Abstract
These YABBY genes are transcription factors (TFs) that play crucial roles in various developmental processes in plants. There is no comprehensive characterization of YABBY genes in a valuable Chinese orchid herb, Dendrobium officinale. In this study, a total of nine YABBY genes were identified in the D. officinale genome. These YABBY genes were divided into four subfamilies: CRC/DL, FIL, INO, and YAB2. Expression pattern analyses showed that eight of the YABBY genes were strongly expressed in reproductive organs (flower buds) but weakly expressed in vegetative organs (roots, leaves, and stems). DoYAB1, DoYAB5, DoDL1, and DoDL3 were abundant in the small flower bud stage, while DoDL2 showed no changes throughout flower development. In addition, DoDL1-3 genes were strongly expressed in the column, tenfold more than in sepals, petals, and the lip. DoYAB1 from the FIL subfamily, DoYAB2 from the YAB2 subfamily, DoYAB3 from the INO subfamily, and DoDL2 and DoDL3 from the CRC/DL subfamily were selected for further analyses. Subcellular localization analysis showed that DoYAB1-3, DoDL2, and DoDL3 were localized in the nucleus. DoYAB2 and DoYAB3 interacted strongly with DoWOX2 and DoWOX4, while DoYAB1 showed a weak interaction with DoWOX4. These results reveal a regulatory network involving YABBY and WOX proteins in D. officinale. Our data provide clues to understanding the role of YABBY genes in the regulation of flower development in this orchid and shed additional light on the function of YABBY genes in plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Transcription factors (TFs) play crucial roles in the regulation of plant growth and development, as well as responses to abiotic stresses, by regulating target genes when binding to cis-acting elements in their promoters (Franco-Zorrilla et al. 2014). The YABBY genes are plant-specific putative TFs that are responsible for various developmental processes in plants, and form a family of small zinc finger TFs. The role of the first YABBY gene FILAMENTOUS FLOWER (FIL) to be identified was in lateral organ formation in the dicotyledonous model plant Arabidopsis (Chen et al. 1999; Sawa et al. 1999a, b). Subsequently, another YABBY gene, CRABS CLAW (CRC), was shown to be involved in carpel development (Alvarez and Smyth 1999). Only six YABBY genes (FIL, CRC, INNER NO OUTER (INO), YABBY2 (YAB2), YAB3, and YAB5) were identified in Arabidopsis (Bowman 2000), and only eight members in the monocotyledonous model plant rice (Oryza sativa) (Toriba et al. 2007). The YABBY proteins contain a zinc finger domain in the N-terminal and a helix-loop-helix domain in the C-terminal (YABBY domain) (Bowman 2000). Phylogenetic and sequence similarity analyses divided the YABBY genes into five subfamilies (CRC/DL, FIL, INO, YAB2, and YAB5) in Arabidopsis (Bowman 2000). However, only four subfamilies (CRC/DL, FIL, INO, and YAB2) were classified in rice (Toriba et al. 2007). YABBY proteins can bind to the promoters of target genes and show a transcriptional regulatory function. For example, the FIL protein showed DNA-binding activity by binding to the FIL binding element 1 (FBE1) in the promoter region of the MYB75 gene in Arabidopsis (Boter et al. 2015). MsYABBY5 from spearmint (Mentha spicata) was shown to bind to the MsWRKY75 promoter and activated MsWRKY75 promoter activity (Wang et al. 2016). These studies indicate that YABBY genes act as TFs in plants.
Besides these functions, YABBY genes regulate target gene expression in plants (Bowman 2000; Franco-Zorrilla et al. 2014). YABBY proteins are located in the nucleus (Dai et al. 2007; Sawa et al. 1999b). Moreover, YABBY TFs are involved in the response to stress, secondary metabolism, and lateral organ (leaf and flower) morphogenesis. Three YABBY genes (GmYABBY3, GmYABBY10, and GmYABBY16) from soybean (Glycine max) are upregulated by the following factors: drought, NaCl, and abscisic acid (ABA) (Zhao et al. 2017). The survival percentage of GmYABBY10-overexpressing transgenic Arabidopsis was lower than that of wild-type plants in both polyethylene glycol (PEG) and NaCl treatments (Zhao et al. 2017), suggesting that the YABBY genes play a negative role in the response to PEG and NaCl stresses. Several studies have shown that YABBY genes are involved in secondary metabolism in plants. For example, the Arabidopsis FIL gene is involved in anthocyanin production by regulating an anthocyanin biosynthesis gene, MYB75 (Boter et al. 2015). Seedlings of the fil8 yab3 double mutant and the fil yab2 yab3 yab5 quadruple mutant showed a lack of anthocyanin accumulation (Boter et al. 2015). MsYABBY5 from spearmint displayed a negative role in the synthesis of terpenes (Wang et al. 2016). YABBY genes also modulate the establishment of polarity in leaf and flower morphogenesis. Arabidopsis FIL subfamily genes are involved in regulating the polarity of lateral organs by specifying abaxial cell fate (Nole-Wilson and Krizek 2006; Eshed et al. 2004; Kumaran et al. 2002; Lugassi et al. 2010). CRC genes from eudicots control nectary and carpel development (Alvarez and Smyth 1999; Bowman and Smyth 1999; Meister et al. 2005; Sun et al. 2013). The DROOPING LEAF (DL) gene is closely related to the Arabidopsis CRC gene, regulating carpel specification, and midrib development in rice (Ohmori et al. 2011; Yamaguchi et al. 2004). In addition, the DL gene regulates leaf development in plants. For example, the ectopic expression of a CRC/DL subfamily gene LiYAB1 from Easter lily (Lilium longiflorum) in the rice dl mutant was able to rescue the drooping leaf phenotype of this mutant (Wang et al. 2009). In addition, transgenic 35S::LiYAB1 Arabidopsis plants produced leaves that curled outwardly (Wang et al. 2009). The YAB2 subfamily genes may possess a similar function as CRC/DL subfamily genes in terms of their ability to regulate leaf and carpel development. Ectopic expression of BoYAB1, a YAB2 subfamily gene of giant timber bamboo (Bambusa oldhamii), induced leaf curling and a late flowering phenotype in Arabidopsis (Liu et al. 2020). The ectopic expression of the YAB2 subfamily gene, OsYAB1, in transgenic rice plants induced extra stamens and carpels (Jang et al. 2004). The INO genes have been implicated in the development of ovules and seeds (di Rienzo et al. 2021; Meister et al. 2005).
Orchids have a fascinating floral morphology and unique flower patterning: their flowers contain a fascinating complex structure consisting of a column (i.e., gynostemium), which is a fused male (stamens) and female (pistils) reproductive organ (Mondragon-Palomino 2013). Dendrobium officinale is a Chinese orchid with high medicinal and ornamental value. In this study, we explored the YABBY gene family in D. officinale and investigated their expression patterns, localization, and interacting proteins. These findings may provide clues to explore the function of this gene family in the development of orchid plants and enrich the knowledge about YABBY genes in plants more broadly.
Materials and methods
Plant materials
D. officinale plants were maintained in the experimental station of South China Botanical Garden, Chinese Academy of Sciences (Guangzhou, China). The expression patterns of YABBY genes in flowers were examined in three developmental stages: small flower buds (FB1, about 0.5 cm long), medium flower buds (FB2, about 1 cm long), and full-bloomed flowers (BF), as well as in the lip, petal, sepal, and column tissues of BF flowers. Each was harvested as three biological replications. All samples were immediately frozen in liquid nitrogen for each treatment and then frozen at − 80 °C as quickly as possible for later use.
A. thaliana Columbia (wild-type, WT) plants were used for protoplast isolation. Arabidopsis seeds were surface-disinfected in a mixture of ethanol (75%, v/v) and Triton X-100 (0.01%, v/v), shaken for 10 min, then washed with absolute ethanol once, 70% ethanol solution (v/v) once, and finally with absolute ethanol once, each time for 1 min. Seeds were sown on half-strength Murashige and Skoog (1/2 MS) medium (Murashige and Skoog 1962) and incubated at 4 °C for 3 days in the dark, then transferred to a 22 °C growth chamber and grown under white light (16-h daily). After a week, seedlings were planted in a mixture of peat soil and vermiculite (2:1, v/v). The leaves of 1-month-old plants were used for protoplast isolation.
Identification of YABBY members from the D. officinale genome
The D. officinale protein sequences were downloaded from the National Center for Biotechnology Information (NCBI, https://ftp.ncbi.nlm.nih.gov) genome database. The Arabidopsis YABBY protein sequences were downloaded from the TAIR database (http://www.arabidopsis.org/). The hidden Markov model (HMM) profile was assessed with the HMMER 3.0 software package under default parameters (http://hmmer.janelia.org/). The HMM profile of the YABBY domain (PF04690) was downloaded from the Pfam protein domain database (http://pfam.xfam.org/), to identify YABBY proteins in the D. officinale genome. D. officinale YABBY proteins that contained multiple termination signals and repeats were removed. Candidate YABBY sequences were verified with the BLASTP program in the NCBI database, using the Conserved Domains website tool (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Finally, a total of nine YABBY proteins were identified from the D. officinale genome.
Phylogenetic analysis and sequence alignment
MAFFT 7 software (Katoh and Standley 2013) was used for multiple sequence alignments of full-length amino acid sequences of the YABBY proteins. MEGA version 7 software (Kumar et al. 2016) was used to construct an unrooted phylogenetic tree of aligned YABBY proteins using the neighbor-joining method (Saitou and Nei 1987) with 1000 bootstrap replicates. Four DoDELLA coding sequences from D. officinale were aligned in DNAMAN version 8.0 (default parameters) to classify their characteristic domains.
Gene expression analysis using RNA-Seq data
Transcriptomic data of different tissues of D. officinale were collected from the NCBI Sequence Read Archive (SRA) database under the following accession numbers: flower buds, SRR4431603; leaves, SRR4431601; stems, SRR4431600; roots, SRR4431598. Fragments per kilobase of exon model per million mapped fragments (FPKM) values were used to calculate the expression of YABBY genes and a heatmap was plotted with TBtools (Chen et al. 2020a) based on the FPKM values.
RNA extraction, cDNA synthesis, and quantitative reverse transcription PCR
The aforementioned D. officinale materials were used to extract total RNA using RNAout2.0 reagent (Tiandz Inc., Beijing, China) according to the manufacturer’s protocol. The integrity of extracted RNA was evaluated by agarose gel electrophoresis, and quality was tested using Nanodrop 2000C (Thermo Scientific, Wilmington, DE, USA) before reverse transcription. Purified total RNA was reverse transcribed using the GoScript™ Reverse Transcription System (Promega, Madison, WI, USA) to synthesize first-strand cDNA in accordance with the manufacturer’s protocol. The cDNA of each sample was diluted three times with ddH2O and used as a template for qRT-PCR analysis. qRT-PCR analysis was performed with the Aptamer™ qPCR SYBR® Green Master Mix Kit (Tianjin Novogene Bioinformatics Technology Co. Ltd., Tianjin, China) and the Roche the LightCycler 480 system (Roche, Basel, Switzerland). The 2−ΔΔCT method (Livak and Schmittgen 2001) was used to calculate relative expression. A D. officinale ACTIN gene (NCBI accession number: JX294908) was used as the reference gene (He et al. 2015) to normalize cDNA concentrations. All qRT-PCR reactions were performed with at least three biological replicates. Each data bar represents the mean ± standard deviation (SD) of three biological replicates (n = 3). Specific primers for qRT-PCR are listed in Supplementary Table 1.
Transcriptional activity and yeast two-hybrid assays
Transcriptional activity of full-length YABBY proteins was assessed using a yeast two-hybrid system. YABBY genes (DoYAB1, DoYAB2, DoYAB3, DoDL2, and DoDL3) were constructed into the yeast BD vector, pGBKT7. The constructed pGBKT7-DoYABBYs (DoYAB1, DoYAB2, DoYAB3, DoDL2, and DoDL3) and the empty vector pGBKT7 were separately transformed into yeast strain AH109 (Weidi Biotechnology Co., Shanghai, China) according to the manufacturer’s instructions. Transcriptional activity was evaluated based on growth on single and triple synthetic dropout nutrient medium (SD/-Trp and SD/-Trp/-His/-Ade) and SD/-Trp/-His-/-Ade + X-α-Gal (Coolaber, Beijing, China) medium to detect β-galactosidase (X-α-Gal) activity.
In this paper, the interaction between YABBY and WOX proteins was estimated by yeast two-hybrid assay. YABBY genes (DoYAB1, DoYAB2, DoYAB3, DoDL2, and DoDL3) were separately constructed into pGBKT7 vector (encoding the GAL4 DNA-binding domain, BD) to generate baits. DoWOX2 and DoWOX4 were separately cloned into pGADT7 vector (encoding the GAL4 activation domain, AD) to generate preys. For yeast two-hybrid screening, DoYAB2-BD was used as a bait to screen a D. officinale cDNA library, which was generated using a Matchmaker™ GAL4 Two-Hybrid System 3 & Libraries kit (Clontech) according to the user manual.
For the two-hybrid assay, prey and bait vectors were co-transformed in yeast strain AH109 for protein–protein interaction analysis. Screening or protein interaction test was estimated based on growth on media defined above. In addition, different concentrations of 3-amino-1,2,4-triazole (3-AT) (Coolaber), which is a histidine analog and competitive inhibitor of the His3 gene product, were added to the screening medium corresponding to different genes with transcriptional activity selected above: DoYAB1 with 80 mmol/L 3-AT, DoYAB3 with 10 mmol/L 3-AT, and DoDL2 with 50 mmol/L 3-AT. The primers used to generate bait and prey vectors are listed in Supplemental Table 2.
Subcellular localization analysis
PCR was used to amplify the DoYAB1, DoYAB2, DoYAB3, DoDL2, and DoDL3 gene fragments without a stop codon. The fragments were separately inserted into the EcoR I site of the pSAT6-EYFP-N1 expression vector (Citovsky et al. 2006). The YABBY-YFP fusion construction was driven by the CaMV 35S promoter and was introduced into Arabidopsis protoplasts. The recombinant plasmid and NLS localization marker (NLS-mCherry) were co-transformed into protoplasts of 1-month-old Arabidopsis mesophylls using a PEG-mediated method (Yoo et al. 2007). After incubation in the dark for 12–18 h, fluorescence signals were detected using a Leica TCS SP8 STED 3 × microscope (Leica Microsystems, Wetzlar, Germany). The primers used to generate the five YABBY-YFP fusion proteins are listed in Supplementary Table 3.
Results
Identification and analysis of YABBY genes in D. officinale
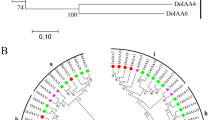
To obtain a comprehensive overview of YBBAY genes in D. officinale, a genome-wide identification was performed in this study. Only nine YABBY genes annotated as YABBY proteins were identified, and these were divided into four subfamilies: YAB2, CRC/DL, INO, and FIL (Fig. 1A). YAB5 subfamily genes were absent in D. officinale (Fig. 1A). Two FIL homologs (DoYAB1 and DoYAB5) were present in the D. officinale genome, with strong bootstrap support (100%) following phylogenetic analysis (Fig. 1A). Four CRC/DL subfamily members were present in D. officinale, and DoDL4 was closely related to AtCRC in Arabidopsis (Fig. 1A). Previous studies showed that YABBY proteins contain a Cys2/Cys2-type zinc finger domain at the N-terminal region and a YABBY domain at the C-terminal region (Sawa et al. 1999b; Toriba et al. 2007). As expected, DoYAB1-3, DoYAB5, DoDL1, and DoDL2 have the two conserved regions (Fig. 1B). However, the sequences of DoDL4 and DoYAB4 protein were diverse, relative to other YABBY proteins: DoDL4 only contained the YABBY domain and DoYAB4 only contained the zinc finger domain (Supplementary Fig. S1). In addition, DoDL3 had a zinc finger domain and an incomplete YABBY domain (Supplementary Fig. S1).
Phylogenetic analyses and sequence alignment of YABBY proteins. A Phylogenetic analysis of YABBY proteins from D. officinale (Do) and Arabidopsis (At). The neighbor-joining (NJ) phylogenetic tree was constructed by MEGA 7 software with 1000 bootstrap replications based on the alignment by MAFFT 7. Bootstrap values higher than 50% are shown. B Sequence alignment of six YABBY proteins from D. officinale using ClustalX 2.1 version. Shaded black and gray indicate identical and similar residues, respectively. Asterisks indicate conserved cysteine residues in the zinc-finger domain, which is indicated by an underline. The YABBY domain is indicated by a red box
Expression pattern of YABBY genes in different organs
To gain insight into the expression and function of the nine YABBY genes, we examined their expression in vegetative organs, including roots, stems, and leaves, as well as in a reproductive organ, flower buds. All nine YABBY genes were absent in the roots (Fig. 2). DoYAB4 was not detected in any organ while the remaining YABBY genes displayed the highest expression in flower buds (Fig. 2). DoDL4 showed a weak signal in flower buds with a FPKM value of 3.2277 but was absent in the three vegetative organs (Fig. 2). The FIL subfamily genes DoYAB1 and DoYAB5 showed similar expression patterns, with the highest expression in flower buds and weak expression in leaves (Fig. 2). These results suggest that YABBY genes may play an important role in flower development in D. officinale.
Expression pattern of YABBY genes in different flower developmental stages and floral organs
The tissues of different developing flowers (Fig. 3A), as well as floral organs (Fig. 4A), were harvested to examine the expression of YABBY genes by qRT-PCR. DoDL4 showed no expression (Figs. 3B and 4B). DoYAB1, DoYAB5, and DoDL1 were strongly detected in FB1 stage and showed downregulated expression during flower development, while DoDL2 showed no differential expression during flower development (Fig. 3B). DoYAB4 and DoDL3 were highly expressed in the BF stage and showed the lowest expression in the FB2 stage (Fig. 3B). One FIL subfamily gene (DoYAB1) showed the highest expression in sepals but weak expression in the lip, whereas the expression of DoYAB5 was strong in the lip (Fig. 4B). DoDL1-3 genes were strongly expressed in the column but showed relatively weak expression in sepals, with the lowest expression in the lip (Fig. 4B).
Expression analysis of YABBY genes from D. officinale in different flower developmental stages by qRT-PCR. A Three stages of flower development used for qRT-PCR analysis. Bar = 1 cm. B Expression pattern of YABBY genes from D. officinale during flower development. Each data bar represents the mean ± standard deviation (SD) of three biological replicates (n = 3). ND, not detected
Expression analysis of YABBY genes in different floral organs. A The floral organs of D. officinale. Se, sepal; pe, petal; co, column. B The expression patterns of YABBY genes in sepals, petals, lip, and column. Each data bar represents the mean ± SD of three biological replicates (n = 3). ND, not detected
To further analyze the function of YABBY genes, DoYAB1 from the FIL subfamily, DoYAB2 from the YAB2 subfamily, DoYAB3 from the INO subfamily, and two genes (DoDL2 and DoDL3) from the CRC/DL subfamily were selected for localization and transcriptional activity analysis, as well as analysis of their interacted proteins.
Localization analysis of YABBY proteins
Since YABBYs are TFs, to examine whether D. officinale YABBY proteins are localized in the nucleus, five YABBY genes (DoYAB1-3, DoDL2, and DoDL3) were cloned into the pSAT6-EYFP-N1 vector to generate YABBY-YFP fusion proteins. Additionally, we used NLS-mCherry as a nuclear marker. The positive control of the free YFP fusion protein was localized throughout the entire cell, including the nucleus and cytoplasm (Supplementary Fig. S2). In contrast, the YABBY-YFP fusion protein and NLS-mCherry signals matched well in the nucleus (Fig. 5). These results suggest that YABBY proteins are localized in the nucleus and might thus be putative transcriptional factors in D. officinale.
Transcriptional activity assay of YABBY proteins in yeast
TFs typically contain a DNA-binding domain and an activation domain, which exhibit transactivation activity. To identify whether YABBY proteins acted as a transcriptional activator, we investigated the transcriptional activity of the five selected YABBY proteins using a yeast two-hybrid assay. The YABBY genes were cloned into the pGBKT7 vector to generate YABBY-GAL4 DNA-binding domain fusion proteins whose transactivation activities were tested in recombinant plasmids transformed into yeast strain AH109. Yeast cells containing pGBKT7 vector (negative control) grew well on SD/-Trp medium but were unable to grow on selective SD/–Trp/-His medium and exhibited no β-galactosidase activity (Fig. 6). Similar to the negative control, DoYAB2-BD and DoDL3-BD displayed slow growth on selective medium and showed no β-galactosidase activity (Fig. 6). In contrast, yeast cells carrying DoYAB1-BD, DoYAB3-BD, and DoDL2-BD grew well on SD/–Trp and SD/–Trp/-His media and displayed β-galactosidase activity (Fig. 6). These data suggest that DoYAB1, DoYAB3, and DoDL2 might act as transcriptional activators in yeast, and probably transcriptional activation activity in D. officinale.
Transcriptional activity analysis of five YABBY proteins (DoYAB1-3, DoDL2, and DoDL3) in yeast. pBD: pGBKT7, as the negative control; DoYAB1-BD: DoYABBY1-pGBKT7; DoYAB2-BD: DoYABBY2-pGBKT7; YAB3-BD: DoYABBY3-pGBKT7; DoDL2-BD: DoDL2-pGBKT7; DoDL3-BD: DoDL3-pGBKT7. SD/-Trp: tryptophan synthetic dropout basic yeast culture medium; SD/-Trp/-His/-Ade: yeast culture medium without tryptophan, histidine, and adenine; SD/-Trp/-His/-Ade + X-α-Gal: yeast culture medium without tryptophan, histidine, and adenine, to which X-α-Gal was added
Interaction of YABBY and WOX proteins
To investigate the protein–protein interaction networks of YABBY in D. officinale, a yeast two-hybrid screen to identify proteins that potentially interact with YABBY was conducted. The DoYAB2 gene showed higher expression (FPKM values = 96) in flower buds than the other YABBY genes (Fig. 2) and showed no transactivation activity (Fig. 6). Consequently, DoYAB2 was selected to identify its interaction with other proteins using yeast two-hybrid screening. Two proteins annotated as WUSCHEL-related homeobox TFs (DoWOX2 and DoWOX4), belonging to the WOX family (Supplementary Fig. S3), were identified. Interestingly, these two WOX genes showed the highest expression in flower buds, similar to YABBY genes (Supplementary Fig. S4). Thus, the interactions of the five YABBY proteins and the two WOX proteins were further explored. The yeast cells carrying either control (pGBKT7 and pGADT7) or fusion plasmids grew well in non-selective SD/–Trp/-Leu medium (Fig. 7A). Only DoYAB1 interacted with DoWOX4, while DoYAB2 and DoYAB3 showed a strong interaction with DoWOX2 and DoWOX4 (Fig. 7B). However, there were no interactions between DoDL proteins and DoWOX proteins (Fig. 7B).
Analysis of the interaction between YABBY and WOX proteins using a yeast two-hybrid assay. An empty vector was used as the control. AD: pGADT7 is a yeast two-hybrid bait expression vector; BD: pGBKT7 is a yeast two-hybrid prey expression vector. Empty: negative control. A SD/-Trp/-Leu: yeast culture medium without tryptophan and leucine; SD/-Trp/-Leu/-His/-Ade: yeast culture medium without tryptophan, leucine, histidine, and adenine. B SD/-Trp/-Leu/-His/-Ade + X-α-Gal: yeast culture medium without tryptophan, leucine, histidine, and adenine, to which X-α-Gal was added; SD/-Trp/-Leu/-His/-Ade + 3 AT: yeast culture medium without tryptophan, leucine, histidine, and adenine, to which different concentrations of 3-AT (DoYAB1 with 80 mmol/L 3-AT, DoYAB3 with 10 mmol/L 3-AT, and DoDL2 with 50 mmol/L 3-AT) were added
Discussion
YABBY gene family in D. officinale
In this study, YABBY genes from D. officinale were identified and characterized. The YABBY gene family is a small family of TFs. For example, six YABBY genes were identified in Arabidopsis (Siegfried et al. 1999) and eight in rice (Toriba et al. 2007). In this study, we identified nine YABBY genes in D. officinale. These D. officinale YABBY genes can be divided into four clades (YAB2, CRC/DL, INO, and FIL). The YAB5 subfamily is present in eudicots, but is absent in monocots and basal angiosperms (Toriba et al. 2007). In this study, the YAB5 subfamily was absent in D. officinale, similar to other orchids such as Apostasia shenzhnica, Phalaenopsis equestris, and Gastrodia elata (Chen et al. 2020b). Chen et al. (2020b) indicated that YAB5 subfamily genes appeared after the divergence of monocots and dicots. Four genes (DoDL1-4) fall into the CRC/DL clade, while only one gene belonged to the CRC/DL subfamily, which has also been found in rice and Arabidopsis (Toriba et al. 2007). Two CRC/DL genes were present in P. equestris and one CRC/DL gene was encountered in A. shenzhnica (Chen et al. 2020b). This indicates that a single ancestral homolog of the CRC/DL gene evolved before the divergence of monocots and dicots, and that the number of DL genes might increase during the development of orchids.
YABBY genes play important roles in leaves, especially during flower development
YABBY genes encode proteins that contain a zinc finger domain and a YABBY domain. For example, all of the YABBY proteins in rice and Arabidopsis have two of these conserved domains (Bowman and Smyth 1999; Toriba et al. 2007). However, DoYAB4 only contained a zinc-finger domain and DoDL4 only had a YABBY domain (Supplementary Fig. S1). In addition, DoYAB4 was not detected in roots, stems, leaves, or flowers and had a FPKM value of zero, while DoDL4 was only detected in flower buds with a FPKM value of 3 (Fig. 2). This suggests that DoYAB4 and DoDL4 might have different functions from other members in the same clade. Except for DoDL4, the other three DoDL genes were strongly expressed in the column (Fig. 4), which is a fused stamen and pistil. In addition, different evidences indicate that DoYABBY genes were highly expressed in flowers, especially in small flower buds (Fig. 2, Fig. 3), which further demonstrated the potential vital role of D. officinale YABBY genes in flower development. These results indicate that YABBY genes play roles during D. officinale flower development. PeDL1 and PeDL2 were strongly detected in the column of another orchid, Phalaenopsis aphrodite (Chen et al. 2021), which was consistent with this study. The AtCRC gene controls carpel development in Arabidopsis (Alvarez and Smyth 1999). A defect of DL in rice causes a homeotic mutation of carpels into stamens in flowers (Nagasawa et al. 2003), suggesting that the CRC/DL subfamily of genes are involved in male and female reproductive organ development. More analyses are needed to reveal the mechanism by which the CRC/DL subfamily of genes controls the development of the gynostemium in orchids.
Analysis of DoYABBY gene expression in different floral organs (Fig. 4) further refined the significance of these genes in flower development. The YAB2 subfamily members play a role in the formation of stamens and carpels. For example, rice OsYAB1, which belongs to the YAB2 subfamily, accumulates in stamen and carpel primordia and causes the formation of supernumerary stamens and carpels when this gene is ectopically expressed (Jang et al. 2004). In contrast, the YAB2 subfamily gene CcYAB2 from Cabomba caroliniana was expressed in vegetative shoots but weakly expressed in floral buds (Yamada et al. 2011). In this study, the YAB2 subfamily gene DoYAB4 was only detected in petals, indicating that YAB2 genes might have undergone functional divergence in plants. The AtFIL gene is implicated in specifying abaxial cell identity and governing floral organ identity in Arabidopsis (Lugassi et al. 2010). OsYAB3 from rice is closely related to the Arabidopsis AtFIL gene, displaying strong signals in leaf primordia, young leaves, and reproductive organs (Dai et al. 2007). The soybean GmFILa gene is primarily distributed in carpel primordia, and in abaxial domains of bracts and sepals. In this study, D. officinale FIL genes were strongly expressed in flower buds (Fig. 1). This suggests that FIL genes play important roles in flower development in plants.
YABBY genes act as transcription factors in plants
The YABBY genes are regarded as putative TFs. Some studies demonstrated that YABBY proteins act as a transcriptional regulator and bind to the cis-element in the promoter of target genes (Boter et al. 2015; Wang et al. 2016). Five of the YABBY proteins (DoYAB1-3, DoDL2, and DoDL3) that were detected were localized in the nucleus, in agreement with previous findings in rice and soybean (Tanaka et al. 2012; Yang et al. 2019). Transcriptional activity analysis showed that DoYAB1, DoYAB3, and DoDL2 acted as transcriptional activators in yeast (Fig. 6). These results suggest that YABBY genes probably function as TFs in D. officinale.
Regulation and interaction networks were present among YABBY and WOX transcription factors
Lateral organs are produced from the shoot apex, including leaves and flowers. YABBY and WOX genes have been shown to control lateral organ formation in plants (Costanzo et al. 2014; van der Graaff et al. 2009; Zhang et al. 2020). Numerous studies have shown that the WOX gene family (such as the WUSCHEL, PRS/WOX3, and MAW/WOX1 subfamily) has a crucial role in plant flower development, including the flower meristem, flower primordium, carpels, sepals, and petals (Besnard et al. 2011; Laux et al. 1996; Nardmann et al. 2004; Vandenbussche et al. 2009). Similarly, the role of YABBY TFs in plant growth and development, especially in flower development, is constantly being emphasized, as evidenced by data obtained thus far (di Rienzo et al. 2021; Nole-Wilson and Krizek 2006; Ohmori et al. 2011; Sun et al. 2013). OsYAB3 showed overlapping expression with OsWOX3 in leaf primordia and young leaves and acts as a transcriptional repressor in regulating the expression of OsYAB3 (Dai et al. 2007). This indicates that a regulatory network between WOX and YABBY is present in plants. In this study, via a yeast two-hybrid assay, YABBY proteins showed an interaction with WOX proteins (Fig. 7). The YABBY genes (excluding DoDL4) and the two WOX genes displayed similar expression patterns, with strong expression in flower buds and weak expression in vegetative organs (roots, stems, and leaves) (Fig. 2 and Supplementary Fig. S4). This suggests that YABBY might interact with WOX to control flower development in D. officinale. This provided a clue that YABBY–WOX interaction networks might be present in D. officinale to regulate flower development. Additionally, these results indicate that a complicated regulatory network involving YABBY and WOX modulates lateral organ development in plants.
In conclusion, we performed a genome-wide identification of YABBY genes in D. officinale and identified nine YABBY genes. These YABBY genes were divided into four subfamilies: YAB2, CRC/DL, INO, and FIL. The expression patterns of YABBY genes in different organs and flower developmental stages, as well as in different floral organs, were analyzed. The localization and transcriptional activity of selected YABBY proteins were estimated. YABBY–WOX interactions were tested by a yeast two-hybrid assay. Our results help to understand the function of YABBY genes in orchids and provide clues that will allow the exploration of the regulatory network in lateral organ development, especially flower development.
Change history
20 June 2023
A Correction to this paper has been published: https://doi.org/10.1007/s00709-023-01875-1
References
Alvarez J, Smyth DR (1999) CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126(11):2377–2386. https://doi.org/10.1242/dev.126.11.2377
Besnard F, Vernoux T, Hamant O (2011) Organogenesis from stem cells in planta: multiple feedback loops integrating molecular and mechanical signals. Cell Mol Life Sci 68(17):2885–2906. https://doi.org/10.1007/s00018-011-0732-4
Boter M, Golz JF, Giménez-Ibañez S, Fernandez-Barbero G, Franco-Zorrilla JM, Solano R (2015) FILAMENTOUS FLOWER is a direct target of JAZ3 and modulates responses to jasmonate. Plant Cell 27(11):3160–3174. https://doi.org/10.1105/tpc.15.00220
Bowman JL (2000) The YABBY gene family and abaxial cell fate. Curr Opin Plant Biol 3(1):17–22. https://doi.org/10.1016/S1369-5266(99)00035-7
Bowman JL, Smyth DR (1999) CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126(11):2387–2396. https://doi.org/10.1242/dev.126.11.2387
Chen QY, Atkinson A, Otsuga D, Christensen T, Reynolds L, Drews GN (1999) The Arabidopsis FILAMENTOUS FLOWER gene is required for flower formation. Development 126(12):2715–2726. https://doi.org/10.1242/dev.126.12.2715
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020a) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13(8):1194–1202. https://doi.org/10.1016/j.molp.2020.06.009
Chen Y-Y, Hsiao Y-Y, Chang S-B, Zhang D, Lan S-R, Liu Z-J, Tsai W-C (2020b) Genome-wide identification of YABBY genes in Orchidaceae and their expression patterns in Phalaenopsis orchid. Genes (Basel) 11(9):955. https://doi.org/10.3390/genes11090955
Chen YY, Hsiao YY, Li CI, Yeh CM, Mitsuda N, Yang HX, Chiu CC, Chang SB, Liu ZJ, Tsai WC (2021) The ancestral duplicated DL/CRC orthologs, PeDL1 and PeDL2, function in orchid reproductive organ innovation. J Exp Bot 72(15):5442–5461. https://doi.org/10.1093/jxb/erab195
Citovsky V, Lee L-Y, Vyas S, Glick E, Chen M-H, Vainstein A, Gafni Y, Gelvin SB, Tzfira T (2006) Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J Mol Biol 362(5):1120–1131. https://doi.org/10.1016/j.jmb.2006.08.017
Costanzo E, Trehin C, Vandenbussche M (2014) The role of WOX genes in flower development. Ann Bot 114(7):1545–1553. https://doi.org/10.1016/10.1093/aob/mcu123
Dai M, Hu Y, Zhao Y, Liu H, Zhou D-X (2007) A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiol 144(1):380–390. https://doi.org/10.1104/pp.107.095737
di Rienzo V, Imanifard Z, Mascio I, Gasser CS, Skinner DJ, Pierri CL, Marini M, Fanelli V, Sabetta W, Montemurro C, Bellin D (2021) Functional conservation of the grapevine candidate gene INNER NO OUTER for ovule development and seed formation. Hortic Res 8(1):29. https://doi.org/10.1038/s41438-021-00467-5
Eshed Y, Izhaki A, Baum SF, Floyd SK, Bowman JL (2004) Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131(12):2997–3006. https://doi.org/10.1242/dev.01186
Franco-Zorrilla JM, Lopez-Vidriero I, Carrasco JL, Godoy M, Vera P, Solano R (2014) DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc Natl Acad Sci USA 111(6):2367–2372. https://doi.org/10.1073/pnas.1316278111
He C, Zhang J, Liu X, Zeng S, Wu K, Yu Z, Wang X, Teixeira da Silva JA, Lin Z, Duan J (2015) Identification of genes involved in biosynthesis of mannan polysaccharides in Dendrobium officinale by RNA-seq analysis. Plant Mol Biol 88(3):219–231. https://doi.org/10.1007/s11103-015-0316-z
Jang S, Hur J, Kim SJ, Han MJ, Kim SR, An G (2004) Ectopic expression of OsYAB1 causes extra stamens and carpels in rice. Plant Mol Biol 56(1):133–143. https://doi.org/10.1007/s11103-004-2648-y
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30(4):772–780. https://doi.org/10.1093/molbev/mst010
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Kumaran MK, Bowman JL, Sundaresan V (2002) YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. Plant Cell 14(11):2761–2770. https://doi.org/10.1105/tpc.004911
Laux T, Mayer KF, Berger J, Jürgens G (1996) The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122(1):87–96. https://doi.org/10.1242/dev.122.1.87
Liu S, Li X, Yang H, Qian Q, Lin X (2020) Ectopic expression of BoYAB1, a member of YABBY gene family in Bambusa oldhamii, causes leaf curling and late flowering in Arabidopsis thaliana. J Hortic Sci Biotechnol 95(2):169–174. https://doi.org/10.1080/14620316.2019.1661289
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Lugassi N, Nakayama N, Bochnik R, Zik M (2010) A novel allele of FILAMENTOUS FLOWER reveals new insights on the link between inflorescence and floral meristem organization and flower morphogenesis. BMC Plant Biol 10.https://doi.org/10.1186/1471-2229-10-131
Meister RJ, Oldenhof H, Bowman JL, Gasser CS (2005) Multiple protein regions contribute to differential activities of YABBY proteins in reproductive development. Plant Physiol 137(2):651–662. https://doi.org/10.1104/pp.104.055368
Mondragon-Palomino M (2013) Perspectives on MADS-box expression during orchid flower evolution and development. Front Plant Sci 4:377. https://doi.org/10.3389/fpls.2013.00377
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Plant Physiol 15(3):473–497. https://doi.org/10.1111/j.1399-3054.1962.tb08052.x
Nagasawa N, Miyoshi M, Sano Y, Satoh H, Hirano H, Sakai H, Nagato Y (2003) SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130(4):705–718. https://doi.org/10.1242/dev.00294
Nardmann J, Ji J, Werr W, Scanlon MJ (2004) The maize duplicate genes narrow sheath1 and narrow sheath2 encode a conserved homeobox gene function in a lateral domain of shoot apical meristems. Development 131(12):2827–2839. https://doi.org/10.1242/dev.01164
Nole-Wilson S, Krizek BA (2006) AINTEGUMENTA contributes to organ polarity and regulates growth of lateral organs in combination with YABBY genes. Plant Physiol 141(3):977–987. https://doi.org/10.1104/pp.106.076604
Ohmori Y, Toriba T, Nakamura H, Ichikawa H, Hirano H-Y (2011) Temporal and spatial regulation of DROOPING LEAF gene expression that promotes midrib formation in rice. Plant J 65(1):77–86. https://doi.org/10.1111/j.1365-313X.2010.04404.x
Saitou N, Nei M (1987) The neighbor-joining method - a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Sawa S, Ito T, Shimura Y, Okada K (1999a) FILAMENTOUS FLOWER controls the formation and development of Arabidopsis inflorescences and floral meristems. Plant Cell 11(1):69–86. https://doi.org/10.1105/tpc.11.1.69
Sawa S, Watanabe K, Goto K, Kanaya E, Morita EH, Okada K (1999b) FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev 13(9):1079–1088. https://doi.org/10.1101/gad.13.9.1079
Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews GN, Bowman JL (1999) Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126(18):4117–4128. https://doi.org/10.1242/dev.126.18.4117
Sun W, Huang W, Li Z, Lv H, Huang H, Wang Y (2013) Characterization of a crabs claw gene in basal eudicot species Epimedium sagittatum (Berberidaceae). Int J Mol Sci 14(1):1119–1131. https://doi.org/10.3390/ijms14011119
Tanaka W, Toriba T, Ohmori Y, Yoshida A, Kawai A, Mayama-Tsuchida T, Ichikawa H, Mitsuda N, Ohme-Takagi M, Hirano H-Y (2012) The YABBY gene TONGARI-BOUSHI1 is involved in lateral organ development and maintenance of meristem organization in the rice spikelet. Plant Cell 24(1):80–95. https://doi.org/10.1105/tpc.111.094797
Toriba T, Harada K, Takamura A, Nakamura H, Ichikawa H, Suzaki T, Hirano H-Y (2007) Molecular characterization the YABBY gene family in Oryza sativa and expression analysis of OsYABBY1. Mol Genet Genomics 277(5):457–468. https://doi.org/10.1007/s00438-006-0202-0
van der Graaff E, Laux T, Rensing SA (2009) The WUS homeobox-containing (WOX) protein family. Genome Biol 10(12):248. https://doi.org/10.1186/gb-2009-10-12-248
Vandenbussche M, Horstman A, Zethof J, Koes R, Rijpkema AS, Gerats T (2009) Differential recruitment of WOX transcription factors for lateral development and organ fusion in Petunia and Arabidopsis. Plant Cell 21(8):2269–2283. https://doi.org/10.1105/tpc.109.065862
Wang A, Tang J, Li D, Chen C, Zhao X, Zhu L (2009) Isolation and functional analysis of LiYAB1, a YABBY family gene, from lily (Lilium longiflorum). J Plant Physiol 166(9):988–995. https://doi.org/10.1016/j.jplph.2008.11.011
Wang Q, Reddy VA, Panicker D, Mao H-Z, Kumar N, Rajan C, Venkatesh PN, Chua N-H, Sarojam R (2016) Metabolic engineering of terpene biosynthesis in plants using a trichome-specific transcription factor MsYABBY5 from spearmint (Mentha spicata). Plant Biotechnol J 14(7):1619–1632. https://doi.org/10.1111/pbi.12525
Yamada T, Yokota SY, Hirayama Y, Imaichi R, Kato M, Gasser CS (2011) Ancestral expression patterns and evolutionary diversification of YABBY genes in angiosperms. Plant J 67(1):26–36. https://doi.org/10.1111/j.1365-313X.2011.04570.x
Yamaguchi T, Nagasawa N, Kawasaki S, Matsuoka M, Nagato Y, Hirano HY (2004) The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16(2):500–509. https://doi.org/10.1105/tpc.018044
Yang H, Shi G, Li X, Hu D, Cui Y, Hou J, Yu D, Huang F (2019) Overexpression of a soybean YABBY gene, GmFILa, causes leaf curling in Arabidopsis thaliana. BMC Plant Biol 19:234. https://doi.org/10.1186/s12870-019-1810-2
Yoo S-D, Cho Y-H, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2(7):1565–1572. https://doi.org/10.1038/nprot.2007.199
Zhang T, Li C, Li D, Liu Y, Yang X (2020) Roles of YABBY transcription factors in the modulation of morphogenesis, development, and phytohormone and stress responses in plants. J Plant Res 133(6):751–763. https://doi.org/10.1007/s10265-020-01227-7
Zhao S-P, Lu D, Yu T-F, Ji Y-J, Zheng WJ, Zhang S-X, Chai S-C, Chen Z-Y, Cui X-Y (2017) Genome-wide analysis of the YABBY family in soybean and functional identification of GmYABBY10 involvement in high salt and drought stresses. Plant Physiol Biochem 119:132–146. https://doi.org/10.1016/j.plaphy.2017.08.026
Funding
This work was supported by the National Natural Science Foundation of China (grant number: 32071819), the Natural Science Foundation of Guangdong Province Projects (grant number: 2021A1515012170), and the Guangdong Forestry Science and Technology Innovation Program (2018KJCX032).
Author information
Authors and Affiliations
Contributions
CH conceived and designed the experiments. DZ performed the experiments. CH and DZ co-drafted the initial manuscript. DZ, CS, and GD participated in sample collection and data analyses. JATS provided scientific guidance and advice. JATS and DZ co-revised the manuscript. JD provided suggestions for the manuscript. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Handling Editor: Pengguo Xia
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zeng, D., Si, C., Teixeira da Silva, J.A. et al. Characterization of YABBY genes in Dendrobium officinale reveals their potential roles in flower development. Protoplasma 260, 483–495 (2023). https://doi.org/10.1007/s00709-022-01790-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-022-01790-x