Abstract
Programmed cell death (PCD) is a crucial process in plant development. In this paper, proteolytically related aspects of kinetin-induced PCD in cortex cells of Vicia faba ssp. minor seedlings were examined using morphological, fluorometric, spectrophotometric, and fluorescence microscopic analyses. Cell viability estimation after 46 μM kinetin treatment of seedling roots showed that the number of dying cortex cells increased with treatment duration, reaching maximum after 72 h. Weight of the apical root segments increased with time and was about 2.5-fold greater after 96 h, while the protein content remained unchanged, compared to the control. The total and cysteine-dependent proteolytic activities fluctuated during 1–96-h treatment, which was not accompanied by the changes in the protein amount, indicating that the absolute protein amounts decreased during kinetin-induced PCD. N-ethylmaleimide (NEM), phenylmethylsulfonyl fluoride (PMSF), and Z-Leu-Leu-Nva-H (MG115), the respective cysteine, serine, and proteasome inhibitors, suppressed kinetin-induced PCD. PMSF significantly decreased serine-dependent proteolytic activities without changing the amount of proteins, unlike NEM and MG115. More pronounced effect of PMSF over NEM indicated that in the root apical segments, the most important proteolytic activity during kinetin-induced PCD was that of serine proteases, while that of cysteine proteases may be important for protein degradation in the last phase of the process. Both NEM and PMSF inhibited apoptotic-like structure formation during kinetin-induced PCD. The level of caspase-3-like activity of β1 proteasome subunit increased after kinetin treatment. Addition of proteasome inhibitor MG-115 reduced the number of dying cells, suggesting that proteasomes might play an important role during kinetin-induced PCD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
According to Conradt and Xue (2005), apoptosis in C. elegans is one of the best-known programmed cell death (PCD) processes in animals. It occurs during embryonic and post-embryonic phases and is related to soma and gonad development of adult hermaphrodites. The process involves three genetically distinguishable phases: specification, killing, and execution. During the specification phase, cells are instructed to undergo PCD, during the killing phase cell death process is activated and during the execution one they are completely degraded.
In animals, most of the PCD events are activated via specific ligands through their appropriate death receptors (Kuranaga 2012), while in plants, it seems that during physiological processes related to tissue and organ development, the specification (signaling) phase of cell death is related to plant hormones, especially to a gaseous hormone, ethylene, which controls formation of tracheary elements (Pesquet and Tuominen 2011), and aerenchyma spaces (Drew et al. 2000; Byczkowska et al. 2013; Doniak et al. 2016). The former process is activated directly via ethylene, whereas the latter one indirectly, as the result of flooding and drought stress and is related to death of cortex cells (Drew et al. 2000). However, in both processes, ethylene is produced by plant tissues and, via ethylene-dependent receptors, PCD is initiated (Iqbal et al. 2013). The ethylene impact on aerenchyma formation can be imitated, also in roots of horse bean seedlings (Byczkowska et al. 2013), by application of its precursor, ACC (1-aminocyclopropane-1-carboxylic acid; Poór et al. 2012). Aerenchyma formation, via ethylene (Doniak et al. 2016), is also induced by kinetin (Kunikowska et al. 2013; Doniak et al. 2016). Thus, it seems that ethylene may be a secondary signaling particle in the kinetin-induced PCD (Kin-PCD; Doniak et al. 2016).
The killing phase of animal apoptosis is regulated by Bcl-2 protein family (Galluzzi et al. 2015) and is activated by apoptosome and caspase cascades (Galluzzi et al. 2016), the cysteine proteases with substrate specificity to aspartate residues (Galluzzi et al. 2016). There are initiating caspases (type 2, 8, 9, 10) and executing ones (type 3, 6, 7) as well as caspases which play an important role in pyroptosis (1, 4, 5, 11, 12, and 14; Vartapetian et al. 2011; Galluzzi et al. 2016). Caspases are synthesised as inactive pro-caspases that are activated by appropriate active ones, which cleave them into small or large subunits and then by their dimerisation or oligomerisation active enzymes are formed (Vartapetian et al. 2011; Galluzzi et al. 2016). In apoptosis, apoptosome, a heptameric structure consisting of Apaf-1 protein, ATP, cytochrome c, and procaspase 9 is the most important factor that leads to the activation of caspases, while in pyroptosis inflammasome consisting of NOD-like receptor, inflammasome adaptor protein activating caspase 1 fulfills the same function as apoptosome (Galluzzi et al. 2016).
In plants, the PCD killing phase is also related to the activity of proteolytic enzymes (Fig. S1), i.e., metacaspases, which transduce death signals in the selected cells (Vartapetian et al. 2011). There are two types of metacaspases, with function and structure similar to caspases, however, different enzyme specificity; metacaspases cleave proteins in specific sites near arginine and lysine residues, but not near aspartate, like caspases (Galluzzi et al. 2016). Metacaspases, similarly to caspases, possess histidine (His) and cysteine (Cys) residues in their catalytic centers (Vercammen et al. 2004). Both types of metacaspases are synthesised as inactive proenzymes. Type II metacaspases, the positive regulators of PCD, like caspases, are autocatalytically activated, while type I, the negative regulators of PCD, are activated by oligomerisation.
The execution phase of PCD described in animals, which is related to the final degradation of cells, is also observed during PCD in plants. During this phase, nuclei degradation is the most important event (van Doorn 2005). There are crucial nucleases which are involved in PCD-dependent condensation and aggregation of nuclear chromatin as well as its transition to the periphery of the nucleus and then its degradation to smaller fragments and/or its breakdown into small and single-stranded (ss)DNA fragments (Domínguez and Cejudo 2012). In apoptosis (Galluzzi et al. 2015) and in apoptosis-like PCD process in plants (Reape and McCabe 2008), DNA is degraded into mono- or oligonucleosomal (140–180; Aleksandrushkina and Vanyushin 2009) or into 50 kbp (Reape and McCabe 2010) fragments. In plant PCD, DNA is degraded by two (I, II) types of endonucleases. The first one is Zn2+-dependent (with the maximum activity at acidic pH) and the other one is Ca2+-dependent (with the maximum activity at basic pH). The type II nucleases, which appear during the first phase of PCD, are specific, mainly, to ssDNA (Sakamoto and Takami 2014). The type I of nucleases, which appear in the second step of PCD, are specific to RNA, ssDNA, and sometimes to double-stranded (ds)DNA (Domínguez and Cejudo 2012). During the kinetin-induced PCD, endonucleolytic activity along with the exonucleolytic one was observed; thus, the internucleosomal DNA degradation was not noted (Doniak et al. 2014).
In the present study, some hallmarks of the killing phase of Kin-PCD were characterised. Using the PCD plant model system involving the kinetin-induced death of cortex cells of 2-cm-long apical parts of horse bean seedling roots, the effects of Z-Leu-Leu-Nva-H (MG115), an inhibitor of proteasomes, N-ethylmaleimide (NEM), an inhibitor of cysteine proteases, and phenylmethylsulfonyl fluoride (PMSF), an inhibitor of serine proteases on cell viability, activity of β5 and β1 proteasome subunits, total and serine- and cysteine-dependent proteolytic activities as well as total protein amounts were studied.
Material and methods
Plant material, germination, treatment, and calculation
Twenty seeds of Vicia faba ssp. minor (horse bean) cv. Nadwiślański were dark-germinated in an incubator for 3 days in Petri dishes (15 cm in diameter and 3 cm high) on two blotting papers moistened with distilled water (2 ml per seed). Then, 6 of the 3-day-old seedlings (Fig. 1A) with nearly equal root length (2.0 ± 0.3 cm) were transferred into a glass container (8 cm in diameter and 4 cm high) with two blotting papers moistened with 10 ml of water or with 10 cm3 of water solution of 46 μM kinetin (Fig. 1B’–E’) or with kinetin together with MG115 (10 μM), NEM (10, 50 and 100 μM), or with PMSF (10, 50 and 100 μM) and cultivated at 23 ± 1 °C and 92 ± 2% of relative humidity.
Three-day-old (A), 24-, 48-, 72-, and 96-h-Ctrl (B–E) and of 24-, 48-, 72-, and 96-h-Kin (B’–E’) series of V. faba ssp. minor seedlings as well as PCD indices (presented as color graphs) of root cortex cells of the root apical segments of 0-h-Ctrl (A”) and of 24-, 48-, 72-, and 96-h-Kin (B”–E”) series; arrows indicate cutting sites on the root apical segments; 0, 50, and 100% indicate the percentage of cells; green, yellow, and red color relates to the number of live, dying, and dead cells, respectively. Scale bars = 20 mm
Analyses were carried out using the 2-cm-long apical parts of seedling roots (root apical segments) in two sets of experiments.
In the first set of experiments, the seedlings were treated for 72 h with Kin alone (72-h-Kin) or with MG115 (72-h-Kin/MG115), NEM (72-h-Kin/NEM), and PMSF (72-h-Kin/PMSF), the viability of cortex cells of root apical segments, protein amounts and activity of proteasome subunits as well as DAPI positive structures (DPSs) formation were characterised and the results were compared to those from the untreated (72-h-Ctrl) ones. In the second set of analyses, the measurements were taken after 1-, 2-, 3-, 6-, 12-, 18-, 24-, 48-, 72- and 96-h treatment with kinetin (1–96-h-Kin); the length and weight of root apical segments, viability of root cortex cells, total and cysteine proteolytic activities, and protein amounts were assessed and the results were compared to those obtained after 0 h (0-h-Ctrl) or to those after 0-, 24-, and 96-h (0–96-h-Ctrl) cultivated without kinetin.
The results of cytomorphologic measurements were statistically verified using the Student’s t test while metabolic ones using Mann-Whitney U test. The P value was applied to show significant differences between the results.
Cell viability estimation
Fresh root apical segments were stained with the mixture of 100 μg ml−1 acridine orange (AO) and 100 μg ml−1 ethidium bromide (EB) in phosphate buffer (PHB, 0.1 M, pH 7.4) for 4 min, then fixed with 2.0% glutardialdehyde (POCH, Poland) in PHB for 15 min. Free-hand longitudinal sections of the segments were prepared using a razor blade and nuclei of the cortex cells were photographed under blue light (400–440 nm of B-2A excitation filter) of the Optiphot-2 epifluorescence microscope (Nikon; Japan) equipped with DDX camera and Act1-software (Precoptic, Poland). The photographs were used to measure resultant fluorescence intensity (R.F.I.) of green and orange fluorescence of AO and EB, respectively, in the nuclei with the Scn Image software according to Byczkowska et al. (2013) and Kunikowska et al. (2013). Cell viability was assessed on the basis of 170–180 nuclei from each of the three independent experiments. Chromatin of living cells was green, while of those undergoing PCD was green-yellow, yellow or yellow-orange, and of dead cells dark orange (Fig. 1A”–E”), and it was reflected in greater R.F.I. values. Their numbers were presented as a PCD index.
Protein amount, activity of proteases, and catalytic subunits of proteasome
The root apical segments were rinsed with deionised water, weighed, and frozen on solid carbon dioxide. To estimate the protein amounts and the total protease activities, the samples were homogenised with 100 mM Tris-HCl (pH 7.4) buffer in Eppendorff-like tubes using a plastic pestle (4–8 °C) and then centrifuged at 5000×g for 10 min (4 °C) and residues were re-extracted. Then, supernatants were combined and protein amounts were measured with the Coomassie G-250 reagent according to Bradford (1976).
The total activity of proteases was spectrophotmetrically determined according to Secades and Guijarro (1999) in the presence or absence of PMSF. The reaction mixture containing 50 μl of the supernatant, 200 μl 60 mM MgCl2, 12 μl 100 mM DTT, 970 μl 100 mM Tris-HCl (pH 7.0), and 30 μl of 1% azocasein was incubated for 30 min at 30 °C. The reaction was stopped with 240 μl of 30% acetic acid and the samples were centrifuged for 15 min (5000×g, 4 °C). Absorbance at 360 nm was measured. To prepare the blank sample, acetic acid was added before the supernatant. Additionally, the total activity of proteases was measured with 10 μl of 100 mM PMSF added to the reaction mixture as well as to the blank sample. The total activities of proteases were expressed in absorbance units (U) per mg of protein and per min.
Cysteine protease activities were determined with α-N-benzyl-DL-arginine-p-nitroanilide (BApNA; Sigma), a synthetic chromogenic substrate, according to Fritz et al. (1974) with modifications. The reaction mixture containing 50 μl of the supernatant, 5 μl of 500 mM EDTA, 520 μl 100 mM Tris-HCl (pH 7.1), 5 μl of 100 mM DTT, 5 μl 100 mM PMSF, and 30 μl of 1% BApNA was incubated for 30 min at 37 °C. The reaction was stopped with 240 μl of 30% acetic acid, samples were centrifuged for 15 min (5000×g, 4 °C), and absorbance at 410 nm was measured. The blank sample was prepared with acetic acid added before the supernatant. Activities of cysteine proteases were expressed in micromole of p-nitroaniline (p-NP), released from the substrate, per milligram protein and per minute.
Activities of catalytic β5 (chymotrypsin-like) and β1 (caspase-like) proteasome subunits with Z-GGL-AMC (Sigma) and Z-LLG-AMC (Sigma), respectively, were fluorometrically determined according to Moravec et al. (2009). The root apical segments were rinsed with deionised water and homogenised on ice with 100 mM Tris-HCl (pH 7.4) containing 0.1 mM EDTA, 0.5 mM DTT, 20% glycerol, 0.04% Nonidet P-40, and 5 mM ATP. The samples were centrifuged for 15 min (5000×g, 4 °C) and residues were re-extracted. Of the combined supernatants, 40 μl were supplemented with 140 μl 50 mM HEPES-Tris (pH 8.0) buffer with 5 mM EGTA and with 10 μl of 1 mM β5 or β1 fluorogenic substrates of proteasome. The mixture was incubated for 30 min at 37 °C and the reaction was stopped with 1350 μl of 1% SDS and fluorescence of samples was measured using the fluorescence/luminescence spectrophotometer F-2500 (Hitachi) at 460 nm after excitation at 380 nm. Activities were expressed in arbitrary units (a.u.) of fluorometrically measured fluorescence intensity (F.I.) per milligram protein and per minute.
DAPI staining, determination of DNA content, and analyses
The root apical segments were fixed in cold Carnoy’s fixative (96% ethanol:glacial acetic acid, 3:1) for 1 h and then washed with 96 and 70% ethanol, hydrated and stained for 15 min with 0.2 μg of 4,6-diamidino-2-phenylindol (DAPI) per 1 ml of McIlvaine buffer (0.1 M citric acid and 0.2 M Na2HPO4·12H2O). Then, 200–300 μm-thick free-hand longitudinal sections of the roots were prepared with a razor blade and washed twice with McIlvaine buffer. The slices were put on glass slides with a drop of the buffer and covered with cover slips. Fluorescence micrographs from the specimens were taken under UV lights of UV-2A excitation filters of Nikon epifluorescence microscope (Optiphot-2). At least 120–130 cells from each of the three independent experiments were used to measure nuclear DNA content and to detect and count the DPSs and to measure their profile area (PA) as well as their total and average (amount of DNA per pxl of DPS profile area) DNA content and to assess the distance between the centers of the nuclei and centers of DPSs. Analyses were done using the Science Image (Byczkowska et al. 2013) and ImageJ Software. DNA content was expressed in a.u. of microcytophotometrically measured F.I.
Results
Kinetin influence on weight and protein content
The roots of 3-day-old seedlings in 0-h-Ctrl (Fig. 1A) were 25 ± 3 mm long, after 96-h cultivation without Kin (96-h-Ctrl; Fig. 1E) they reached the length 76.0 ± 8 mm and were longer by about 20 mm than those in 96-h-Kin (Fig. 1E’), whose lengths reached 56.8 ± 8 mm.
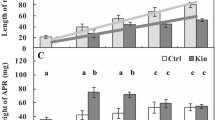
The weight of 3-day-old-seedling roots in 0-h-Ctrl was about 51.7 ± 8 mg. During the cultivation without Kin (0–96-h-Ctrl), the weight of the root apical segments decreased after 24- and 96-h treatment to about 38 and 34.5 mg, respectively. In 1–96-h-Kin, the weight of the root apical segments increased at all times starting from 6 to 96 h and it was greater than in 0–96-h-Ctrl by about 30 and 70%, respectively (Fig. 2a). The average weight of the root apical segments of 1–96-h-Kin was about 2.5-fold greater compared to 0–96-h-Ctrl (Fig. 2a).
Weight of the root apical segments (a) and protein amounts in (b) root apical segments of 0-, 24-, and 96-h-Ctrl and of 1-, 2-, 3-, 6-, 12-, 18-, 24-, 48-, 72-, and 96-h-Kin V. faba ssp. minor seedlings. Means ± SE (n = 8) of two replicates of four independent experiments. * or ** indicate significant differences at P < 0.05 between the results of 1–96-h-Kin and 0-h-Ctrl or 0–96-h-Ctrl, respectively
Amount of proteins in the root apical segments in the 1–96-h-Kin was similar to the 0–96-h-Ctrl throughout the experiment (Fig. 2b).
Effect of kinetin and MG115, NEM, and PMSF on cortex cell viability as well as activities of catalytic subunits and proteases
When the seedlings were treated with 46 μM kinetin for 72 h about 45% of root cortex cells were dying or dead (Fig. 1D”; Fig. 3a). However, after 0, 24, 48, and 96 h, the respective values were 4, 17.5, 39.0, and 36.0% (Fig. 1A”–E”). In the 72-h-Kin/MG115, the number of dying cells was similar to that in the 72-h-Ctrl (Fig. 3a). At the same time, in the 72-h-Kin/MG115, the amount of proteins was higher by about 30% than in the 72-h-Ctrl and 72-h-Kin (Fig. 3b).
PCD indices in cortex (a), amount of proteins (b), and activities of catalytic subunits of proteasome (c) in the root apical segments of 72-h-Ctrl, 72-h-Kin, and 72-h-Kin/MG115 of V. faba ssp. minor seedlings. Means ± SE (n = 6) of two replicates of three independent experiments. * or ** indicate significant differences at P < 0.05 between the results of 72-h-Kin and 72-h-Ctrl or 72-h-Kin/MG115, respectively
Activity of β5 subunit of the proteasome in the extracts from the 72-h-Ctrl was about 0.4 a.u. of F.I. and was similar to that in the 72-h-Kin (Fig. 3c), while activity of β1 subunit of the proteasome in the 72-h-Kin was about 0.3 a.u. of F.I. and was greater by about two times compared to the 72-h-Ctrl (Fig. 3c). In the 72-h-Kin/MG115, the activity of β5 subunits was lower by about 30%, while that of β1 subunits was lower by about 50% compared to the 72-h-Kin (Fig. 3c).
Kin together with 10 or 50 μM NEM (72-h-Kin/NEM10 or 72-h-Kin/NEM50) decreased PCD index by about 30% as compared to the 72-h-Kin (Fig. 4a). However, Kin and 100 μM NEM (72-h-Kin/NEM100) did not significantly change the number of dying cortex cells as compared to the 72-h-Kin (Fig. 4a).
PCD indices in cortex (a, c) and amount of proteins (b, d) in the root apical segments of 72-h-Ctrl and 72-h-Kin or of 72-h-Kin/NEM at 10, 50, and 100 μM (a, b) or of 72-h-Kin/PMSF at 10, 50, and 100 μM (c, d) V. faba ssp. minor seedlings. Means ± SE (n = 6) of two replicates of three independent experiments. * or ** indicate significant differences at P < 0.05 between the results of 72-h-Kin, 72-h-Kin/NEM, 72-h-Kin/PMSF and 72-h-Ctrl or 72-h-Kin/NEM, 72-h-Kin/PMSF and 72-h-Kin, respectively
In the 72-h-Kin/NEM100, the amount of proteins was greater by about 20% than in the 0-h-Ctrl, 72-h-Kin, 72-h-Kin/NEM10, and 72-h-Kin/NEM50 (Fig. 4b).
In the 72-h-Kin/PMSF10 and 72-h-Kin/PMSF50, the number of dying cortex cells was three and five times lower, respectively, as compared to the 72-h-Kin (Fig. 4c) while there were no differences compared to the 72-h-Ctrl. After PMSF treatment, the amount of proteins was similar in all analyzed series (Fig. 4d).
Total activities of proteases in the 0–96-h-Ctrl were constant (Fig. 5a) and were by about 20% higher in 1-, 2-, 18-, and 72-h-Kin and by about 50% lower in 24- and 96-h-Kin than in the 0–96-h-Ctrl (Fig. 5a). PMSF applied to the reaction mixture reduced, on average, by about 2-fold the total activity of proteases in comparison to the 0-h-Ctrl and 1–96-h-Kin (Fig. 5b). Activities in the 1- and 6-h-Kin were greater than in the 0–96-h-Ctrl by about 10%, while in the 24–48- and 72–96-h-Kin, they were lower than in the 0–96-Ctrl by about 20 and 50%, respectively.
Total activity of proteases measured without (a) or with (b) 100 μM PMSF and activity of cysteine proteases (C) in the root apical segments of 0-, 24-, and 96-h-Ctrl as well as of 1-, 2-, 3-, 6-, 12-, 18-, 24-, 48-, 72-, and 96-h-Kin V. faba ssp. minor seedlings. Means ± SE (n = 4) of two replicates of two independent experiments. * or ** indicate significant differences at P < 0.05 between the results of 1–96-h-Kin and 0-h-Ctrl or 0–96-h-Ctrl, respectively
Cysteine protease activities in the 0–96-h-Ctrl were constant (Fig. 5c), while in the 1–96-h-Kin, they were variable. In the 2-h- and 3-h-Kin, they were greater by about 20% while in the 1-h- and 24-h-Kin, they were lower by about 40 and 20% than in respective Ctrl (Fig. 5c).
Effect of kinetin, NEM, and PMSF on DPSs
Staining of the root apical segments with DAPI allowed for the identification of nuclei and DPSs in cortex cells (Fig. 6A–F, A’, B′). In the 72-h-Ctrl, there were 2 DPSs per cell but after kinetin treatment (72-h-Kin, Fig. 6B–D), their number was 3-fold greater (Fig. 7a), while their size was 2-fold larger (Fig. 7b) compared to the 72-h-Ctrl (Fig. 6A, A’). In the 72-h-Kin/NEM10 (Fig. 6C) and 72-h-Kin/NEM50 (Fig. 6D) or 72-h-Kin/PMSF10 (Fig. 6E) and 72-h-Kin/PMSF50 (Fig. 6F), the number of DPSs was lower than in the 72-h-Kin and similar to the 72-h-Ctrl and was on average 2, while their average PA was not changed (Fig. 7b). In the 72-h-Kin (Fig. 6B, B’), 72-h-Kin/NEM10 (Fig. 6C) and 72-h-Kin/NEM50 (Fig. 6D) or 72-h-Kin/PMSF10 (Fig. 6E) and 72-h-Kin/PMSF50 (Fig. 6F), compared to the 72-h-Ctrl (Fig. 6A, A’), DPSs with variable sizes (Fig. 6A’, B’; Fig. 7b) expressed in pxl of PA were observed. Analyses of the distance between the centers of nuclei and the centers of DPSs showed that it was 2-fold longer in the cortex cells of the 72-h-Kin compared to the 72-h-Ctrl, while in the 72-h-Kin/NEM10 and 72-h-Kin/NEM50 or 72-h-Kin/PMSF10 and 72-h-Kin/PMSF50, the distance was on average 3-fold shorter than in 72-h-Kin and on average 2-fold shorter than in the 72-h-Ctrl (Fig. 7c). In the 72-h-Kin, DPSs contained 2-fold greater total amount of DNA than in the 72-h-Ctrl (Fig. 8a). In the 72-h-Kin/NEM10 and 72-h-Kin/NEM50 or 72-h-Kin/PMSF10 and 72-h-Kin/PMSF50, total amount of DNA in DPSs was similar to the 72-h-Ctrl (Fig. 8a). However, in the 72-h-Ctrl and 72-h-Kin, the average DNA content in the DPSs was similar. In the 72-h-Kin/NEM10 and 72-h-Kin/NEM50 or 72-h-Kin/PMSF10 and 72-h-Kin/PMSF50, this value was lower (Fig. 8b).
Microphotographs of the nuclei and DPSs in cortex cells of the root apical segments of control 72-h-Ctrl (A, A’) and kinetin-treated (B–F, B’) V. faba ssp. minor seedlings after DAPI staining: (B, B’) 72-h-Kin, (C) 72-h-Kin/NEM10, (D) 72-h-Kin/NEM50, (E) 72-h-Kin/PMSF10, (F) 72-h-Kin/PMSF50. Arrows indicate DPSs. Scale bars = 20 μm
Number (a), size (b), and distance between nuclei and DPSs (c) in cortex cells of the root apical segments of 72-h-Ctrl, 72-h-Kin, Kin/PMSF10, Kin/PMSF50 and Kin/NEM10, Kin/NEM50 V. faba ssp. minor seedlings. * and ** indicate significant differences at P < 0.05 between the results of 72-h-Kin, 72-h-Kin/NEM, 72-h-Kin/PMSF and 72-h-Ctrl or 72-h-Kin/NEM, 72-h-Kin/PMSF and 72-h-Kin, respectively
DPSs total (a) and average (b) as well as nuclear (c) DNA content in cortex cells of the root apical segments of 72-h-Ctrl and 72-h-Kin, Kin/PMSF10, Kin/PMSF50 and Kin/NEM10, Kin/NEM50 V. faba ssp. minor seedlings. * and ** indicate significant differences at P < 0.05 between the results of 72-h-Kin, 72-h-Kin/NEM, 72-h-Kin/PMSF and 72-h-Ctrl or 72-h-Kin/NEM, 72-h-Kin/PMSF and 72-h-Kin, respectively
The total DNA content in nuclei in the 72-h-Kin was 2-fold lower than in those of the 72-h-Ctrl while in the 72-h-Kin/NEM10 and 72-h-Kin/NEM50 or 72-h-Kin/PMSF10 and 72-h-Kin/PMSF50, the amount of DNA was similar to the 72-h-Ctrl (Fig. 8c).
Discussion
Proteolytic activities in plants and animals, concerning proteasomes and proteases, play the crucial role in cellular metabolism (Beynon and Bond 1999). In plants and animals, they participate in protein degradation (Beynon and Bond 1999) and in transduction of signals of the killing phase of PCD (Coffeen and Wolpert 2004; Galluzzi et al. 2016).
In plants, the proteolytic activities during PCD are related to proteasome subunits, subtilases (saspases and phytaspases), and VPEases. The structure of these enzymes differs from that of caspases but their specificity is similar; thus, these enzymes are plant aspartate-specific proteases with caspase-like activity. Proteasomes related to the ubiquitine-dependent protein degradation are involved in animal apoptosis and in plant PCD (Collazo et al. 2006). Especially one of the three proteasome β subunits with proteolytic activity, β1, which exhibits caspase-3-like activity with specificity to DEVD, a peptide substrate, is susceptible to the inhibitors of caspase-3 (Cai et al. 2014).
The present research showed that proteasomes might play an important role during the kinetin-induced PCD, because proteasome inhibitor MG-115 reduced the number of dying cells and the process was accompanied by a slight increase in the amount of proteins. However, Kurepa and Smalle (2008) reported that inhibition of proteasome activity evoked opposite effect; it did not inhibit cell death but enhanced xylem formation. Inhibiting proteasome activity by MG-132 induced autophagy in HCT116 cells (Williams et al. 2013). The Kin-PCD probably resulted from the fact that caspase-3-like β1 subunit (Moravec et al. 2009), like an initiating caspase (Vartapetian et al. 2011; Galluzzi et al. 2016), can transduce death signals. In plants, metacaspases and caspase-like enzymes are responsible for PCD transduction pathway (Fig. S1; Vartapetian et al. 2011). The suggestion that caspase-3-like β1subunit may play the signaling role in Kin-induced PCD was confirmed by the fact that the level of β1 subunit activity increased after kinetin treatment while that of chymotripsin-like β5 one was not changed. The caspase-3-like activities, in plants, were identified during (i) physiological PCD related to the formation of tracheary elements in Arabidopsis thaliana (Kurepa and Smalle 2008; Fukuda 2016), Zinnia elegans (Twumasi et al. 2010), and Populus tomentosa (Han et al. 2012); (ii) PCD related to biotic stress during response of Solanum tuberosum to Phytophthora infestans (Fernández et al. 2012), of A. thaliana to Pseudomonas syringae pv. tomato (Hatsugai et al. 2009) and of Lycopersicon esculentum to Alternaria sp. (Wang et al. 1996); and (iii) abiotic stress, related to Ce4+ ions in Taxus cuspidata (Ge et al. 2005), to cadmium in A. thaliana (Ye et al. 2013) and to high temperature in tobacco Bright-Yellow 2 cell lines (Vacca et al. 2007).
Constant caspase-like activities in the root apical segments of the 0–96-h-Ctrl are related to physiological PCD leading to differentiation of tracheary elements (Gunawardena 2008), while elevation of these activities after Kin treatment is related to the kinetin-PCD-dependent effect, i.e., aerenchyma formation (Doniak et al. 2016).
In addition, the activities of both proteasome subunits, compared to the 72-h-Kin, and β5 activity compared to the 72-h-Ctrl, were lower when MG-115 was added to the reaction mixture. Chymotripsin-like activity of β5 and caspase-3-like activity of β1 subunit were susceptible to MG-115 proteasome inhibitor, indicating specificity of the applied inhibitor to both subunits.
Subtilases are serine proteases with the amino-acid triad (Asp-His-Ser) in the catalytic center and showing functional similarities to caspases (Schaller et al. 2012). One type/class of the subtilases, saspases, shows specificity to caspase-6- (VEHD, VKMD, VNLD) and caspase-8- (IETD) substrates while another one, phytaspases, shows the specificity to caspase-6- (VEID), caspase-1- (YVAD), caspase-9- (LEHD), and caspase-8- (IETD) substrates (Chichkova et al. 2010; Vartapetian et al. 2011). VPEases are another group of caspase-like proteases in plants which are similar to caspases because of their specificity for aspartate and susceptibility to caspase inhibitors and the fact that in the catalytic center, there are Cys and His residues (Cacas 2010).
Determination of the total activities of proteases showed their fluctuation during 1–96-h treatment with kinetin compared to the 0–96-h-Ctrl, with the maximum activity after 1–2 and 18 h and with the minimum activity after 24 and 96 h of treatment. Determination of cysteine proteases, however, indicated that their activities were rather constant, with the minimum activity observed after 1 and 24 h of treatment and the maximum activity observed after 2–3 h of treatment. The changes of protease activities in 1–96-h-Kin were not accompanied by the changes in the protein amount. This fact indicated that during Kin-PCD, the absolute protein amounts decreased, because the weight of the root apical segments increased but due to the cell death, the number of cells decreased (Kunikowska et al. 2013; Doniak et al. 2016). Dead cell replacement by aerenchyma spaces was accompanied by greater amount of cellulose in 2-fold thicker cell walls of non-dying cortex cells in the root apical segments (Doniak et al. 2016).
Effect of NEM on the kinetin-induced-PCD was less pronounced than that of PMSF while protein amounts in the root apical segments of kinetin-treated seedlings were rather similar, after application of either inhibitor. Only in the series with the highest concentration of NEM the higher amount of proteins were observed. In addition, application of PMSF to the reaction mixture reduced the total protease activities on average by about 50%. In the root apical segments during PCD induced by kinetin, the most important proteolytic activities were that of serine proteases. In plant PCD, these enzymes play an important role as signal transduction factors (Vartapetian et al. 2011), specific to aspartic acid, similarly as caspases and caspase-like proteases (Kacprzyk et al. 2011). Thus, during PCD induced by kinetin, cysteine proteases may be important for protein degradation in the last phase of the process, because their activities were rather constant throughout the experiment and after treatment with the highest concentration of NEM, the amount of proteins was greater while the inhibition of cell death was slightly more effective than after PMSF application.
Animal as well as plant proteases are important transducers of PCD signals (Beynon and Bond 1999). They can activate nucleases which lead to chromatin condensation and nucleic acid degradation (Domínguez and Cejudo 2012; Doniak et al. 2014). It is known that degradation of DNA in Kin-PCD is related to heterochromatin condensation, euchromatin decondensation, and nuclei fragmentation (Kunikowska et al. 2013) without DNA-laddering (Kunikowska et al. 2013; Doniak et al. 2014).
One of the specific hallmarks of the kinetin-induced PCD is the formation of so-called micronuclei (Kunikowska et al. 2013). V. faba ssp. minor is widely used to study plant Kin-PCD (Kunikowska et al. 2013) as well as to perform a micronucleus test (Dhyèvre et al. 2014). In Ctrl, about 2 DPSs per 100 cells were observed. After kinetin treatment, the number of these structures was 3-fold greater, while after the application of proteolytic inhibitors, their number was similar to the Ctrl. Moreover, in the Kin, Kin/PMSF, and Kin/NEM DPSs, they were bigger by their average PAs and DNA content, compared to the Ctrl. However, in all Kin variants, both small and large DPSs were observed. These facts suggested that in the Ctrl during division of meristematic cells, some of the nuclear DNA was exported as chromosomal and extrachromosomal fragments (Shimizu 2011) or as typical micronuclei (Dhyèvre et al. 2014). During Kin-PCD, nuclei of cortex cells can also use apoptotic-like bodies to export DNA. Formation of the apoptotic-like bodies may be related to removal of damaged DNA from nuclei during PCD (Reape and McCabe 2008). Our results may also indicate two processes of the apoptotic-like bodies formation, one connected with cell protection against death and the other with nuclei fragmentation, because after Kin treatment, lower amount of nuclear DNA was observed and the inhibitors of protein synthesis decreased the number of DPSs while their DNA amount was similar to the Ctrl. These results confirmed the fact that kinetin can play a dual role, killing some cells while protecting others. Such role of Kin has been already mentioned (Barciszewski et al. 2007; Doniak et al. 2017), suggesting an interaction of cytokinins with cell membranes as a protection against oxidative damage (Barciszewski et al. 2007).
Our results clearly confirmed that PMSF and NEM inhibited the apoptotic-like structure formation during Kin-PCD because in the Kin/PMSF and Kin/NEM, like in the Ctrl, only micronuclei were observed. However, their sizes were similar to those in Kin and average DNA content was lower than in the apoptotic-like bodies. It was also observed that in Kin-PCD, the distance between DPSs and nuclei was greater compared to the Ctrl whereas in Kin/PMSF and Kin/NEM series that distance was reduced and was significantly smaller than in the Ctrl. The presented results indicated that formation and migration of micronuclei and apoptotic-like bodies in the cortex of horse bean roots depended on cysteine and serine proteases.
Conclusion
The Kin-induced-PCD is vacuolar (Kunikowska et al. 2013) and/or autolytic (Doniak et al. 2014) and is hallmarked with programmed necrosis (Doniak et al. 2016). In the process, the dead and destroyed root cortex cells are replaced with aerenchyma spaces (Doniak et al. 2016). This process may consist of signaling, killing, and execution/degradation phases (Box S1; Fig. S1).
The unchanged amount of proteins, fluctuations of serine- and cysteine-dependent proteases activities; influence of NEM, PMSF, and MG115 on the number of dying cells; relationship between NEM concentration and protein amounts; changes in β1 proteasome subunit activity; and influence of MG115 on its activity indicated that proteolytic activities played important role in the signal transduction and course of Kin-PCD.
On the basis of fluctuations of proteolytic activities, the killing phase of Kin-PCD can be divided into two main steps, i.e., the early (4–24 h) and late (24–96 h). The former seems to be controlled by serine proteases while the latter by cysteine proteases. These results indicated that plant serine-dependent aspartate-specific proteases together with caspase-like ones controlled the initiation of the killing phase of PCD while cysteine ones, with specificity to arginine and lysine, the execution/degradation phase (Box S1; Fig. S1).
Serine and cysteine proteolytic enzyme activities also controlled micronuclei and apoptotic-like body formation, size and DNA content as well as their movement, the most important Kin-PCD hallmarks, related to degradation of cortex cell nuclei during the execution/degradation phase.
These findings will be the starting point to study signaling phase of Kin-PCD, in which the role of cytokinin (HK4) receptors will be analyzed, and which can lead to elucidation of the execution/degradation (Doniak et al. 2014) phase of this process, in which the nucleolytic activities will be characterised. For these studies, the microcytophotometric, spectrophotometric, fluorescence microscopic, and morphologic analyses will be applied.
Abbreviations
- Ctrl:
-
control
- Kin:
-
Kinetin
- MG115:
-
Z-Leu-Leu-Nva-H
- NEM:
-
N-ethylmaleimide
- PCD:
-
Programmed cell death
- PMSF:
-
Phenylmethylsulfonyl fluoride
References
Aleksandrushkina NI, Vanyushin BF (2009) Endonucleases and their involvement in plant apoptosis. Russ J Plant Physiol 56:291–305
Barciszewski J, Massino F, Clark BFC (2007) Kinetin—a multiactive molecule. Inter J Biol Macromol 40:182–192
Beynon R, Bond JS (1999) Proteolytic enzymes, IInd edn. Oxford University press, Oxford
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Byczkowska A, Kunikowska A, Kaźmierczak A (2013) Determination of ACC-induced cell-programmed death in roots of Vicia faba ssp. minor seedlings by acridine orange and ethidium bromide staining. Protoplasma 250:121–128
Cacas JL (2010) Devil inside: does plant programmed cell death involve the endomembrane system? Plant Cell Environ 33:1453–1473
Cai YM, Yu J, Gallois P (2014) Endoplasmic reticulum stress-induced PCD and caspase-like activities involved. Front Plant Sci 5:41–46
Chichkova NV, Shaw J, Galiullina RA, Drury GE, Tuzhikov AI, Kim SH, Kalkum M, Hong TB, Gorshkova EN, Torrance L, Vartapetian AB, Taliansky M (2010) Phytaspase, a relocalisable cell death promoting plant protease with caspase specificity. The EMBO J 29:1149–1161
Coffeen WC, Wolpert TJ (2004) Purification and characterization of serine proteases that exhibit caspase-like activity and are associated with programmed cell death in Avena sativa. Plant Cell 16:857–873
Collazo C, Chacon O, Boras O (2006) Programmed cell death in plants resembles apoptosis of animals. Biotecnol Aplic 23:1–10
Conradt B, Xue D (2005) Programmed cell death. In: Wormbook. The C. elegans Research Community, Wormbook. doi:10.1895/wormbook.1.32.1
Dhyèvre A, Foltête AS, Aran D, Muller S, Cotelle S (2014) Effects of soil pH on the Vicia-micronucleus genotoxicity assay. Mutation Res 774:17–21
Domínguez F, Cejudo FJ (2012) A comparison between nuclear dismantling during plant and animal programmed cell death. Plant Sci 197:114–121
Doniak M, Barciszewska MZ, Kaźmierczak J, Kaźmierczak A (2014) The crucial elements of the ‘last step’ of programmed cell death induced by kinetin in root cortex of V. faba ssp. minor seedlings. Plant Cell Rep 33:2063–2067
Doniak M, Byczkowska A, Kaźmierczak A (2016) Kinetin-induced programmed cell death of cortex cells is mediated by ethylene and calcium ions in roots of Vicia faba ssp. minor. Plant Growth Regul 78:335–343
Doniak M, Kaźmierczak A, Byczkowska A, Glińska S (2017) Reactive oxygen species and sugars may be the messengers in kinetin-induced death of root cortex cells of V. faba ssp. minor seedlings. Biol Plant. doi:10.1007/s10535-016-0654-y
Drew MC, He CJ, Morgan PW (2000) Programmed cell death and aerenchyma formation in roots. Trends Plant Sci 5:123–127
Fernández MB, Daleo GR, Guevara MG (2012) DEVDase activity is induced in potato leaves during Phytophthora infestans infection. Plant Physiol Biochem 61:197–203
Fritz H, Rrautshold I, Werle E (1974) Protease inhibitors. In: Bergmeyer HU (ed) Methods enzymatic analysis. T. 2. Acad. Press, New York
Fukuda H (2016) Signaling, transcriptional regulation, and asynchronous pattern formation governing plant xylem development. Proc Jpn Acad Ser B 92. doi:10.2183/pjab.92.98
Galluzzi L, Bravo-San Pedro JM, Vitale I, Aaronson SA, Abrams JM et al (2015) Essential versus accessory aspects of cell death: recommendations of the NCCD 2015. Cell Death Differ 22:58–73
Galluzzi L, López-Soto A, Kumar S, Kroemer G (2016) Caspases connect cell-death signaling to organismal homeostasis. Immunity 44:221–231
Ge ZQ, Yang S, Cheng JS, Yuan YJ (2005) Signal role for activation of caspase-3-like protease and burst of superoxide anions during Ce4+-induced apoptosis of cultured Taxus cuspidata cells. Biometals 18:221–232
Gunawardena AH (2008) Programmed cell death and tissue remodelling in plants. J Exp Bot 59:445–451
Han JJ, Lin W, Oda Y, Cui KM, Fukuda H, He X (2012) The proteasome is responsible for caspase-3-like activity during xylem development. Plant J 72:129–141
Hatsugai N, Iwasaki S, Tamura K, Kondo M, Fuji K, Ogasawara K, Nishimura M, Hara-Nishimura I (2009) A novel membrane fusion-mediated plant immunity against bacterial pathogens. Genes Dev 23:2496–2506
Iqbal N, Trivellini A, Masood A, Ferrante A, Khan NA (2013) Current understanding on ethylene signaling in plants: the influence of nutrient availability. Plant Physiol Biochem 73:128–138
Kacprzyk J, Daly CT, McCabe PF (2011) The botanical dance of death: programmed cell death in plants. Advances Bot Res 60:169–271
Kunikowska A, Byczkowska A, Kaźmierczak A (2013) Kinetin induces cell death in root cortex cells of Vicia faba ssp. minor seedlings. Protoplasma 250:851–861
Kuranaga E (2012) Beyond apoptosis: caspase regulatory mechanisms and functions in vivo. Genes Cells 17:83–97
Kurepa J, Smalle JA (2008) Structure, function and regulation of plant proteasomes. Biochimie 90:324–335
Moravec R, O’Brien MA, Daily WJ, Scurria MA, Bernad L, Riss TL (2009) Cell-based bioluminescent assays for all three proteasome activities in a homogeneous format. Anal Biochem 387:294–302
Pesquet E, Tuominen H (2011) Ethylene stimulates tracheary element differentiation in Zinnia elegans cell cultures. New Phytol 190:138–149
Poór P, Kovács J, Szopkó D, Tari I (2012) Ethylene signaling in salt stress- and salicylic acid-induced programmed cell death in tomato suspension cells. Protoplasma 250:273–284
Reape TJ, McCabe PF (2008) Apoptotic-like programmed cell death in plants. New Phytol 180:13–26
Reape TJ, McCabe PF (2010) Apoptotic-like regulation of programmed cell death in plants. Apoptosis 15:249–256
Sakamoto W, Takami T (2014) Nucleases in higher plants and their possible involvement in DNA degradation during leaf senescence. J Exp Bot 65:3835–3843
Schaller A, Stintzi A, Graff L (2012) Subtilases—versatile tools for protein turnover, plant development, and interactions with the environment. Physiol Plant 145:52–66
Secades P, Guijarro J (1999) Purification and characterization of an extracellular protease from the fish pathogen Yersinia ruckeri and effect of culture conditions on production. Appl Environ Microbiol 65:3969–3975
Shimizu N (2011) Molecular mechanisms of the origin of micronuclei from extrachromosomal elements. Mutagenesis 26:119–123
Twumasi P, Iakimova ET, Qian T, van Ieperen W, Schel JHN, Emons AMC, van Kooten O, Woltering EJ (2010) Caspase inhibitors affect the kinetics and dimensions of tracheary elements in xylogenic Zinnia (Zinnia elegans) cell cultures. BMC Plant Biol 10:162
Vacca RA, Valenti D, Bobba A, de Pinto MC, Merafina RS, De Garab L, Passarella S, Marra E (2007) Proteasome function is required for activation of programmed cell death in heat shocked tobacco Bright-Yellow 2 cells. FEBS Lett 581:917–922
Van Doorn WG (2005) Plant programmed cell death and the point of no return. Trends Plant Sci 10:478–483
Vartapetian B, Tuzhikov AI, Chichkova NV, Taliansky M, Wolpert TJ (2011) A plant alternative to animal caspases: subtilisin-like proteases. Cell Death Differ 18:1289–1297
Vercammen D, van de Cotte B, De Jaeger G, Eeckhout D, Casteels P, Vandepoele K, Vandenberghe I, Van Beeumen J, Inze D, Van Breusegem F (2004) Type II metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana cleave substrates after arginine and lysine. J Biol Chem 279:45329–45336
Wang H, Li J, Bostock RM, Gilchrista DG (1996) Apoptosis: a functional paradigm for programmed plant cell death induced by a host-selective phytotoxin and invoked during development. Plant Cell 8:375–391
Williams JA, Hou Y, Ni HM, Ding WX (2013) Role of intracellular calcium in proteasome inhibitor-induced endoplasmic reticulum stress, autophagy, and cell death. Pharm Res 30:2279–2289
Ye Y, Li Z, Xing D (2013) Nitric oxide promotes MPK6-mediated caspase-3-like activation in cadmium-induced Arabidopsis thaliana programmed cell death. Plant Cell Environ 36:1–15
Acknowledgments
We thank Ms. M. Fronczak for the help in preparing the manuscript in English. This work was supported by a grant from the University of Łódź, no. 506/1141 and no. B1551000000988.02.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Pavel Dráber
Electronic supplementary material
ESM 1
(RTF 16 kb)
Fig. S1
Simple exemplification of existence signalling, killing and execution phase of Kin-induced of death of cortex cells of the root apical segments of V. faba ssp. minor seedlings in a triangular form. The scheme shows that kinetin induced death signal via cytokinin receptors, then the proteolytic machinery is activated activating the nuclei and cell wall degradation. (GIF 36 kb)
Rights and permissions
About this article
Cite this article
Kaźmierczak, A., Doniak, M. & Kunikowska, A. Proteolytic activities in cortex of apical parts of Vicia faba ssp. minor seedling roots during kinetin-induced programmed cell death. Protoplasma 254, 2273–2285 (2017). https://doi.org/10.1007/s00709-017-1119-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-017-1119-7