Abstract
Recently, we have demonstrated the possibility to transfer the light-induced hyperpolarization occurring on select 13C nuclei of photosynthetic cofactors due to the solid-state photochemically induced dynamic nuclear polarization (photo-CIDNP) effect into the proton pool via inverse 13C–1H cross-polarization Bielytskyi et al. (J Magn Reson 293: 82–91, 2018). Such approach allowed us to observe the selective response from the protons belonging to photochemically active cofactors in their native protein environment. In the present study, we demonstrate that also 15N nuclei can be used as a source of polarization in a similar type of experiments. We present the 2D photo-CIDNP 15N–1H heteronuclear correlation (HETCOR) experiment acquired on uniformly 15N-labeled quinone-blocked reaction center (RC) from Rhodobacter sphaeroides R26. Obtained 1H chemical shifts match with previously observed ones on selectively 13C-labeled RC from Rhodobacter sphaeroides WT. We expect that using 15N as a source of polarization in potential heteronuclear spin-torch experiments could be advantageous for systems where the procedure for selective 13C labeling is not yet established.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Photochemically induced dynamic nuclear polarization (photo-CIDNP) magic-angle-spinning (MAS) NMR is an analytical technique used for studying electronic structures of photosynthetic cofactors embedded in protein membrane complexes [1, 2]. It is based on solid-state photo-CIDNP effect [3], a nuclear hyperpolarization method that manifests itself in a strong signal enhancement (by a factor of 10,000–80,000 for 13C) observed for 13C and 15N nuclei in MAS NMR experiments. The effect relies on the evolution of spin-correlated radical pair (SCRP) which, in photosynthetic reaction centers (RCs), is formed between electron donor and acceptor after initial photo-induced electron transfer. During the lifetime of the SCRP, up to three different solid-state mechanisms operate in parallel to generate a nuclear spin hyperpolarization [4], known as three-spin mixing (TSM) [5], differential decay (DD) [6] and differential relaxation (DR) [7]. These mechanisms were recently generalized with a concept of level crossings and level anti-crossings [8].
Beyond its classical applications using direct 13C and 15N information, the solid-state photo-CIDNP effect can allow for spin-torch experiments, in which the hyperpolarization created on 13C nuclei of the photosynthetic cofactors forming the SCRP is transferred to the protein vicinity via natural spin diffusion. Thus, some 13C signals that do not originate from the cofactors participating in SCRP have been previously reported in photo-CIDNP MAS NMR experiments on 13C-labeled bacterial RCs [9,10,11]. Alternatively, the heteronuclear spin-torch approach in which hyperpolarization created on 13C or 15N nuclei of the active cofactors is selectively transferred to protons of the close protein environment is more appealing, as it does not require the presence of isotope labels beyond the cofactors participating in the SCRP. Moreover, the potential information about the proton chemical shifts of the nearby amino acids might allow for new insights into the role of hydrogen bonding networks around the electron donor in both ground and charge-separated state. Recently, we demonstrated the applicability of such an approach by transferring the hyperpolarization generated by the solid-state photo-CIDNP effect on 13C nuclei to the nearby protons in the selectively 13C-labeled RCs of Rhodobacter (R.) sphaeroides WT [12]. Such experiment allowed us to determine proton chemical shifts of the electron donor and acceptor in their native protein environment. The 13C isotope labeling procedures of the photosynthetic RCs usually imply using expensive 13C-labeled-δ-aminolevulinic acid (ALA) [10]. These procedures are still a challenge for the majority of photosynthetic organisms. On the other hand, uniform 15N labeling is usually rather straightforward to implement and requires utilizing relatively inexpensive compounds. Thus, successful 15N labeling was reported for RCs from purple bacteria (R. sphaeroides) [13, 14], heliobacteria (Heliobacillus mobilis) [15], plant’s photosystems (PS) I and II from spinach (Spinacia oleracea) [11] and duckweed (Spirodella oligorrhiza) [16]. With 15N labeling, also more “exotic” RCs have been addressed, such as RCs from acidobacterium (Chloracidobacterium thermophilum) [17] and the diatom (Phaeodactylum tricornutum) [18]. This makes 15N nucleus attractive for heteronuclear spin-torch experiments as it could be utilized for plenty of photosynthetic organisms where the 13C isotope labeling procedure is challenging or not yet been established. In the present study, we show by using u-15N-labeled quinone-blocked RCs of mutant R. sphaeroides R26 that 15N nuclei can be effectively used as an alternative source of hyperpolarization in such type of experiments by recording the 2D photo-CIDNP 15N–1H HETCOR MAS NMR spectra.
2 Materials and Methods
2.1 Sample Preparation
Rhodobacter sphaeroides R26 was grown under anaerobic conditions in a medium containing 95% 15N labeled NH4Cl (VEB Berlin Chemie, Berlin-Adlershof, Germany). The extent of 15N incorporation has been determined previously to be ~ 60% [14]. The RCs were isolated according to the procedure described in Ref. [19]. The quinones in the RCs were reduced with 0.05 M sodium dithionite in Tris buffer prior to experiments. Approximately, 5 mg of the RC protein complex embedded in LDAO micelles was used for NMR measurements.
2.2 NMR Measurements
All NMR experiments were carried out at magnetic field of 9.4 T (400 MHz 1H Larmor frequency) with an AVANCE III NMR spectrometer equipped with 4-mm double resonance MAS probe (Bruker, Karlsruhe, Germany). The sample was loaded into an optically transparent 4-mm sapphire rotor, inserted into the MAS probe and frozen in the dark conditions at slow spinning frequency of 500 Hz. This was done to ensure a homogeneous sample distribution against the rotor walls [20]. Once frozen, the sample was spun at MAS frequency of 8176 ± 10 regulated by pneumatic control unit at a constant temperature of 247 K, which was maintained by a temperature control unit. The idle time of several hours was needed to equilibrate the temperature of the probe electronics and to ensure the stable 1H radio frequency circuit, which is crucial for a proper calibration of 1H chemical shift axis. The sample was illuminated with the continuous illumination setup [21, 22], which comprises a 1000-W xenon arc lamp with collimation optics, set of IR and UV filters, a focusing element and a light fiber bundle. Optimized 1H and 15N 90° pulse lengths were 2.5 and 4.8 μs, respectively. The 15N NMR spectra were referenced using the response from solid standard 15NH4NO3 at 23.5 ppm. Appropriately scaled 1H dimension was referenced by assigning the midpoint of two methylene proton peaks of solid glycine to 3.52 ppm.
1D photo-CIDNP 15N experiments were acquired using Hahn-echo pulse sequence with 1 k scans and 4 s relaxation delay. Swept-frequency two-pulse phase modulation (SWf-TPPM) heteronuclear decoupling [23] with 100 kHz RF field was applied during acquisition.
The inverse CP experiments were carried out with the Lee–Goldburg CP (LGCP) [24] optimized to satisfy Hartmann–Hahn (HH) n = − 1 condition with 30 kHz effective 1H RF lock field and 70–100% ramp on 15N channel [25]. The solvent suppression was achieved with repeating alternating X and Y pulses on-resonance with the water peak, with RF frequency of 20 kHz and total length of the saturation block of 100 ms. The u-13C,15N l-histidine hydrochloride monohydrate sample was bought from Cambridge Isotope Laboratories (Andower, USA).
Acquisition in analog mode using windowed version of supercycled-phase-modulated Lee-Goldburg homonuclear decoupling (wPMLG3-S2) [26, 27] was optimized by monitoring the splitting between the methylene protons of unlabeled glycine while keeping the MAS frequency at 8176 Hz and changing the following parameters: length of individual PMLG pulse, window duration, RF amplitude and the carrier offset [28, 29]. In wPMLG3 decoupling, the basic unit consists of PMLG3 block (τPMLG3) and the acquisition window τw resulting in a cycle period of τc = τPMLG3 + τw. Each PMLG3 block consisted of 6 pulses with the following phases: 214.64°, 283.92°, 353.21°, 173.20°, 103.92° and 34.64° (m3p shape in TopSpin 3.2 library). A consecutive PMLG3 block is then repeated with 180° phase shift to complete S2 supercycle [26]. The optimal pulse length of each pulse in PMLG3 block was found to be 2.45 μs (τPMLG3 = 14.70 μs), and the window duration τw was 4.95 μs. The overall length of decoupling cycle time τc was equal to 19.65 μs, resulting in the characteristic frequency νc = 50.9 kHz. In order to avoid line broadening due to undesirable destructive PMLG interferences, it is required that 3τc < τr and 4τc ≠ τr where τr is the rotor period [30, 31]. In the present setup, the rotor period τr = 122.3 μs and τr/τc = 6.2, being far off of the obvious degeneracies. The RF amplitude during decoupling was set to 88 kHz. The 1H RF offset frequency was set to − 2900 Hz during windowed acquisition to avoid RF-rotary-frequency lines and other artifacts falling over the spectral region of interest [32]. The scaling factor of 0.52 was determined directly on the protein sample by plotting the dependence of the apparent offset shift of the water signal against the actual offset change (see Fig. 1) and was further used to correct the 1H spectral width.
a Calculation of the 1H chemical shift scaling factor by plotting the apparent offset change against the actual offset change as determined on the water peak in frozen protein sample. The slope of the line gives the scaling factor of windowed homonuclear decoupling sequence. The 0.52 scaling factor is constant over the wide range of the offset frequencies. b The scaled 1H MAS NMR spectrum of solid glycine recorded with optimized wPMLG-S2 decoupling sequence at 9.4 T and MAS frequency of 8176 Hz. The midpoint position of two methylene proton peaks was set to 3.52 ppm for 1H reference. The signal labeled with an asterisk refers to a rotary-resonance artifact rising from the homonuclear decoupling
2D photo-CIDNP 15N–1H experiments were recorded with 64 t1 increments, accumulating 960 scans in each indirect slice with relaxation delay of 4 s, resulting in 3 days of experimental time. Frequency discrimination during the evolution period was achieved with STATES-TPPI [33]. A 45° shifted squared sine bell window function (qsine SSB = 4 in TopSpin) was applied in the indirect dimension, and further zero-filled to 2048 points prior to Fourier transformation. A 90° shifted squared sine bell window function (qsine SSB = 2) was applied in the direct dimension and zero-filled to 1024 data points.
The data were processed with Bruker TopSpin 3.2 and plotted with MNova 12 (Mestrelab Research, S. L. Santiago de Compostela, Spain).
2.3 Pulse Sequence
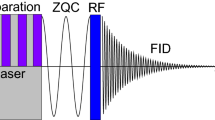
The pulse sequence used to record 2D photo-CIDNP 15N–1H HETCOR MAS NMR spectra is described in detail in Ref. [12] and is shown in Fig. 2.
Pulse sequence used to record 2D photo-CIDNP 15N–1H HETCOR MAS NMR spectra. It consists of the water suppression block, a direct 15N 90° excitation pulse, the evolution period t1 during which the high power heteronuclear decoupling is applied, two spin-lock pulses to satisfy HH condition and 1H windowed acquisition with wPMLG3-S2; ϕ1 = + y−y; ϕ2 = + x; ϕ3 = + x +x –x –x + y +y –y – y; ϕrec = + x – x –x + x + y – y – y + y
3 Results and Discussion
3.1 Efficiency of 15N–1H Inverse LGCP
The performance of the pulse sequence shown in Fig. 2 was tested on the solid u-13C,15N labeled l-histidine hydrochloride monohydrate. The 1D 15N-to-1H MAS NMR spectra recorded with long and short contact times are shown in Fig. 3a. 200 μs contact time is sufficiently long to complete one-bond 15N–1H magnetization transfers between nitrogen atoms of the imidazole ring and adjacent protons, while longer contact times are required to perform 15N–1H transfers for the amino group, as a result of its mobile nature that causes intrinsic decoupling of 15N–1H dipolar interactions. On the other hand, the contact time of 4500 μs allows for performing multiple-bond transfers, which is reflected by the presence of the 1H signals on 1D spectrum that do not have direct 15N partners. The 1D 15N–1H MAS NMR spectrum recorded with 4500 μs contact time resembles the pattern of the 1D MAS NMR spectrum recorded with simple pulse-acquire experiment and windowed detection (Fig. 3b). The locking of protons under LG condition [24] provides long T H1ρ relaxation times due to the reduced strength of 1H–1H homonuclear dipolar couplings [34]. The 2D 15N–1H correlation MAS NMR spectra recorded with different LGCP contact times are presented in Fig. 3c. The spectrum recorded with 200 μs contact time contains solely correlation peaks between covalently bonded 15N–1H pairs, while the spectrum obtained with 4500 μs provides many more correlation peaks between distant pairs. Thus, the nitrogen of the amino group NH3+ has correlations to all protons. Careful estimation of the distances based on the neutron diffraction data [35] allows to assume that the transfers beyond > 3.9 Å are possible with this contact time (Fig. 3d). Such effective transfer is surprising considering that the inverse cross-polarization from 15N nucleus to 1H is expected to be inefficient due to a low gyromagnetic ratio of 15N nucleus and small 15N–1H heteronuclear dipolar couplings. Prevalence of the signal originating from the nitrogen of NH3+ group and weak intensities from nitrogen atoms of the imidazole ring in Fig. 3c is a result of a relatively short relaxation delay time used to record 2D spectrum (15 s). Such delay was used for the sake of the experimental time and implies incomplete recovery of the 15N of the imidazole ring due to longer 15N longitudinal relaxation time T1 as compared to NH3+ group.
a 1D 15N–1H MAS NMR spectra of solid u-13C,15N labeled l-histidine hydrochloride monohydrate recorded with the pulse sequence from Fig. 2 with t1 = 0, 60 s relaxation delay and contact time of 200 μs (red dashed) and 4500 μs (black). b 1D 1H MAS NMR spectrum recorded with simple pulse-acquire experiment and wPMLG3-S2 analog detection. The signals labeled with an asterisk refer to a rotary-resonance artifact rising from the homonuclear decoupling. c 2D 15N–1H correlation MAS NMR spectra of solid u-13C,15N labeled l-histidine hydrochloride monohydrate acquired using pulse sequence from Fig. 2 at 9.4 T and MAS frequency of 8176 Hz with LGCP contact time of 4500 μs (top) and 200 μs (bottom). The spectra were obtained with 64 t1 increments, accumulating 64 scans in each indirect slice with relaxation delay of 15 s. The sample was packed in standard 4-mm zirconia rotor without CRAMPS stoppers. d The distances between selected 15N and 1H of solid u-13C,15N labeled l-histidine hydrochloride monohydrate
Overall, application of the pulse sequence shown in Fig. 2 on the solid u-13C,15N labeled l-histidine hydrochloride monohydrate demonstrated that the 15N nucleus can be used as a source of polarization in inverse CP experiments. Such calibration experiment suggests that the LGCP contact time of 4500 μs could be sufficient to perform the transfers between the nitrogen atoms of the special pair and protons of the nearby amino acids, with certain 15N–1H distances being < 3.5 Å (axial histidines, see further in text).
3.2 15N-to-1H Transfer of Nuclear Hyperpolarization in Frozen Photosynthetic RCs
The 1D 15N photo-CIDNP MAS NMR spectrum of R. sphaeroides R26 is presented in Fig. 4. It consists of 11 light-induced emissive (negative) signals between 180 and 300 ppm which have been assigned to the response from 15N labels of two bacteriochlorophylls BChl a (PL and PM) and one bacteriopheophytin BPhe a (ФA) [36]. For the structures and numbering of BChl a and BPhe a molecules, see Fig. 5. The emissive character of the light-induced signals is interpreted in terms of the dominance of TSM mechanism over DD during the build-up of photo-CIDNP [37]. Also, the contribution from the DR mechanism is present in carotenoid-less mutant R26 and contributes to the emissive (negative) signal of 15N nuclei on the donor site [14, 38]. The broad absorptive (positive) signal at ~ 120 ppm corresponds to the Boltzmann-polarized response of the amide nitrogens of the protein backbone, which also appears in the spectrum acquired in dark (shown in black). In the light-induced spectrum shown in red, 11 emissive signals have been grouped into two sets of 4 and 1 set of 3 signals, each belonging to a particular cofactor forming the SCRP [14]. Thus, the more intense set of BChl a donor signals appears at 260 (N-II), 255 (N-IV), 197 (N-III) and 187 ppm (N-I) and is assigned to PL. Such assignment is based on the intensity ratio between two halves of the special pair PL and PM, which was previously determined to be 2:1 by 1H ENDOR [39] and 3:2 by 13C photo-CIDNP MAS NMR [37]. The asymmetry in distribution of the electron spin density in the electronic ground state of the special pair in favor of the PL cofactor is referred to as “symmetry break” and is assumed to be relevant for the selectivity of the light-induced electron transfer into one of the two branches of the cofactors, despite of a high structural symmetry [40] (see Fig. S2 for the structure of the RC). The second set of weaker intensity shows up at 258 (N-IV), 253 (N-II), 192 (N-III) and 191 ppm (N-I) and is assigned to PM. Finally, the set belonging to BPhe a consists of two overlapping signals at ~ 298 ppm (N-II and N-IV) and the signal at 129 ppm (N-I). Previously, one more signal corresponding to (N-III) at 137 ppm was detected, however, because its low intensity is hardly observed in this work.
1D 15N photo-CIDNP MAS NMR spectrum of u-15N labeled R. sphaeroides R26 recorded with Hahn-echo pulse sequence at 9.4 T and MAS frequency of 8176 Hz at 247 K in dark (top black) and under continuous illumination (bottom red) with 1024 scans and 4 s relaxation delay. The color code of the individual labels refers to the assignment to the three cofactors forming the SCRP: green, red and blue refer to the two BChl a molecules of the donor (PL and PM) and BPhe a molecule of the acceptor (ФA), respectively. The assignment is based on the Ref. [36]. Spinning sidebands are labeled with asterisks (color figure online)
The 2D photo-CIDNP 15N–1H HETCOR MAS NMR spectra of u-15N labeled R. sphaeroides R26 acquired in dark and under continuous illumination with LGCP contact time of 4500 μs are presented in Fig. 6. The spectrum acquired in dark (Fig. 6a) contains the absorptive correlation signals between the amide backbone nitrogens grouped at ~ 120 ppm and corresponding amide protons at ~ 8 ppm, as well as aromatic and Hα protons located at 6.5 and 4 ppm, respectively. The spectrum obtained under continuous illumination (Fig. 6b) contains additional emissive correlation peaks located between 180 and 300 ppm in 15N dimension and between 8.5 and -2 ppm in 1H dimension. These light-induced correlation signals suggest the efficiency of distant 15N–1H transfers, as all nitrogen atoms in the following chemical shift range are deprotonated. We first discuss the correlation pattern belonging to the BChl a donor. The group of 1H signals between 7 and 8.5 ppm originates from protons H-5, H-10 and H-20, as was assigned previously [12]. The distances between N-II and H-5, N-II and H-10, N-IV and H-20 are about 3.3 Å [41]; therefore, the presence of such correlation peaks implies that the 15N–1H transfer occurs at this distance. The 2D approach allows for differentiating these signals from the amide backbone response with similar chemical shifts, which would otherwise overlap in 1D proton spectra. Interestingly, only N-II and N-IV from the donor have correlations with protons H-5, H-10 and H-20, while N-I and N-III located within similar distances do not have similar correlations. This could be attributed to the insufficient signal intensity on N-I and N-III, as a consequence of a low electron spin density distributed over these nuclei as compared to positions N-II and N-IV [36]. Indeed, the polarization arising due to the solid-state photo-CIDNP effect at the end of the photo-cycle is roughly proportional to the square of the electron spin density on a particular atom [42, 43]. Thus, the most efficient positions for “intensity pumping” are N-II and N-IV from where the polarization is transferred into the proton pool.
2D photo-CIDNP 15N–1H HETCOR MAS NMR spectra of u-15N labeled R. sphaeroides R26 with tentative assignment of 1H resonances, recorded with pulse sequence from Fig. 2 at 9.4 T and MAS frequency of 8176 Hz at 247 K a in dark and b under continuous illumination. The spectra were obtained with LGCP contact time 4500 μs and 64 t1 increments, accumulating 960 scans in each indirect slice with relaxation delay of 4 s. Red contours represent emissive character of the light-induced correlation signals and blue contours represent absorptive backbone signals. The color code of the individual 15N labels refers to the assignment to the three cofactors forming the SCRP: green, red and blue refer to the two BChl a molecules of the donor (PL and PM) and the BPhe a of the acceptor (ФA), respectively (color figure online)
The region of proton signals between – 2 and 5 ppm correlated with N-II and N-IV of the donor is crowded and lacks resolution. Therefore, we can only tentatively assign the groups of signals based on the previous 13C-1H transfer experiment [12] and the data obtained for BChl a and BPhe a in solution [44]. The 1H signals between 3 and ~ 4.5 ppm are tentatively assigned to H-7, H-8, H-17, H-18. The correlations from 0 to 3 ppm probably belong to the feedback from protons H-81, H-171. Finally, the most shielded region between − 2 and 0 ppm is assigned to the aliphatic protons from the periphery of the cofactors, located close to N-II and N-IV, i.e., H-71, H-82, H-172 and H-181. Such correlations suggest the possibility of the polarization transfer beyond 3.5 Å. Then, the correlations between donor signals N-I, N-III and protons between -2 and 0 ppm might be due to the transfers to protons H-21 and H-121 located in the vicinity or rings I and III. The 1H signal at 1 ppm correlated to N-I of either PL or PM remains puzzling. It is unlikely that it belongs to either H-81 or H-171 considering the distances between N-I and C-81 and C-171 (~ 6.2 Å). Assuming that this proton might be the H-32 also implies the possibility of the long transfer beyond 5 Å which is hardly possible since the transfers at shorter distances between N-I and H-5/H-20 or NIII and H-10 are not observed. On the other hand, two axial histidines, His-L173 and His-M202, which are coordinated to magnesium atoms of PL and PM, respectively, are located in close vicinity of the cofactors (~ 3 Å) [41]. This allows for making an assumption about the possibility of intermolecular transfers between these pairs. Previously, the ε and δ protons of the axial histidines in the RC complex from R. sphaeroides were found to resonate at ~ 2 ppm [45]. Such strong shielding occurs due to the ring currents generated by the conjugated porphyrin ring system of BChl a in a close proximity [46]. To estimate this effect, we performed DFT calculation of nucleus independent chemical shift (NICS) (see Supplementary table S1). Such estimation shows that the ε and δ protons belonging to His-L173 and His-H202 are expected to resonate in the upfield region at about 1.5–3.0 ppm, which would match results obtained in Ref. [45]. Based on the model of the special pair with optimized geometry (Fig. 7), the distances between N-I and N-III of PL and PM and ε-, δ -1H of the respective axial histidines His-L173 and His-M202 could be as short as 2.6–3.1 Å. Following previous discussion, the 15N–1H transfers within such distances were assumed to be possible. The intensity of the correlation peak at ~ 187/1.0 ppm suggests that it belongs to N-I of PL rather than PM. Then, its closest partner would be ε-1H of His-L173, which is expected to resonate at about 1.5 ppm. Since the error in determining 1H chemical shifts is estimated to be at least ± 0.5 ppm, such assignment would be reasonable. Further discussion on potential intermolecular transfer, however, requires knowledge about the chemical shifts of protons directly bonded to the cofactors, which would allow for discriminating them from the signals of the environment. Presently, in our laboratory, a map showing chemical shifts of protons covalently bound to PL, PM and ФA is under construction.
Selected intermolecular distances between 15N of the BChl a cofactors PL and PM and 1H of coordinated histidines His-L173 and His-M202. The coordinates are taken from the model structure after geometry optimization (see Fig. S1). The distance between N-I of PL and ε-1H of His-L173 is 2.6 Å; the distance between N-III of PL and δ-1H of His-L173 is 2.8 Å; the distance between N-I of PM and ε-1H of His-M202 is 3.0 Å; the distance between N-III of PM and δ-1H of His-M202 is 3.4 Å.
The correlation signals belonging to the protonated nitrogens N-III and N-I of BPhe a acceptor ФA are missing. The intensities of N-III and N-I signals of the acceptor are extremely weak; thus, the transfer of polarization to the adjacent protons is expected to be inefficient. Moreover, the close broad response from the backbone is not photo-CIDNP induced and has the opposite sign as compared to the light-induced signals. Therefore, backbone signals might superimpose the weak correlation signals between N-III and N-I and their adjacent protons, which are expected to be located in a very close proximity. Noticeably, the correlations between N-II and N-IV of the acceptor with protons H-5, H-10 and H-20 are missing. Otherwise, the correlation pattern is similar to the donor. We do not exclude, however, that the absence of certain correlation peaks might be due to the unequal efficiency of the 15N–1H LGCP transfer for different 15N positions. This issue could be addressed by implementing adiabatic passage through HH condition, which is generally more efficient as compared to HH CP for polarization transfer between 1H and 15N [47, 48]. Moreover, due to the strong magnetic-field-dependence of the solid-state photo-CIDNP effect [49], the efficiency of such experiments might be greatly improved by applying the optimum external magnetic field. Thus, for RCs from R. sphaeroides R26 the 15N photo-CIDNP-induced signals are the most pronounced at the field of 4.7 T [14].
The photo-CIDNP 15N–1H correlation experiment allows to obtain information about the chemical shifts of protons around cofactors forming the SCRP in their native protein environment with help of simple illumination setup and relatively short experimental time. 1H chemical shifts obtained in this work match previously observed for the selectively 13C-labeled RC from R. sphaeroides WT [12]. This is in agreement with data from 15N and 13C photo-CIDNP MAS NMR experiments suggesting that both R26 and WT RCs have identical electronic structures in the electronic ground state [14, 50]. The uniformly isotope labeling does not complicate the resulting 15N–1H spectra since the solid-state photo-CIDNP is generated on 15N positions exclusively belonging to the cofactors that participate in the SCRP. As a result, solely 1H signals in the vicinity of the electron donor and acceptor are highlighted and have been distinguished from the protein backbone feedback.
4 Conclusions
In the present study, we performed 15N–1H heteronuclear transfer of photo-CIDNP generated hyperpolarization in the core of the frozen photosynthetic RC via inverse CP. The use of 15N nuclei as a source of hyperpolarization allows for highlighting the protons at a distance of up to 3.5 Å at moderate contact time. On that basis, we observed similarities in 1H chemical shift pattern of R26 mutant and the WT organism. Estimated ring current effects induced by the conjugated porphyrin ring system of BChl a on axial histidines suggest strong shielding effects experienced by the protons of the corresponding amino acids. Thus, we assumed the potential intermolecular transfer between the N-I of PL and ε-1H of axial histidine His-L173.
Overall, since uniform 15N isotope labeling is generally straightforward to achieve as compared to 13C labeling, this approach provides an alternative way to perform heteronuclear spin-torch experiments on the photosynthetic organisms for which the selective 13C-labeling procedure has not yet been established. Therefore, we are planning to apply such experiments on u-15N labeled PSII to reveal the state of protons in plant’s photosynthetic machinery.
References
B.E. Bode, S.S. Tamarath, K.B. Sai Sankar Gupta, G. Jeschke, J. Matysik, in Hyperpolarization Methods in NMR Spectroscopy, ed. by L.T. Kuhn (Springer, Berlin, Heidelberg, 2012), pp. 105–121
M. Najdanova, G.J. Janssen, H.J.M. de Groot, J. Matysik, A. Alia, J. Photochem. Photobiol. B 152, 261 (2015)
M.G. Zysmilich, A. McDermott, J. Am. Chem. Soc. 116, 8362 (1994)
G. Jeschke, J. Matysik, Chem. Phys. 294, 239 (2003)
G. Jeschke, J. Am. Chem. Soc. 120, 4425 (1998)
T. Polenova, A.E. McDermot, J. Phys. Chem. B 103, 553 (1999)
A. McDermott, M.G. Zysmilich, T. Polenova, Solid State Nucl. Magn. Reson. 11, 21 (1998)
D. Sosnovsky, G. Jeschke, J. Matysik, H.-M. Vieth, K.L. Ivanov, J. Phys. Chem. 144, 144202 (2016)
J. Matysik, E. Schulten, A. Alia, P. Gast, J. Raap, J. Lugtenburg, A.J. Hoff, H.J.M. de Groot, Biol. Chem. 382, 1271 (2001)
E.A.M. Schulten, J. Matysik, A. Alia, S. Kiihne, J. Raap, J. Lugtenburg, P. Gast, A.J. Hoff, H.J.M. de Groot, Biochemistry 41, 8708 (2002)
S. Paul, B.E. Bode, J. Matysik, A. Alia, J. Phys. Chem. B 119, 13897–13903 (2015)
P. Bielytskyi, D. Gräsing, K.R. Mote, K.B. Sai Sankar Gupta, S. Vega, P.K. Madhu, A. Alia, J. Matysik, J. Magn. Reson. 293, 82 (2018)
M.G. Zysmilich, A. McDermott, J. Am. Chem. Soc. 118, 5867 (1996)
E. Daviso, K.B. Sai Sankar Gupta, S. Prakash, P. Gast, A. Alia, G. Jeschke, J. Matysik, in Photosynthesis. Energy from the Sun, ed. by J.F. Allen, E. Gantt, J.H. Golbeck, B. Osmond (Springer, Dordrecht, 2008), pp. 25–28
S.S. Tamarath, A. Alia, E. Daviso, D. Mance, J.H. Golbeck, J. Matysik, Biochemistry 51, 5763 (2012)
G.J. Janssen, E. Roy, J. Matysik, A. Alia, Appl. Magn. Reson. 42, 57 (2012)
J.C. Zill, Z. He, M. Tank, B.H. Ferlez, D.P. Canniffe, Y. Lahav, P. Bellstedt, A. Alia, I. Schapiro, J.H. Gobeck, D.A. Bryant, J. Matysik, Photosynth. Res. 137, 295 (2018)
J.C. Zill, M. Kansy, R. Goss, A. Alia, C. Wilhelm, J. Matysik, Photosynth. Res. (2018). https://doi.org/10.1007/s11120-018-0578-9
G. Fehler, M.Y. Okamura, in The Photosynthetic Bacteria, ed. by R.K. Clayton, W. Sistrom (Plenum Press, New York, 1978), pp. 349–378
M.R. Fischer, H.J.M. de Groot, J. Raap, C. Winkel, A.J. Hoff, J. Lugtenburg, Biochemistry 31, 11038 (1992)
J. Matysik, A. Alia, J.G. Hollander, T. Egorova-Zachernyuk, P. Gast, H.J.M. de Groot, Indian J. Biochem. Biophys. 37, 418 (2000)
E. Daviso, G. Jeschke, J. Matysik, in Biophysical Techniques in Photosynthesis, vol. II, ed. by T. Aartsma, J. Matysik (Springer Netherlands, Dordrecht, 2008), pp. 385–399
R.S. Thakur, N.D. Kurur, P.K. Madhu, Chem. Phys. Lett. 426, 459 (2006)
M. Lee, W.I. Goldburg, Phys. Rev. A 140, 1261 (1965)
O.B. Peersen, X. Wu, S.O. Smith, J. Magn. Reson., Ser. A 106, 127 (1994)
S. Paul, R.S. Thakur, M. Goswami, A.C. Sauerwein, S. Mamone, M. Concistrè, H. Förster, M.H. Levitt, P.K. Madhu, J. Magn. Reson. 197, 14 (2009)
M. Leskes, P.K. Madhu, S. Vega, Chem. Phys. Lett. 447, 370 (2007)
K.R. Mote, V. Agarwal, P.K. Madhu, Prog. Nucl. Magn. Reson. Spectrosc. 97, 1 (2016)
C. Coelho, J. Rocha, P.K. Madhu, L. Marfa, J. Magn. Reson. 194, 264 (2008)
E. Vinogradov, P.K. Madhu, S. Vega, J. Chem. Phys. 115, 8983 (2001)
E. Vinogradov, P.K. Madhu, S. Vega, Chem. Phys. Lett. 354, 193 (2002)
M. Leskes, P.K. Madhu, S. Vega, J. Chem. Phys. 125, 124506 (2006)
D. Marion, M. Ikura, R. Tschudin, A. Bax, J. Magn. Reson. 85, 393 (1989)
B.-J. van Rossum, C.P. de Groot, V. Ladizhansky, S. Vega, H.J.M. de Groot, J. Am. Chem. Soc. 122, 3465 (2000)
H. Fuess, D. Hohlwein, S.A. Mason, Acta Cryst. B 33, 654 (1977)
E. Daviso, S. Prakash, A. Alia, P. Gast, G. Jeschke, J. Matysik, Appl. Magn. Reson. 37, 49 (2010)
S. Prakash, A. Alia, P. Gast, H.J.M. de Groot, G. Jeschke, J. Matysik, J. Am. Chem. Soc. 127, 14290 (2005)
S.S. Thamarath, B.E. Bode, S. Prakash, K.B. Sai Sankar Gupta, A. Alia, G. Jeschke, J. Matysik, J. Am. Chem. Soc. 134, 5921 (2012)
F. Lendzian, M. Huber, R.A. Isaacson, B. Endeward, M. Plato, B. Bonigk, K. Möbius, W. Lubitz, G. Feher, Biochim. Biophys. Acta 1183, 139 (1993)
K.B. Sai Sankar Gupta, A. Alia, H.J.M. de Groot, J. Matysik, J. Am. Chem. Soc. 135, 10382 (2013)
M.H. Stowell, T.M. McPhillips, D.C. Rees, S.M. Soltis, E. Abresch, G. Feher, Science 276, 812 (1997)
A. Diller, S. Prakash, A. Alia, P. Gast, J. Matysik, G. Jeschke, J. Phys. Chem. B 111, 10606 (2007)
E. Daviso, S. Prakash, A. Alia, P. Gast, J. Neugebauer, G. Jeschke, J. Matysik, Proc. Natl. Acad. Sci. USA 106, 22281 (2009)
T. Egorova-Zachernyuk, B.J. van Rossum, C. Erkelens, H.J.M. de Groot, Magn. Reson. Chem. 46, 1074 (2008)
A. Alia, P.K. Wawrzyniak, G.J. Janssen, F. Buda, J. Matysik, H.J.M. de Groot, J. Am. Chem. Soc. 131, 9626 (2009)
C. Giessner-Prettre, B. Pullman, J. Theor. Biol. 31, 287 (1971)
M. Ernst, B.H. Meier, in Encyclopedia of NMR, vol. 1, ed. by R.K. Harris, R.E. Wasylishen (Wiley, Chichester, West Sussex, 2012), pp. 44–55
P. Palmas, S. Ilas, E. Girard, Magn. Reson. Chem. 49, 788 (2011)
D. Gräsing, P. Bielytskyi, I.F. Céspedes-Camacho, A. Alia, T. Marquardsen, F. Engelke, J. Matysik, Sci. Rep. 7, 12111 (2017)
S. Prakash, P. Gast, H.J.M. de Groot, J. Matysik, G. Jeschke, J. Am. Chem. Soc. 128, 12794 (2006)
Acknowledgements
Generous support by the Deutsche Forschungsgemeinschaft is acknowledged (MA-4972/2-1 and MA 4972/11-1). PB would like to thank Prof. P.K. Madhu (TIFR Hyderabad) for exciting discussions.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bielytskyi, P., Gräsing, D., Zahn, S. et al. 15N–1H Transfer of Light-Induced Nuclear Hyperpolarization in Frozen Photosynthetic Reaction Centers. Appl Magn Reson 50, 695–708 (2019). https://doi.org/10.1007/s00723-019-1110-x
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00723-019-1110-x