dedicated Convolutional Neural Network (CNN) architecture to remove motion artifacts from the binned (using uniform and adaptive binning) DCE golden-angle-radial liver perfusion data. The results of the proposed method are compared with XD-GRASP reconstruction. The results demonstrate that the proposed method takes significantly less computation time and provides similar quality of the reconstructed images as compared to the XD-GRASP method. Furthermore, receiver coil sensitivity information is required in XD-GRASP to reconstruct the MR image that may be difficult to estimate in some applications, whereas the proposed method does not require any such information.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Magnetic Resonance Imaging (MRI) is a medical imaging procedure that is safe but has long scan time. MRI examinations are sensitive to motion (due to long scan time) and artefacts may originate because of the patient motion, cardiac motion or respiratory motion [1]. Therefore, it is often desirable to introduce motion correction techniques to deal with the motion artifacts.
Dynamic Contrast-Enhanced MRI (DCE-MRI) [1,2,3] is an imaging method where MRI scans are acquired after the injection of a contrast agent. The movement of the contrast agent helps to differentiate between the healthy and defective tissues [2]. Respiratory motion in abdominal DCE-MRI creates clinical challenges. Prospective motion management in MRI requires breath-hold acquisition or external sensors [4] that involve high cost and complex setup. Another approach to deal with motion is the extraction of self-gated respiratory signal [5] from the raw k-space data. This self-gated signal helps to analyze the respiratory motion in free breathing acquisition without breath-hold or external sensors for motion management [4]. The self-gated signal is extracted by taking one-dimensional (1D) Fourier Transform (FT) of central k-space for each acquired spoke that provides the projection profiles of the entire imaging volume. Principal Component Analysis (PCA) is applied on these projection profiles to get the most common signal variation among all the elements that depicts existing signal, e.g., respiratory motion [5].
The literature shows that non-Cartesian trajectory is more beneficial than Cartesian trajectory as it is less sensitive to motion artifacts [6,7,8]. In Cartesian sampling, the center of the k-space is covered by only a few phase encoding steps, whereas in radial sampling, the center of the acquired k-space is oversampled as each profile incorporates data from the center of the k-space [8], therefore, the center of the k-space carries more information. There are two ways to acquire radial data: (i) Uniform sampling and (ii) Golden-angle sampling [9, 10]. The angle between the acquired spokes is constant in both cases. Uniform sampling is distributed uniformly between [0, π]. In golden-angle-radial sampling, angle between the acquired spokes is ϑ ≈ 111.25° [9]. Radial trajectory acquired with Golden angle has several advantages [10] e.g., every radial spoke fills the largest gap between the previously acquired spokes, no two spokes are acquired twice, and the spokes will be uniformly distributed if they belong to a Fibonacci number [6, 7].
In DCE-MRI, the target volume is imaged before, during and after the injection of contrast agent. The major challenge in DCE-MRI is its long scan time (about 30–40 min in DCE-MRI of abdomen) [6]. The long scan time makes MRI examinations sensitive to patient motion causing motion artifacts. Golden-angle-radial data are used to extract the self-gated signal that divides the acquired free-breathing data into different respiratory phases also called motion states or bins [5]. Different types of binning techniques have been used in literature such as uniform binning and adaptive binning [9,10,11].
Feng et al. used uniform binning [10] in which the range of respiratory signals is divided into fixed intervals such that each interval has an equal number of samples/spokes. Usman et al. [11] used adaptive binning where instead of dividing spokes in equal number in each bin like uniform binning, there are overlapping spokes in each bin; overlapping the spokes decreases the under-sampling factor (i.e., fewer aliasing artifacts expected in the solution image).
Feng et al. [10] proposed XD-GRASP, a reconstruction framework for free-breathing DCE golden-angle-radial MRI. XD-GRASP sorts the acquired dynamic data into extra motion-state dimensions using the self-navigation properties of radial trajectory and reconstructs the multi-dimensional dataset using Compressed Sensing [12]. This technique has some limitations such as it is a time-consuming method, and it cannot preserve noise-like textures and fine-scale details that may hold diagnostically important information for clinicians.
In recent years, deep learning has made significant developments in many fields such as image processing, speech recognition and natural language processing [13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29]. Initially, deep learning was used for image segmentation [15] and classification [16] but recently different deep learning-based architectures have been designed for image reconstruction [19, 20]. In case of MR image reconstruction using deep learning, the performance of deep neural networks was limited due to the size of the available data set [18] as it is not possible to train the network with a smaller number of images. Transfer learning [18, 21, 22] has been introduced in literature to deal with this problem. In transfer learning, the weights of a pre-trained network are updated based on new data set and it overcomes the limitation of availability of a large dataset.
The literature shows that deep learning can be used to reconstruct MR image from the under-sampled MRI data acquired from MRI scanner [19, 22,23,24,25,26,27,28,29]. Chang Min Hyun [19] presented a deep learning method for brain MR image reconstruction by reducing the k-space data with sub-Nyquist sampling strategies. This work shows a remarkable performance, retaining only 29% of the original data and produces good reconstruction results. Emmanuel Ahishakiye [20] presented an overview of the deep learning methods used in medical image reconstruction. The analysis of the authors [20] shows that Convolution Neural Network (CNN) is the most common network used for MR image reconstruction, but it requires a large set of training data [20].
Deep learning is also used to reduce the computation time to reconstruct the MR image. Christopher M. Sandino et al. [13] used deep CNN to accelerate MRI. In this paper [13], the results are compared with CS reconstruction algorithm that show that CNN is approximately 150 × faster without losing quality of the image.
Deep learning is a set of algorithms that parse data, learn from it, and then apply what has been learnt to make intelligent decisions. CNN is a class of deep, artificial neural networks that is applied to analyze visual imagery. The architecture of CNN comprises of different specific layers; each layer performing a different function.
In this paper, a dedicated CNN architecture is proposed to remove motion artifacts from MR images. Following layers are present in the CNN architecture used in this research work:
Convolutional layer: In the proposed CNN architecture, the first layer is convolution layer. This layer takes an image matrix and a kernel (filter) as input and provides convoluted image as output. The kernel size used in first 17 layers of the proposed method is 3 \(\times 3\) and in the 18th layer is 1 \(\times 1\). This kernel size is selected empirically. This layer determines the important features (such as information of different tissues) in the input image.
Activation layer: Rectified Linear Unit (ReLU) is used as an activation function (in the proposed CNN) that performs pixel wise operation. It replaces all the non-negative values by zero. The activation layer acts as a “gate” between the two convolutional layers. It can be a simple function that turns the output on and off, depending on a rule or threshold.
Batch normalization: Batch normalization layer [26] normalizes the activations of each layer to zero-mean and a constant standard deviation. This has the effect of stabilizing the learning process and reducing the number of training epochs required to train deep networks [19, 20]. In the proposed architecture, batch normalization layer significantly reduces the number of epochs.
Regression layer: Regression layers are widely employed to solve tasks where the goal is to predict output (solution image) values based on the estimated weights. In the proposed architecture, regression layer predicts responses (solution image) of a trained regression network using weights at the final stage.
The results of the proposed method are compared with the state-of-the-art XD-GRASP reconstruction [10]. The rest of the paper is organized as follows: materials and methods used in this work are mentioned in Sect. 2, whereas Sects. 3 and 4 summarize the results and discussion respectively. Finally, the conclusion is presented in Sect. 5.
2 Materials and Methods
2.1 Methodology
In this research work, a dedicated CNN architecture is proposed to reconstruct MR images from the under-sampled binned golden-angle-radial liver perfusion data. Figure 1 shows the architecture of the proposed CNN, where 18 convolution layers are used to estimate the weights followed by Rectified linear unit (ReLU) as an activation function and Batch Normalization (BN) to avoid over-fitting. The last layer is regression layer that provides the final reconstructed image. The number of layers is empirically chosen to be 18; lesser number of layers provide less accurate image, and no significant improvement was achieved in the output image with more than 18 layers.
All the experiments and analysis have been performed using MATLAB (2019a) on Intel(R) core (TM) i7 4th generation CPU with 16 GB RAM and NVIDIA Titan XP GPU with 3840 CUDA cores and 12GB Memory.
The availability of golden-angle-radial liver perfusion data is limited, therefore, initial CNN training is performed on cardiac data [30] at the first stage and transfer learning is applied in the second stage which uses golden-angle radial liver perfusion data. Figure 2 shows the main steps of the initial training.
The initial training is performed using 1600 images obtained from an open-source database [10, 30]. Each label image (402 spokes in each image) is first converted into golden-angle-radial k-space data using golden-angle (111.25°) radial sampling and retrospectively under-sampled with Acceleration Factor (AF) equals to 4 for the training purposes. Total number of spokes in each image of the under-sampled data is 101. The under-sampled golden-angle-radial k-space data are first gridded with non-Uniform Fast Fourier Transform (NUFFT) [30,31,32] to get the Cartesian k-space followed by inverse FFT (iFFT) operator [31] to obtain the aliased MR images. The proposed CNN architecture is based on supervised learning and is trained using the label (fully-sampled) MR images and the aliased MR images. For the training purpose, “rmsprop” [14] optimizer with a learning rate of 0.0001 (empirically selected) is used. We have used “mean square error” loss function in this work.
In the second stage of the proposed method, the CNN is fine-tuned with binned golden-angle radial liver perfusion data separately for the uniform and adaptive binning [5, 9]. Figure 3 shows a flow diagram for the reconstruction of binned data. To reconstruct the uniformly binned data, the trained network is fine-tuned (transfer learning) with the uniform binned liver data i.e., transfer learning [18, 21, 22] is applied on the initially trained CNN architecture with cardiac MR images.
The golden-angle-radial liver perfusion data used in this work have 1100 spokes with 502 sampling points along each spoke. Firstly, 1100 spokes are distributed into 11 contrast enhancement phases called frames, so the number of spokes in each frame are 1100/11 = 100. The frames are formed to track contrast enhancement changes. For 100 spokes in each contrast enhancement phase of the liver perfusion data, 4 bins are created (the number of bins chosen heuristically). Therefore, the number of spokes in each bin is 25 in our experiments. More than four bins reduce the number of spokes in each bin, and do not provide good reconstruction results whereas less than four bins do not provide correct information about the motion. The binned data cannot be directly used for image reconstruction, as the radial data are not sampled on a regular Cartesian grid, therefore, it requires gridding. NUFFT [32] is applied to grid the golden-angle-radial data onto Cartesian grid and then iFFT is applied on the gridded data to obtain the aliased MR images. The weights of the trained CNN are updated with the aliased and the labelled MR images. The updated CNN network is used to reconstruct MR images from the uniformly binned liver data.
For adaptive binning, all steps are the same as for uniform binning but instead of having four bins, five bins are formed that makes the number of spokes in each bin equal to 20. Different experiments have been performed that show the number of spokes less than 25 do not give good reconstruction results; therefore, 5 spokes are overlapped in each bin that makes total number of spokes equals to 25 in each bin. Similarly, we can take 6 bins, in that case the number of overlapped spokes will be 9 that makes total number of spokes equal to 25 in each bin.
The MR dataset used (both cardiac and golden-angle-radial Liver perfusion data) is complex, therefore two networks are used for reconstruction. Using first network, the magnitude part of the complex data sets is trained and reconstructed, and using the second network, the imaginary part of the complex data sets is trained and reconstructed in the proposed method using CNN. In the final stage, results of both the networks are combined to obtain the final reconstructed MR image.
2.2 Dataset
For initial training of the CNN, cardiac data are used that have been obtained from an open-source database [30]. It has 1600 fully sampled cardiac images that are converted to golden-angle-radial data and under-sampled with AF = 4 retrospectively to train the CNN.
The dataset with golden-angle-radial sampling is obtained from “Center for Advanced Imaging Innovation and Research (CAI2R)” [30], NYU school of Medicine, USA. Two 3D axial liver perfusion MRI datasets have been used in this work downloaded from CAI2R [30]: The acquisition parameters for both the dataset-1 and dataset-2 are 512 readouts, 1100 spokes, and 20 receiver coils. In this data, 100 adjacent spokes are grouped into one time point, resulting in 11 contrast enhancement phases. Each phase is then subdivided into further respiratory motion states or bins (using adaptive binning and uniform binning).
2.3 Quantifying Parameters
The quality of the reconstructed images is assessed visually and using different quantifying parameters i.e., Mean Signal-to-Noise Ratio (mean SNR), Central line profile and computation time.
Signal-to-noise ratio is defined as the ratio of the power of a signal to the power of background noise. The mean SNR [33] can be found as:
where, \(\mu R\) is the mean and \(\sigma R\) is the standard deviation of the reconstructed image containing both the signal and noise. Mean Signal-to-Noise Ratio is calculated using pseudo multiple replica method [33].
Central line-profiles of the CNN based reconstructed image and the XD-GRASP based reconstructed images are compared. Liver edge sharpness, clarity of hepatic vessel and streaking artifacts are examined visually. Comparison of computation time is performed between XD-GRASP reconstruction and the proposed method.
3 Results
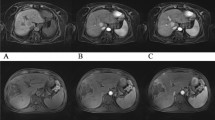
Two datasets (dataset-1 and dataset-2) [30] have been used in this work to verify the results of the proposed method. Figures 4 and 5 show the results of the reconstructed uniformly binned DCE golden-angle-radial liver perfusion DCE-MRI using the proposed method and using the XD-GRASP method for dataset-1 and dataset-2, respectively. The red circles indicate that resolving an image into respiratory phases helps to identify the change of the contrast agent.
Table 1 shows the mean SNR of the reconstructed images of the uniformly binned DCE golden-angle-radial liver perfusion data using the proposed method and XD-GRASP method at different motion states for both dataset-1 and dataset-2. The proposed method and XD-GRASP method show similar results in terms of mean SNR values as shown in Table 1. The averaged mean SNR value of the reconstructed images for dataset-1 using the proposed method is 21.19 and using XD-GRASP method is 21.99. Similarly, the averaged mean SNR value of the reconstructed images for dataset-2 using the proposed method is 30.56 and using XD-GRASP method is 31.02 as shown in Table 1.
Figures 6 and 7 compare the central-line profiles of the reconstructed images (uniformly binned) obtained by XD-GRASP method and the proposed method for dataset-1 and dataset-2, respectively. It can be seen in Figs. 6 and 7 that both methods provide almost comparable central line profiles.
Central-line profile comparison for the reconstructed images using the XD-GRASP method and proposed method for uniform binning (dataset-1). a Central-line profiles of the reconstructed images from XD-GRASP and proposed method, respectively, for motion state-1, b Central-line profiles of the reconstructed images from XD-GRASP and proposed method respectively for motion state-2, c Central-line profiles of the reconstructed images from XD-GRASP and proposed method respectively for motion state-3, d Central-line profiles of the reconstructed image from XD-GRASP and proposed method, respectively, for motion state-4
Central-line profile comparison using the XD-GRASP and the proposed method for uniform binning (dataset-2). a Central-line profiles of the reconstructed images from XD-GRASP and proposed method respectively for motion state-1, b Central-line profile of the reconstructed image from XD-GRASP and proposed method respectively for motion state-2, c Central-line profile of the reconstructed image from XD-GRASP and proposed method, respectively, for motion state-3, d Central-line profile of the reconstructed image from XD-GRASP and proposed method, respectively, for motion state-4
Table 2 shows a comparison of the computation time between the proposed method and the XD-GRASP method. Table 2 shows that there is a significant difference in reconstruction time of the two algorithms. The proposed method took approximately 99.95% less computation time for dataset-1 and 99.92% lesser time for dataset-2 as compared to XD-GRASP reconstruction.
Figures 8 and 9 show the results of the reconstructed adaptively binned DCE golden-angle-radial liver perfusion data using the proposed method and XD-GRASP method for dataset-1 and dataset-2, respectively. The averaged mean SNR value of the reconstructed images for dataset-1 using the proposed method is 28.23 and using XD-GRASP method is 28.50. Similarly, the averaged mean SNR value of the reconstructed images for dataset-2 using the proposed method is 42.42 and using XD-GRASP method is 42.66. The red circles indicate that resolving an image into respiratory phases helps to identify the change of the contrast agent.
Table 3 shows the mean SNR of the reconstructed images for the adaptively binned DCE golden-angle-radial liver perfusion data (dataset-1 and dataset-2) using the proposed method and XD-GRASP method at different motion states. Table 3 demonstrates that both the methods show almost similar mean SNR values.
Figures 10 and 11 provide a comparison of the central-line profiles of the reconstructed images using XD-GRASP method and the proposed method for the adaptively binned DCE golden-angle-radial liver perfusion dataset-1 and dataset-2, respectively. The results show that both the methods provide almost similar line profiles.
Central-line profile comparison for the reconstructed images using XD-GRASP and proposed method for adaptive binning for dataset-1: a central-line profiles of the reconstructed image using XD-GRASP and proposed method for motion state-1, b central-line profiles of the reconstructed image using XD-GRASP and proposed method for motion state-2, c central-line profiles of the reconstructed image using XD-GRASP and proposed method for motion state-3, d central-line profiles of the reconstructed image using XD-GRASP and proposed method for motion state-4, e central-line profiles of the reconstructed image using XD-GRASP and proposed method for motion state-5
Central-line profile comparison using XD-GRASP and proposed method for adaptive binning for dataset-2. a Central-line profiles of the reconstructed image using XD-GRASP and proposed method for motion state-1, b Central-line profile of the reconstructed image using XD-GRASP and proposed method for motion state-2, c Central-line profile of the reconstructed image using XD-GRASP and proposed method for motion state-3, d Central-line profile of the reconstructed image using XD-GRASP and proposed method for motion state-4, e Central-line profile of the reconstructed image using XD-GRASP and proposed method for motion state-5
Table 4 shows a comparison of the computation time for the proposed method and XD-GRASP method. It can be seen from Table 4 that the proposed method took approximately 99.95% less computation time for dataset-1 and 99.61% less computation time for dataset-2 as compared to XD-GRASP reconstruction.
Our experiments show that both methods (XG-GRASP and proposed method) provide almost similar reconstruction results; however, the proposed method takes significantly less computation time as compared to XD-GRASP. The proposed method can be used as an alternative method for reconstructing the binned golden-angle-radial perfusion data with significantly less computation time. Moreover, the proposed method does not require receiver coil sensitivity information (as required in the XD-GRASP method).
4 Discussion
Golden-angle-radial acquisition detects and removes blurring artifacts caused by respiratory motion. In this work, self-gated signal is extracted from the center of k-space of golden-angle-radial data. This self-gated signal is divided into different respiratory phases using two binning techniques: uniform binning and adaptive binning. The proposed CNN architecture reconstructs the binned data, and the results are compared with the gold standard XD-GRASP reconstruction algorithm.
The results show that the proposed method successfully removes blurring artifacts and helps to visualize the information embedded in different motion states. The mean SNR values of the reconstructed images by the proposed method are comparable with the XD-GRASP results but the proposed method takes significantly less computation time as compared to XD-GRASP. The results in Tables 2 and 4 show that the proposed method takes significantly less computation time as compared to the XD-GRASP reconstruction. Furthermore, the receiver coil sensitivity information [12, 34,35,36] is required in the XD-GRASP method to reconstruct the MR image that may be difficult to estimate in some applications, whereas the proposed method does not require any such information.
The number of bins in both the binning (uniform and adaptive) techniques are selected heuristically. The number of bins should be optimal for better reconstruction e.g., for fewer bins, the respiratory motion may not be resolved properly, and the motion blurring artifacts will remain in the reconstructed images. On the other hand, if the number of bins is too many then lesser numbers of spokes go into each bin and the final images will have under-sampling artifacts.
In this research work, it is observed that adaptive binning outperforms the uniform binning because adaptive binning contains some overlapping spokes in each bin (this is not possible in uniform binning) that increases the reconstruction data and reduces the artifacts that makes the reconstruction of the binned images easy. More bins can be formed using adaptive binning that reduces intra-bin motion in the final reconstructed images.
The proposed CNN architecture has eighteen layers, and the learning rate equals to 0.0001. Number of layers and learning rate are selected empirically to get good quality MR images. However, the results of the proposed method can be further improved with the availability of more data as the weights will be learned with more accuracy.
5 Conclusion
In this research, a deep learning algorithm based on transfer learning approach has been proposed to reconstruct the free breathing Golden-angle-radial liver perfusion data. As a proof-of-concept, a dedicated CNN architecture was implemented to provide solutions to the complex problems. The results demonstrate that the proposed method provides almost similar reconstructed image quality as of XD-GRASP method; moreover, the proposed method shows significant improvement in computation time (up to 99% in our experiments) as compared to XD-GRASP method. The receiver coil sensitivity information is required in the XD-GRASP reconstruction which may be difficult to estimate in some applications, however, the proposed method does not require any such information.
6 Competing interests
There is no conflict of interest.
7 Ethical approval
Not applicable.
Availability of data and materials
The data sets were obtained from an open-source database of Center for Advanced Imaging Innovation and Research (CAI2R). Website: http://cai2r.net/resources/software.
References
R. Grimm et al., Self-gated MRI motion modeling for respiratory motion compensation in integrated PET/MRI. Med. Image Anal. 19(1), 110–120 (2015)
V. Hamy et al., Respiratory motion correction in dynamic MRI using robust data decomposition registration – Application to DCE-MRI. Med. Image Anal. 18(2), 301–313 (2014)
J. Zhang, F. Najeeb, X. Wang, P. Xu, H. Omer, J. Zheng, C. Wang, Improved dynamic contrast-enhanced mri using low rank with joint sparsity. IEEE Access 10, 121193–121203 (2022)
C. Forman, D. Piccini, R. Grimm, J. Hutter, J. Hornegger, M. Zenge, High-resolution 3D whole-heart coronary MRA: a study on the combination of data acquisition in multiple breath-holds and 1D residual respiratory motion compensation. Magn. Reson. Mater. Phys., Biol. Med. 27(5), 435–443 (2014)
I. Shahzadi, M. Siddiqui, I. Aslam, H. Omer, Respiratory motion compensation using data binning in dynamic contrast enhanced golden-angle radial MRI. Magn. Reson. Imaging 70, 115–125 (2020)
L. Feng, R. Grimm, K.T. Block, H. Chandarana, S. Kim, J. Xu, R. Otazo, Golden-angle radial sparse parallel MRI: combination of compressed sensing, parallel imaging, and golden-angle radial sampling for fast and flexible dynamic volumetric MRI. Magn. Reson. Med. 72(3), 707–717 (2014)
I. Shahzadi, I. Aslam, S. Qazi, H. Omer, Golden-Angle Radial Sparse Parallel MR Image Reconstruction Using SC-GROG Followed by Iterative Soft Thresholding. Appl. Magn. Reson. 50(8), 977–988 (2019)
J. Weiss, C. Ruff, U. Grosse, G. Grözinger, M. Horger, K. Nikolaou, S. Gatidis, Assessment of hepatic perfusion using GRASP MRI: bringing liver MRI on a new level. Invest. Radiol. 54(12), 737–743 (2019)
F. Najeeb, M. Usman, I. Aslam, S.A. Qazi, H. Omer, Respiratory motion-corrected, compressively sampled dynamic MR image reconstruction by exploiting multiple sparsity constraints and phase correlation-based data binning. Magn. Reson. Mater. Phys., Biol. Med. 33, 411–419 (2020)
L. Feng, L. Axel, H. Chandarana, K. Block, D. Sodickson, R. Otazo, XD-GRASP: Golden-angle radial MRI with reconstruction of extra motion-state dimensions using compressed sensing. Magn. Reson. Med. 75(2), 775–788 (2015)
M. Usman et al., Motion corrected compressed sensing for free-breathing dynamic cardiac MRI. Magn. Reson. Med. 70(2), 504–516 (2012)
I. Aslam, F. Najeeb, H. Omer, Accelerating MRI using GROG gridding followed by ESPIRiT for non-Cartesian trajectories. Appl. Magn. Reson. 49, 107–124 (2018)
Sandino, C., Dixit, N., Cheng, J.Y., and Vasanawala, S.S. Deep convolutional neural networks for accelerated dynamic magnetic resonance imaging, preprint, 2017.
A.S. Lundervold, A. Lundervold, An overview of deep learning in medical imaging focusing on MRI. Z. Med. Phys. 29(2), 102–127 (2019)
Development of Deep Learning Algorithm for Brain Tumor Segmentation, Int. J. Eng. Adv. Technol vol. 9, no. 1, pp. 2800–2803, 2019.
M. Huang, S. Huang, Y. Zhang, U. Bhatti, Medical image segmentation using deep learning with feature enhancement. IET Image Process (2020). https://doi.org/10.1049/iet-ipr.2019.0772
L. Sun, Z. Fan, Y. Huang, X. Ding, J. Paisley, Compressed Sensing MRI Using a Recursive Dilated Network. Proc. AAAI Conf Artif. Intell. (2018). https://doi.org/10.1609/aaai.v32i1.11869
F. Knoll, K. Hammernik, E. Kobler, T. Pock, M. Recht, D. Sodickson, Assessment of the generalization of learned image reconstruction and the potential for transfer learning. Magn. Reson. Med. 81(1), 116–128 (2018)
C. Hyun, H. Kim, S. Lee, S. Lee, J. Seo, Deep learning for undersampled MRI reconstruction. Phys. Med. Biol. 63(13), 135007 (2018)
A. Emmanuel, Deep learning in medical image reconstruction: a general overview 2019.
S.A. Madiha et al., Transfer learning in deep neural network based under-sampled MR image reconstruction. Magn Reason Imag 76, 96–107 (2021)
S.U.H. Dar, M. Özbey, A.B. Çatlı, T. Çukur, A transfer-learning approach for accelerated MRI using deep neural networks. Magn. Reson. Med. 84(2), 663–685 (2020)
He, Juncai, Lin Li, Jinchao Xu, and Chunyue Zheng. "ReLU deep neural networks and linear finite elements." arXiv preprint arXiv:1807.03973, 2018.
V. Ghodrati, J. Shao, M. Bydder, Z. Zhou, W. Yin, K.L. Nguyen, P. Hu, MR image reconstruction using deep learning: evaluation of network structure and loss functions. Quant. Imaging Med. Surg. 9(9), 1516 (2019)
S. Wang, J. Hong, M. Yang, Sensorineural hearing loss identification via nine-layer convolutional neural network with batch normalization and dropout. Multimedia Tools Appl. 79(21–22), 15135–15150 (2018)
Pal, A., & Rathi, Y. A review and experimental evaluation of deep learning methods for MRI reconstruction. J. Mach. Learn. Biomed Imag. 2022.
Singh D, Monga A, de Moura HL, Zhang X, Zibetti MVW, Regatte RR. Emerging trends in fast MRI using deep-learning reconstruction on undersampled k-space data: a systematic review, Bioengineering, 2023.
Liu, X., Wang, J., Liu, F., Zhou, S.K. (2021). “Universal Undersampled MRI Reconstruction. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part VI 24, pp. 211–221. Springer International Publishing, 2021.
Ramzi, Z. Advanced deep neural networks for MRI image reconstruction from highly undersampled data in challenging acquisition settings. PhD diss., Université Paris-Saclay, 2022.
"Software Downloads | CAI2R", Cai2r.net, 2020. [Online]. Available: http://cai2r.net/resources/software. Accessed 05 May 2023.
A. Fertner, Computationally efficient methods for analysis and synthesis of real signals using FFT and IFFT. IEEE Trans. Signal Process. 47(4), 1061–1064 (1999)
J. Fessler, B. Sutton, Nonuniform fast Fourier transforms using min-max interpolation. IEEE Trans. Signal Process. 51(2), 560–574 (2003)
P. Robson, A. Grant, A. Madhuranthakam, R. Lattanzi, D. Sodickson, C. McKenzie, Comprehensive quantification of signal-to-noise ratio and g-factor for image-based and k-space-based parallel imaging reconstructions. Magn. Reson. Med. 60(4), 895–907 (2008)
A. Irfan, A. Nisar, H. Shahzad, H. Omer, Sensitivity maps estimation using eigenvalues in sense reconstruction. Appl. Magn. Reson. 47(5), 487–498 (2016)
A. Fariha et al., GROG-pCS: GRAPPA Operator Gridding with CS-based p-thresholding for under-sampled radially encoded MRI. Int. J. Emerg. Multidiscip 1, 1 (2023)
M. Uecker, P. Lai, M.J. Murphy, P. Virtue, M. Elad, J.M. Pauly, M. Lustig, ESPIRiT—an eigenvalue approach to autocalibrating parallel MRI: where SENSE meets GRAPPA. Magn. Reson. Med. 71(3), 990–1001 (2014)
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Authors A, C, D and E were involved in the research work. Authors A, B, C and E have contributed in writing and all authors have reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Aslam, T., Najeeb, F., Shahzad, H. et al. Motion Corrected DCE-MR Image Reconstruction Using Deep Learning. Appl Magn Reson 55, 509–526 (2024). https://doi.org/10.1007/s00723-024-01642-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00723-024-01642-2