Abstract
In this work, we have overexpressed a vesicle trafficking protein, Rab7, from a stress-tolerant plant, Pennisetum glaucum, in a high-yielding but stress-sensitive rice variety Pusa Basmati-1 (PB-1). The transgenic rice plants were tested for tolerance against salinity and drought stress. The transgenic plants showed considerable tolerance at the vegetative stage against both salinity (200 mM NaCl) and drought stress (up to 12 days after withdrawing water). The protection against salt and drought stress may be by regulating Na+ ion homeostasis, as the transgenic plants showed altered expression of multiple transporter genes, including OsNHX1, OsNHX2, OsSOS1, OsVHA, and OsGLRs. In addition, decreased generation and maintenance of lesser reactive oxygen species (ROS), with maintenance of chloroplast grana and photosynthetic machinery was observed. When evaluated for reproductive growth, 89–96 % of seed setting was maintained in transgenic plants during drought stress; however, under salt stress, a 33–53 % decrease in seed setting was observed. These results indicate that PgRab7 overexpression in rice confers differential tolerance at the seed setting stage during salinity and drought stress and could be a favored target for raising drought-tolerant crops.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Throughout their life, plants come across perturbations in environmental conditions which hamper their growth and development. In crop plants, such fluctuations, mostly in the form of soil salinity and drought, reduce yield. Recently, transgenic plants have been raised that have improved survival in the saline and dehydrated environments (Singla-Pareek et al. 2007). However, most abiotic stresses being multigenic traits involve intricate molecular pathways in metabolic configuration which operate in parallel to maintain fitness and enhance tolerance against stress under perturbed environmental conditions (Radomiljac et al. 2013). Therefore, identifying potential candidate gene for stress management becomes a Herculean task.
Salinity stress ultimately leads to cellular dehydration, imbalance in ion homeostasis, and generation of reactive oxygen species (ROS). In response, plant cells usually accumulate compatible osmolytes in the cytoplasm and salt in the vacuole to avoid dehydration and ion imbalance. Antioxidant enzymes are upregulated to defend against the oxidative stress (Niu et al. 1995, Hasegawa et al. 2000, Munns and Tester 2008, Hamaji et al. 2009). Similarly, drought stress inevitably converges in the enhanced generation of ROS from different cellular compartments particularly chloroplasts, mitochondria, and peroxisomes. Under a certain level of drought stress, amplified ROS is maintained under tight control through a cooperative action of cells antioxidant machinery (Cruz de Carvalho 2008). Plant cells effort to minimize cellular ROS has been proposed as one of survival strategy during desiccation stress (Singh et al. 2015).
Vesicle trafficking, in particular endocytosis, is involved in adaptation against environmental stresses like drought (O’Mahony and Oliver 1999, Nahm et al. 2003, Liu et al. 2012, Sui et al. 2013), salinity (Bolte et al. 2000, Nahm et al. 2003, Mazel et al. 2004, George and Parida 2011, Liu et al. 2012, Sui et al. 2013), and elicitor-induced oxidative stress (Kieffer et al. 1997). Rab proteins, which constitute the largest sub-family of the small monomeric GTP-binding proteins, play an important role in intracellular vesicle trafficking and membrane organization (Zerial and McBride 2001). They cycle between membrane and cytosolic compartments and between GTP/GDP bound states. In yeast and animal cells, Rab7 GTPase is localized to late endosomes and lysosomes/vacuoles (Schimmoller and Reizman 1993, Bruckert et al. 2000, Balderhaar et al. 2010) and is involved in the formation of lytic compartments (Bucci et al. 2000, Rosales et al. 2009). In rice, OsRab7 is localized to the vacuolar membrane, suggesting OsRab7 is involved in vesicular transport to the vacuole and plays a role in vacuole biogenesis (Nahm et al. 2003). In Arabidopsis, Rab7-related proteins are located on the vacuolar membrane and regulate vesicle fusions with the vacuole (Randall and Crowell 1999, Mazel et al. 2004), while in Medicago truncatula, the presence of Rab7-related protein was found on both endosomes and tonoplast (Limpens et al. 2009), indicating that Rab7 multi-vesicular bodies are involved in endocytosis.
Reports on the pattern of Rab7 expression at differential developmental stages suggest that OsRab7 is not tissue specific in rice. It expresses at similar levels in shoots and roots of seedlings, and at the stage of maturation. Imposition of salt stress (150 mM) gradually induced Rab7, whereas during dehydration stress, Rab7 levels dramatically increased initially for 4 h and then declined 10 h after stress was imposed (Nahm et al. 2003). Publicly available expression data also indicate that during drought and NaCl stress, OsRab7B1 is highly expressed in all plant tissues such as young leaf, mature leaf, shoot apical meristem, panicle developmental stages (P1–P6), and different stages of seed development (S1–S5) (http://www.ricearray.org).
We have previously reported characterization of PgRab7 in tobacco during salt and osmotic stress (Agarwal et al. 2008). In the present communication, we are validating its role in rice with reference to environmental stresses like salinity and drought with an objective to ascertain its suitability as a candidate gene for raising crops that are tolerant to abiotic stress.
Materials and methods
Plant material
Salt and drought-sensitive rice (Oryza sativa L.) cultivar Pusa Basmati-1 (PB-1) was used as experimental material. The seeds were obtained from Indian Agricultural Research Institute, Pusa, New Delhi, India.
Generation of transgenic rice plants
For rice plant transformation, pCAMBIA1301-PgRab7 recombinant plasmid (Agarwal et al. 2008) was mobilized into Agrobacterium tumefaciens (LBA4404) using the freeze-thaw method (Singla-Pareek et al. 2003). Agrobacterium-mediated transformation of PB-1 rice was carried out using the method by Garg et al. (2002). The un-transformed (WT) plants were also generated at the same time and under same growth conditions. Transgenic rice plants from T3 generation were used for all experiments.
Confirmation of putative transgenic lines by PCR analysis
Total genomic DNA was isolated from leaf tissue of WT plants and putative transformants using a CTAB method (Dellaporta et al. 1983). The putative lines were confirmed by PCR analysis using DNA as template and three pairs of primers. The first pair was PgRab7 specific primer (F′-5′TGGAGAGGACGGATCCATGG3′ and R′-5′CCAAGCTTGCATGCCTGCAG3′). For the second pair, forward primer (F′-5′AACATGGTGGAGCACGACACTCT3′) was designed from the 5′ end of CaMV 35S promoter and reverse primer (R′-5′CTAGCATTCGCAGCCTGACGAAC3′) from 3′ end of PgRab7 while third pair primer was hptII gene specific forward (F′-5′ATGAAAAAGCCTGAACTCACC3′) and reverse (R′-5′CTATTTCTTTGCCCTCGGAC3′) primer. The PCR was performed in a total volume of 25 μl in a My Cycler ™ thermo cycler (Bio-Rad, USA) for 30 cycles. The PCR products were run on 0.8 % agarose gel in 1× TAE buffer, stained with ethidium bromide (EtBr), and visualized under UV light.
Western blot analysis
Total soluble proteins were isolated from the leaf of WT and transgenic rice plants and quantified following Bradford (1976). Total proteins (50 μg) were separated on 12 % SDS-PAGE and transferred onto nitrocellulose membrane. Anti-PgRab7 polyclonal antibodies (1:10,000 dilution) were used as primary antibody while alkaline phosphatase linked anti-rabbit IgG antibodies as secondary antibodies (Sigma Chemicals, USA), and the signals were developed and detected using NBT and BCIP. The anti-PgRab7 polyclonal antibodies were developed as described by Agarwal et al. (2008).
Leaf disc senescence assay for salinity tolerance
The leaf disc senescence assay and chlorophyll measurement were carried out as described by Singla-Pareek et al. (2003). Healthy and fully expanded second leaf from the top of WT and transgenic plants (90-days old) were briefly washed in deionized water, and leaf segments of 1.2 cm length were chopped down and floated in a 10 ml NaCl solution (0, 100, 200, and 400 mM) for 72 h. The chlorophyll content of leaf segments was estimated as described (Arnon 1949).
Seed germination test under salinity stress
Seeds of transgenic and WT plants were surface sterilized. The sterilized seeds were germinated on half strength MS medium (Murashige and Skoog 1962; Caisson Labs, USA) supplemented with 0, 100, 175, and 200 mM NaCl. The seedlings were grown in culture room (16 h light/8 h dark and 28 °C day/21 °C night temperature). After differential growth, these were photographed, and the percentage of seed germination, root and shoot lengths were measured. Values obtained for each parameter in WT plants were taken as 100 %, and the value of transgenic lines was calculated with respect to WT plants.
Analysis of transgenic lines in the presence of 200 mM NaCl and drought stress
Seeds of transgenic and WT plants were germinated on MS medium with or without 200 mM NaCl. The surviving seedlings were transferred to earthen pots and grown in water or in a 200 mM NaCl solution in greenhouse (16 h light/8 h dark and 28 °C day/21 °C night temperature) till the tillering and seed setting stage.
In another set of experiments, seeds of transgenic and WT plants were germinated in earthen pots and grown in a greenhouse for 2 months. Then, 200 mM NaCl solution was applied and maintained up to seed harvest. To maintain 200 mM NaCl concentration (conductivity of 20–22 dS/m) in culture, regular monitoring of electrical conductivity was made through a hand-held conductivity meter (WTW GmbH, Germany), and the medium in the earthen pots was topped up with water in the earthen pots to maintain the concentration of NaCl. For drought stress, watering was completely withdrawn for 12 days in 2-month-old transgenic and WT plants, and then re-watered until the seed maturation stage.
Na+ ion content measurement
Mature (120-days old) WT and transgenic plants grown under water or in a 200 mM NaCl in green house were used for Na+ ion measurement as described by Tripathy et al. (2012). Leaves were collected from three different plants of each type and thoroughly rinsed in deionized water, and the fresh weight of each sample was determined. The samples were dried at 70 °C for 48 h, and the dry weight was measured. The samples were ground to a fine powder and digested in concentrated HNO3 overnight at 120 °C. Samples were then dissolved in HNO3/HClO4 (1:1, v/v) at 220 °C, re-suspended in 5 % (v/v) HNO3, and analyzed for Na+ ion content by using simultaneous inductively coupled argon-plasma emission spectrometry (ICP trace analyzer; Labtam, Australia).
Transmission electron microscopy study
Segments (3 cm away from the tip) of young vegetative leaves (second leaves from top) and root were cut from WT and transgenic plants (2 months old) at 0, 12, and 72 h after salt (200 mM NaCl) treatment and fixed with primary fixative (2.5 % glutaraldehyde; 2.5 % para formaldehyde; 0.1 M sodium phosphate buffer, pH 7.2). After several buffer rinses, the tissue were post fixed in 1 % OsO4 in 0.1 M PBS buffer, for 2 h at 4 °C. The tissues were dehydrated in an ethanol series followed by several changes of propylene oxide and embedded in Spurr’s resin (Botha et al. 2008; Jia et al. 2008). Ultra-thin sections (80 nm) were mounted on copper grids and viewed under transmission electron microscope (JEOL TEM-2100F, Japan) at an accelerating voltage of 120 kV. The leaf and root cells vacuole sizes were measured using ImageJ software (NIH, USA).
Confocal studies in root tip of transgenic plants using CoroNa Green and propidium iodide fluorescent dyes
The CoroNa Green staining was performed according to Park et al. (2009). The CoroNa Green AM (Molecular Probe, USA), a cell-permeable sodium indicator, was used to visualize intracellular Na+ ion accumulation. Seeds of WT and transgenic plants were grown hydroponically in BOD incubator (SANYO Electric Co. Ltd., Japan) at 28 ± 1 °C for 10 days. Ten-day-old seedlings were transferred to Yoshida nutrient solution (Yoshida et al. 1972) supplemented with 150 mM NaCl for 48 h. Root tips were cut into 1 cm and stained with 10 μM CoroNa Green for 3 h at room temperature under dark. Detection of Na+ ion using CoroNa Green was observed under a confocal laser-scanning microscope (Nikon A1R; Tokyo, Japan). The confocal images were taken at 10× magnification at fixed focal length and constant filter intensity. This dye has absorption and fluorescence emission maxima at ~492 and ~516 nm, respectively. A 488 nm excitation laser source was used and emission wavelengths between 505 and 525 nm passed through a fluorescence isothiocyanate green fluorescence filter. To prevent visualization artifacts for each sample, images were captured with equal photomultiplier tube settings, where control sample (0 mM NaCl) showed no green fluorescence. In the same way, propidium iodide (2 μM) was used to visualize the dead cells.
DAB staining of leaf and root tissue under control and different stress condition
The diaminobenzidine (DAB) staining assay was performed according to Deunff et al. (2004). To measure the production of H2O2, seeds of WT and transgenic plants were grown hydroponically in BOD incubator at 28 ± 1 °C for 14 days. Fourteen-day-old seedlings were transferred to Yoshida nutrient solution supplemented either with 150 mM NaCl for 24 h or 1 μM of methyl viologen (MV) for 12 h. The seedlings kept in the Yoshida solution were treated as control. The accumulation of H2O2 was visualized in 1 cm leaf segments and root tissue after incubation for 12 h in citrate buffer (10 mM, pH 6) containing 2.5 mM DAB. After development of the brown color, the chlorophyll was washed by hot 96 % ethanol. The images were taken with microscope, and the intensity of brown color was quantified by using Adobe Photoshop software (Adobe Systems Incorporated, Park Avenue, USA).
RNA isolation, cDNA synthesis, and quantitative real-time PCR
Total RNA from leaf tissues of WT and transgenic plants (P36 and P40) that had been exposed to various stress treatments was isolated using RaFlexTM solutions (Bangalore Genei, India) according to the manufacturer’s protocol. Approximately 5 μg of total RNA was used for the first strand cDNA synthesis using RevertAid™ RNase H minus cDNA synthesis kit (Fermentas Life Sciences, USA) following manufacturer’s protocol. Real-time PCR primers were designed from the 3′ UTR regions using NCBI Primer-BLAST (NIH, USA). cDNA was further diluted ten times, and reaction mixture was prepared adding 5 μl of diluted cDNA, 0.25 μl of 10 μM of each primer, 10 μl of 2× SYBR Green master mix (Applied Biosystems, USA), and 4.5 μl RNAase free water in a final volume of 20 μl. ABI Prism 7500 Sequence Detection System was used for qRT-PCR as follows: 95 °C for 10 min follow by 40 cycles of 95 °C for 30 s, 58 °C for 30 s, 72 °C for 30 s in 96-well optical reaction plates (Applied Biosystems, USA). The rice elongation factor 1α gene (OsEF1α) was used as an internal control. For each sample, three biological replicates and three technical replicates were analyzed. Amplification was checked using dissociation graph and relative expression ratio was calculated using the 2−ΔΔCt method (Livak and Schmittgen 2001). Primer sequences for qRT-PCR are listed in Table S1.
Measurement of photosynthetic performance
Photosynthetic performance of fully expanded leaves of WT and transgenic plants grown under water control, saline (200 mM NaCl), and drought stress (12 days) was measured by using an infrared gas analyzer (IRGA; Licor, Lincoln, USA) following instructions provided by the manufacturer. Data were recorded on a sunny day in the greenhouse conditions. Results were represented as net photosynthesis rate (μmol CO2 m−2 s−1) and electron transport rate (ETR) across PSII.
Statistical analysis
All the statistical significance of wild-type and transgenic lines treated with control and different stresses was performed by one-way ANOVA (Dunnett’s multiple comparison test) and two-way ANOVA (Bonferroni post tests) using GraphPad Prism Software ver. 5 (San Diego, CA, USA).
Results
Over-expression of PgRab7 in rice
The previous studies in model plants A. thaliana and Nicotiana tabacum, demonstrated the involvement of Rab7 in response to salinity and osmotic stress (Mazel et al. 2004, Agarwal et al. 2008, George and Parida 2011). To verify these reports in a crop plant and ascertain the ability of Rab7 to rescue productivity during salinity and drought, the complete ORF of PgRab7 (GenBank Acc No. AY829438) was overexpressed in rice PB-1 genetic background using Agrobacterium-mediated transformation.
The PgRab7 driven by the CaMV 35S promoter was cloned in pCAMBIA1301 vector (Fig. 1a). The preliminary screening of T0 hygromycin-resistant putative rice transformants by genomic PCR analysis using PgRab7 specific primers yielded a 660 bp amplification product (Fig. 1b). Use of CaMV 35S forward and PgRab7 reverse primer produced a 1.04 kb amplicon (Fig. 1c), and hptII specific primers generated a 1 kb amplification product (Fig. 1d). All the transgenic plants showed a single product of expected size. Transgenic status of 22 putative PgRab7 transformants was confirmed by PCR analysis, and the lines were grown to maturity. Immunoblot analysis of WT and transgenic plants (with equal protein, 50 μg) immunodecorated a 22.7 kDa polypeptide with anti-PgRab7 polyclonal antibodies. The transgenic lines showed 10–17-fold higher protein than the WT plants (Fig. 1e, f). Five transgenic lines (P24, P27, P36, P40, and P42) from the transgenic plants pool were randomly selected for the subsequent analysis. As representative, two transgenic lines (P36 and P40) are presented.
Analysis of PgRab7 overexpressing transgenic plants. a Structure of T-DNA region of PgRab7-pCAMBIA1301 construct used for plant transformation. b PCR confirmation of the transgenic plants using gene specific primers. c PCR confirmation of the transgenic plants with CaMV 35S forward and PgRab7 reverse primers. d PCR confirmation of the transgenic plants with hptII specific primers. e SDS-PAGE gel (12 %) profile to show equal amount (50 μg) of total proteins loading. f Western blot analysis of WT plant and transgenic plants using anti-PgRab7 to quantify protein expression. PC positive control (plasmid DNA template), NC negative control (no DNA template). WT1 and WT2, un-transformed control PB-1 sample; lanes P24, P27, P36, P40, and P42 represent putative transformed PB-1 samples; M, 1 kb DNA ladder or protein molecular weight marker
Rice plants overexpressing PgRab7 showed salinity tolerance at vegetative stage
In accordance with our previous report, where enhanced expression of PgRab7 improved the salinity tolerance in N. tabacum (Agarwal et al. 2008), rice plants overexpressing PgRab7 were also tolerant to salinity stress. The salt tolerance of transgenic plants was assessed by a leaf disc senescence assay. Under control conditions, leaf segments of both PgRab7 transgenic and WT plants appeared equally green. Under salt stress, leaf segments of WT plants were completely bleached and turned yellow whereas leaf segments of transgenic plants remained green until 72 h (Fig. 2a). Figure 2b revealed that the total chlorophyll content in the WT plants was 1.56 mg g−1 fr. wt., whereas in transgenic plants P36 and P40, it was 1.57 and 1.6 mg g−1 fr. wt., respectively, in 72 h control conditions. The chlorophyll content was reduced to 0.32 mg g−1 fr. wt. in the WT plants, whereas in the transgenic lines (P36 and P40), the value was 0.78 and 0.6 mg g−1 fr. wt., respectively, under salt stress (100 mM NaCl). Further increasing the salt stress to 200 mM NaCl, the chlorophyll content decreased further to 0.3 mg g−1 fr. wt. in WT and 0.54 and 0.45 mg g−1 fr. wt, in P36 and P40, respectively. Notably, in 400 mM NaCl, the chlorophyll content of WT plants reduced further, whereas in transgenics, it remained unaltered.
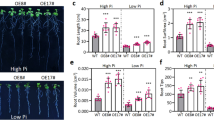
Relative salinity tolerance, morphology, and leaf disc senescence assay of wild-type and PgRab7-transgenic plants under salinity stress. a Representative picture to show phenotypic differences in leaf segments of WT and transgenic plants under 0, 100, 200, and 400 mM NaCl solutions up to 72 h. b Chlorophyll content (mg g−1 fresh weight) of the corresponding leaf segments 72 h after salt stress. Transgenic plants and WT seedlings were grown under control (c and d), 100 mM NaCl (c and e), 175 mM NaCl (c and f), and 200 mM NaCl (c and g). c Diagram showing number of seed germination (15-day-old seedlings), (d, e, f, g) shoot and root length (10-day-old seedlings), respectively, of WT and transgenic seedlings subjected to salt stress as mentioned above. Values are mean ± SD (n = 3). An asterisk indicates that the mean value was significantly different from that of the WT plants when analyzed by the two-way ANOVA (Bonferroni post tests) and one-way ANOVA (Dunnett’s multiple comparison test). *P < 0.05, **P < 0.01, ***P < 0.001
To further check the salt tolerance of transgenic seedlings, seeds of WT and transgenic plants (P36 and P40) were germinated and grown in MS medium supplemented with 0, 100, 175, and 200 mM NaCl. A total of 12 seeds were taken from each line, and the germination was observed till 15 days. Under control and 100 mM NaCl, all transgenic plants and WT seeds showed 100 % germination (Fig. S1A, B and Fig. 2c). In 175 mM NaCl stress, germination of WT seeds was reduced to 33 % where as for the transgenic plants it was 91–100 % (Fig. S1C and Fig. 2c). Further increasing the salt stress (200 mM NaCl), seeds of WT plants did not germinate whereas seeds of transgenic plants germinated up to 80–91 % (Fig. S1D and Fig. 2c).
Morphological growth parameters such as root and shoot length were also recorded in transgenic and WT plants 10 days after exposure to different levels of salt stress. Under untreated control conditions, transgenic lines showed up to 29 % longer shoot and 10 % longer root as compared to WT plants (Fig. S1E and Fig. 2d). Under saline (100 mM NaCl) stress, transgenic plants showed up to 1.5 % longer shoot and 29 % longer roots compared to WT plants (Fig. S1F and Fig. 2e). Further increase in the salt stress (175 and 200 mM NaCl), enhanced the difference in length of shoot and root between transgenic and WT seedlings (Fig. S1G, H and Fig. 2f, g).
Sodium accumulation in transgenic rice seedlings under saline stress
Salt stress induced accumulation of Na+ ion was measured in the root of transgenic plants (P36 and P40) and WT seedlings using CoroNa Green fluorescent dye, which fluoresces upon binding with Na+ ion. The roots from rice seedlings hydroponically grown for 10 days followed by salt (150 mM NaCl) exposure for 48 h were taken for Na+ measurement. Propidium iodide (PI) was used to assay for non-viable cells. While cells of roots in WT plants showed more fluorescence with both the dyes indicating higher Na+ ion accumulation and correspondingly higher cell death, transgenic plants showed significantly reduced accumulation of Na+ (less fluorescence) and fewer dead cells were observed in the root tip region (Fig. 3a). In control condition, there was no significant difference in green fluorescence in WT plants and transgenic lines. Under salt stress, transgenic lines showed 71–75 % less green fluorescence as compared to WT plants (Fig. 3b). Similarly, in control condition, root tips of transgenic lines showed 32–68 % less PI binding than the WT plants and maintained a lower number (35–46 % lower compared to WT plants) of dead cells even under saline condition (Fig. 3c), suggesting that transgenic lines accumulated less Na+ ion in the root tips in comparison with WT plants under saline conditions.
Status of intracellular sodium in wild-type and PgRab7-transgenic plants under salt stress. Intracellular Na+ ion and dead cell accumulation was characterized using CoroNa Green fluorescence dye and PI. a Root tips of WT and transgenic plants stained with CoroNa Green and PI showing accumulation of more Na+ ion inside the root cells of WT and less in transgenic cells. b Densitometric quantification of green florescence color in root tips showed enhanced accumulation of Na+ ion in WT seedlings under 150 mM NaCl stress for 48 h as compared to transgenic plants. c Densitometric quantification of red color in root tips showed enhanced cell death in WT seedlings under 150 mM NaCl stress for 48 h as compared to transgenic plants. d Na+ ion content in the leaves of WT and transgenic plants exposed to water control or 200 mM NaCl after 2 months of salinity treatment. BF bright field, PI propidium iodide. Values are mean ± SD (n = 3). An asterisk indicates that the mean value was significantly different from that of the WT plants when analyzed by the one-way ANOVA (Dunnett’s multiple comparison test). *P < 0.05, *** P < 0.001
The Na+ ion accumulation was also measured in whole leaves of WT and PgRab7 transgenic plants. Under control condition, Na+ ion concentrations were not significantly different in leaves of WT plants and transgenic plants (P36 and P40), whereas, in 200 mM NaCl, Na+ ion concentration in the leaves of PgRab7 overexpressing transgenic plants was twofold less than WT plants (Fig. 3d).
Leaf and root cells in transgenic rice maintain the integrity of vacuole at 200 mM NaCl stress
A comparative study of ultrastructure of rice leaf and root cells was carried out between WT plants and transgenic plants (P36 and P40) at different time periods following 200 mM NaCl stress to check the effect of salt stress on vacuolar size, if any. In leaf cells harvested just after imposition of salt stress (0 h), the vacuole size of WT plants and transgenic plants was nearly equal. However, with increase in the duration of imposed stress, the vacuole size of WT plants increased up to 12 h followed by a significant decrease at 72 h, and at this time point the vacuoles in the majority of cells were completely ruptured. In transgenic plants, the vacuole size continued to increase and stayed intact up to 72 h (Fig. 4a). In root cells, at 0 and 12 h of NaCl stress, the vacuole size of WT plants was smaller as compared to transgenic lines and showed complete disruption at 72 h, whereas in transgenic plants, the vacuole size remained intact (Fig. 4b).
Effect of salt stress (200 mM NaCl) on the leaf and root cells vacuole size of wild-type and PgRab7-transgenic plants at different time intervals. a Leaf cell vacuole size of WT and transgenic plants under control and 200 mM NaCl at 0, 12, and 72 h. b Root cell vacuole of WT and transgenic plants under control and 200 mM NaCl at the time points indicated above. Values are mean ± SD (n = 5). An asterisk indicates that the mean value was significantly different from that of the WT plants when analyzed by the one-way ANOVA (Dunnett’s multiple comparison test). **P < 0.01, ***P < 0.001
Transgenic seedlings accumulate less H2O2 under salinity and oxidative stress
Abiotic stresses cause ROS accumulation. To check cellular ROS status, DAB staining of leaf and root tissues was done during salt and oxidative stress induced by NaCl and MV, respectively. Equal-sized green leaf were taken from 14-day-old plants (control) and from plants treated for 24 h with 150 mM NaCl and for 12 h with 1 μM MV and stained with DAB solution for 12 h. Under control conditions, leaf tissue of transgenic plants showed 12–13 % lesser brown color than the WT plants. However, after salt and MV stress, transgenic plants showed 14–20 and 20–23 % less brown color with respect to WT plants (Fig. 5a, c). This result suggests that transgenic lines experience less oxidative stress in comparison with WT plants under stress. Similar results were obtained with root tissues also (Fig. 5b).
Salinity (NaCl) and oxidative (MV) stress induced H2O2 accumulation in wild-type and PgRab7-transgenic seedlings. a A single representative leaf segment from WT and transgenic seedlings for control, 150 mM NaCl for 24 h, and 1 μM of MV for 12 h treatments is shown. A significantly lower reddish brown color development in transgenic seedlings is indicative of less H2O2 accumulation. b A significantly less reddish brown color developed in root tissue of transgenic lines under different stresses mentioned above. c Densitometric quantification of color in corresponding leaves showed enhanced accumulation of H2O2 in WT seedlings under all types of stress as compared to transgenic plants. MV methyl viologen. Values are mean ± SD (n = 3). An asterisk indicates that the mean value was significantly different from that of the WT plants when analyzed by the two-way ANOVA (Bonferroni post tests). ***P < 0.001
PgRab7 transgenic maintain the chloroplast grana intact at 200 mM NaCl stress
Reduction in primary production is directly linked to photosynthetic performance, particularly structural integrity of photosynthetic apparatus. Therefore, the chloroplast ultrastructure was compared at different time points of salt stress (200 mM NaCl) in WT and transgenic plants P36 and P40 (Fig. 6). In both WT and transgenic plants, the grana stacking was intact up to 12 h of salt stress; however, prolonged salt incubation (72 h) resulted in complete loss in the grana stacking in WT plants whereas in transgenic plants the grana stacking was intact, which probably sustained active photosynthesis.
Effect of salt (NaCl) stress on chloroplast morphology. Transmission electron micrograph images of chloroplast grana stacking of wild-type and transgenic plants (P36 and P40) under 200 mM NaCl at 0, 12, and 72 h intervals. Under 200 mM NaCl stress at 72 h, grana stacking of WT plants was completely disturbed whereas in transgenic plants the grana stacking was maintained intact. Arrows indicates the grana stacking of chloroplast
Growth and yield response of PgRab7 transgenic plants under saline condition
To check the growth and grain productivity of different transgenic plants (P24, P27, P36, P40, and P42) under salinity stress, plants were grown under water control and 200 mM NaCl stress. The germinated seedlings were allowed to grow until the tillering and seed setting stages. Morphologically, the transgenic plants in 200 mM NaCl stress showed shorter height in comparison to the transgenic plants growing in water (Fig. 7a). To confirm that the transgenic plants show better tolerance under salinity, in another set of experiments, 2-month-old WT plants and transgenic lines were supplied with 200 mM NaCl solution up to seed harvest (Fig. 7b). The WT plants did not set seed and showed 100 % yield penalty in NaCl stress as compared to their water control. Transgenic plants showed 33–53 % yield penalty as compared to their respective water controls (Fig. 9).
Growth and photosynthetic response of wild-type and PgRab7-transgenic plants (P24, P27, P36, P40, and P42) grown under control and 200 mM NaCl solution. As representative, two transgenic lines (P36 and P40) were presented. a Comparison of growth of WT and transgenic plants completed life cycle on water control and 200 mM NaCl solution. b Comparison of growth of WT and transgenic plants grown on water control and 200 mM NaCl solution. c Rate of photosynthesis in terms of net carbon assimilation. d Electron transport rate. Values are mean ± SD (n = 5). An asterisk indicates that the mean value was significantly different from that of the WT plants when analyzed by the two-way ANOVA (Bonferroni post tests). *P < 0.05, ***P < 0.001
To investigate the photosynthesis efficiency of WT plants and transgenic lines (P36 and P40) under salinity stress, photosynthetic parameters such as total photosynthesis, and ETR were measured by IRGA. In water controls, transgenic plants showed up to 13 % higher photosynthesis, 26 % higher ETR than the WT plants. During the 200 mM NaCl stress, transgenic lines recorded up to 344 % higher photosynthesis, 40 % higher ETR than the WT plants (Fig. 7c, d). Results indicate that PgRab7 transgenic lines maintained enhanced photosynthesis efficiency under both control and 200 mM NaCl stress compared to WT plants.
Growth and yield response of PgRab7 transgenic plants under drought condition
Rab7 is a house-keeping gene that is primarily involved in intra-cellular docking, a function that may serve as protective machinery during several biotic and abiotic perturbations. To check if the transgenic lines overexpressing PgRab7 also show tolerance to drought stress, watering was completely withdrawn for 12 days in 2-month-old WT and transgenic plants (P24, P27, P36, P40, and P42) and then re-watered until seed maturation stage (Fig. 8a, b). WT plants showed 82.5 % yield penalty in terms of seed weight per plant in drought stress as compared to plants that were irrigated throughout. Transgenic plants showed only 4–11 % yield penalty as compared to their respective water controls (Fig. 9).
Growth and photosynthetic response of wild-type and PgRab7-transgenic plants (P24, P27, P36, P40, and P42) grown under control and drought stress. As representative, two transgenic lines (P36 and P40) were presented. a WT and transgenic plants grown in water and under drought stress for 12 days. b WT and transgenic plants growing in water and after 15 days continuous watering followed by 12 days drought stress. c Rate of photosynthesis in terms of net carbon assimilation. d Electron transport rate. Values are mean ± SD (n = 5). An asterisk indicates that the mean value was significantly different from that of the WT plants when analyzed by the two-way ANOVA (Bonferroni post tests). **P < 0.01, ***P < 0.001
Comparison of seed weight (g) per plant of wild-type and PgRab7 overexpressing transgenic plants (P24, P27, P36, P40, and P42) grown with 200 mM NaCl and 12 days drought stress. WT plants did not survive until harvest under 200 mM NaCl. Each value represents the mean of five replicates ±SD. An asterisk indicates that the mean value was significantly different from that of the WT plants when analyzed by the two-way ANOVA (Bonferroni post tests). *P < 0.05, ***P < 0.001
The photosynthetic efficiency of WT and transgenic plants (P36 and P40) in drought stress was measured by IRGA. In water control, transgenic plants showed up to 13 % higher photosynthesis, 26 % higher ETR than the WT plants. During drought stress, transgenic lines showed up to 150 % higher photosynthesis, 63 % higher ETR than the WT plants (Fig. 8c, d), suggesting higher photosynthetic efficiency under both control and drought stress in PgRab7 transgenic lines as compared to WT plants.
PgRab7 overexpression altered the expression of native rice genes in salt and drought stress
To determine how PgRab7 over-expresser lines modulate native rice genes in control conditions, and in salinity (200 mM NaCl) and drought (12 days) stress, microarray analyses were performed. Our microarray data (unpublished results) suggest that ion transporters (OsNHX1, OsNHX2, OsHKT1, OsHKT2, OsGLRs, OsSOS1, and OsVHA), genes for OsWD domain protein, OsChlA-B binding protein, and genes involved in ROS pathway were differentially regulated at the level of significance. To validate the finding, qRT-PCR analysis were performed. The PgRab7 transgenic lines showed four-fold higher OsRab7 transcript under control and drought stress conditions, whereas there was 2.5-fold more transcript under salinity stress as compared to WT plants (Fig. 10a). Quantification of the transcript of different transporters and anti-porters indicated 2.5–7.5-fold increase in the transcript level of OsNHX1, OsNHX2, OsHKT1, OsHKT2, OsSOS1, and OsVHA under control conditions, whereas the transcript of OsGLRs decreased three-fold as compared to WT plants (Fig. 10b). During salinity stress, OsNHX1, OsSOS1, and OsVHA transcripts increased two- to three-fold, whereas OsNHX2, OsHKT1, OsHKT2, and OsGLRs transcripts decreased three-fold compared to WT plants (Fig. 10c). During drought stress the OsNHX2, OsSOS1, and OsVHA transcript increased 2–2.5-fold, whereas the OsNHX1, OsHKT1, OsHKT2, and OsGLRs transcript decreased 2–3.5-fold in comparison with the WT plants (Fig. 10d). Transcripts encoding the OsWD domain protein increased 7–13-fold in control, salinity, and drought stress as compared to WT plants (Fig. 10e). Transcripts encoding the OsChlA-B binding protein increased by two- to four-fold in control, salinity, and drought stress as compared to WT plants (Fig. 10f). When the transcript level of genes involved in ROS detoxification pathway (OsSOD1, OsAPX, OsCAT1A, and OsCAT1B) were quantified, the transcript level of all four genes was two times higher in transgenic lines under control conditions as compared to WT plants (Fig. 10g). During salinity stress, the transcript level increased 2.5–3.5-fold (Fig. 10h) and under drought stress the transcript level increased 2–2.5-fold as compared to WT plants (Fig. 10i).
Quantitative RT-PCR analysis of key genes involved in different cellular processes in wild-type and transgenic (P36 and P40) plants under salinity (200 mM) and drought stress (12 days). The rice elongation factor 1α gene (OsEF1α) was used as internal control. The mRNA levels for each gene in transgenic plants were calculated relative to the expression in wild-type plants. a OsRab7 gene. b–d Genes involved in ion transport/exchange (OsNHX1, OsNHX2, OsHKT1, OsHKT2, OsGLRs, OsSOS1, and OsVHA1). e WD domain containing gene. f Chlorophyll A-B binding gene. g–i Reactive oxygen species pathway genes (OsSOD1, OsAPX, OsCAT1A, and OsCAT1B). Data is presented as the mean (±SE) of relative transcript level for three independent experiments
Discussion
Our finding that the overexpression of Rab7 both reduced the yield penalty and improved the survival of rice under saline and drought conditions provides further evidence that Rab7 plays an important role in signaling pathways that help protect plants against abiotic stresses. The rationale behind selecting rice for these studies is its cultivation at the global level for staple food. Rab7 overexpression could make a high yielding cultivar tolerant against multiple stresses without compromising on yield, this result both confirmed a function for Rab7 in stress pathways as well as argued for its field applicability.
Tolerance of PgRab7 transgenic plants to salinity stress
The role of vesicle trafficking in relation to various environmental stresses has been analyzed at the mechanistic level (Leyman et al. 1999, Levine et al. 2001, Mazel et al. 2004, Hamaji et al. 2009, George and Parida 2011). To meet the criteria prevailing in field conditions, in our experiments stress was applied from seed germination until seed set stage. In most of the reports on salt stress, particularly in rice, a sensitive cultivar has been shown to senesce within 72 h after 200 mM NaCl treatment (Munns and Tester 2008, Kawano et al. 2009). Our finding on survival of PgRab7 transgenic plants treated with up to 200 mM NaCl from germination to the seed set stage provides first-hand evidence for their value as crops that are tolerant to abiotic stresses (Fig. 2a–g).
Plants overexpressing PgRab7 suffer less yield penalty under salt and drought stress
When transgenic plants constitutively expressing PgRab7 were evaluated for seed setting during salt and drought stress, they were found to have a higher seed yield than similarly stressed WT plants. While a significant level of tolerance was recorded at vegetative stage in both the stresses, imposition of drought stress resulted in a much smaller penalty on seed production than did NaCl stress. Possible interpretation for this observation could be that under severe ionic imbalance during salinity stress, PgRab7 protected the photosynthetic apparatus through reduced Na+ ion accumulation thus allowing plants to sustain photosynthesis, but it was unable to promote the translocation of photosynthate to the sink with optimal efficiency, and this would result in a yield penalty. Under drought stress, the extent of ionic imbalance would be much less as compared to salinity. Thus, under such conditions, overexpression of Rab7 protected the plants at both the vegetative as well as seed set stage. Moreover, other mechanisms like accumulation of osmolytes during drought stress might also protect the transgenic lines from decreased seed set.
Probable mechanism of tolerance to high NaCl stress at vegetative stage
The differential performance of transgenic plants harboring PgRab7 under saline and drought stress led us to explore the mechanistic details of this differential protection. Here, we deduce inferences from experimental observations to propose an explanation for this result, as described below.
Regulating Na+ ion partitioning and maintenance of vacuole
Na+-induced ion toxicity is the primary effect of salt stress in plants, but this problem can be overcome by excluding and/or sequestering sodium. During exclusion, Na+ export to extracellular spaces is controlled by plasma membrane Na+/H+ antiporters such as SOS1 (Wu et al. 1996, Munns 2002, Shi et al. 2003, Cheong and Yun 2007, Park et al. 2008). In sequestration, Na+ ion storage in vacuoles is accomplished by vacuolar Na+/H+ antiporters, e.g., NHX1, and the vacuolar H+-ATPase subunit VHA, which utilizes energy derived from the proton electrochemical gradient across vacuolar membranes (Apse et al. 1999, Zhang and Blumwald 2001, Kader et al. 2006, Duan et al. 2007, Verma et al. 2007, Baisakh et al. 2012). A number of recent reports also suggest that NHX-type of Na+/H+ antiporters play an important role in the regulation of vesicular trafficking, cell expansion, growth, and development in plants (Bassil et al. 2011, 2012).
Membrane trafficking is an essential process in plants that involves the transport of various macromolecules and proteins to their target destination within the cells (Cheung and de Vries 2008). Apart from transporting macromolecules, vesicle trafficking is also involved in driving cellular intoxicants to vacuoles. Being a component of trafficking, Rab7 is involved in membrane docking (Stenmark 2009). Ectopic expression of AtRab7 led to higher accumulation of Na+ in the vacuole of transgenic plants, with higher sodium accumulation in shoots compared to roots (Mazel et al. 2004). An earlier report documented that when a knockout of the Qa-SNARE protein VAM3/SYP22 in Arabidopsis was challenged with salt stress, there was differential accumulation of sodium in roots and shoots (Hamaji et al. 2009). Consistent with this report, our results also show that root tips of hydroponically grown PgRab7 transgenic lines had significantly lower Na+ ion accumulation compared to WT plants (Fig. 3a–c). Our Na+ ion measurement data also reveal that WT plants accumulate more Na+ ion as compared to that of PgRab7 transgenic lines (Fig. 3d), perhaps because a lower amount of Na+ ions enters the root cell of transgenic plants through the GLRs (non-selective cation channels) or SOS1 (Fig. 10b–d).
During salinity stress of suspension-cultured cells of mangrove, there is a rapid increase in vacuolar volume without any change in cellular volume (Mimura et al. 2003). Similarly, the comparative analysis of vacuolar size in WT and transgenic rice plants (at 0, 12, and 72 h) following 200 mM NaCl stress, the vacuole size of transgenic plants increased up to 12 h and the vacuole remained intact even under 200 mM NaCl stress (Fig. 4a, b). The upregulation of NHX1, SOS1, and VHA transcript in PgRab7 transgenic plants suggests that these antiporters may have role in reducing the toxicity inside the cell through selective exclusion and/or sequestering of Na+ ions (Fig. 10b–d).
Maintenance of low cellular ROS level
Intracellular generation of ROS is a key stress signature of plants’ response towards stress (Fedoroff 2006, Gill and Tuteja 2010). There are reports that ectopic expression of antioxidant enzymes enhances the tolerance of plants to salinity stress (Diaz-Vivancos et al. 2013). ROS accumulation in the leaf sections from WT and PgRab7 transgenic plants following salt (NaCl) and oxidative (MV) stress showed, a significant reduction in the ROS pool in the transgenic plants (Fig. 5a–c), indicating better ROS scavenging ability in transgenic plants via upregulation of scavenging enzymes (Fig. 10g–i).
Maintenance of chloroplast structure integrity and photosynthesis
Productivity of a plant is directly correlated with its photosynthetic efficiency, which is tightly linked to the maintenance of structural integrity and organization of chloroplast membrane. Our observation that chloroplasts in the transgenic lines maintain grana stacks up to 72 h during the 200 mM NaCl stress provides strong evidence for better and prolonged photosynthetic performance of transgenic plants expressing PgRab7. Maintenance of the integrity of photosynthetic membranes in PgRab7 transgenic plants during salinity stress may be due to efficient compartmentalization of Na+ into the vacuole, reducing its toxic effects on the chloroplasts. Similarly, efficient scavenging of stress-induced ROS accumulation, which may have resulted from a higher expression of transcripts encoding different antioxidant enzymes during salinity and drought (Fig. 10g–i), could help explain the lesser damage to photosynthetic membranes and consequent maintenance of photosynthetic efficiency in the transgenic lines.
Overall our findings indicate that transgenic rice plants overexpressing PgRab7 were protected against salt and drought stress up to the extent where un-transformed plants did not survive at all. Thus, Rab7 could be an ideal candidate for generating crops tolerant to multiple stresses. We also propose that overexpression of PgRab7 could better protect plants from salinity and drought stress through controlled entry of Na+ ions into the cell involving ion transporters like HKT1, HKT2, and GLRs or selective exclusion of Na+ ions through SOS1 and/or their transportation and storage in the vacuoles through NHX1 and VHA. The upregulation of WD domain protein and chlorophyll A-B binding protein during imposed stress could maintain the structural integrity of photosynthetic apparatus and thus overall photosynthetic activity. The overproduction of ROS during salinity and drought stress was actively scavenged in transgenic lines through improving antioxidant defense machinery, including SOD1, APX, CAT1A, and CAT1B.
Abbreviations
- DAB:
-
3,3-diaminobenzidine
- ETR:
-
electron transport rate
- IRGA:
-
infrared gas analyzer
- MS:
-
Murashige and Skoog
- MV:
-
methyl viologen
- qRT-PCR:
-
quantitative real time-PCR
- ROS:
-
reactive oxygen species
- TEM:
-
transmission electron microscopy
- UV-light:
-
ultra violet light
- WT:
-
wild-type
References
Agarwal PK, Agarwal P, Jain P, Jha B, Reddy MK, Sopory SK (2008) Constitutive overexpression of a stress-inducible small GTP binding protein PgRab7 from Pennisetum glaucum enhances abiotic stress tolerance in transgenic tobacco. Plant Cell Rep 27:105–115
Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 258:1256–1258
Arnon DI (1949) Copper enzymes in isolated chloroplasts. Poly-phenoloxidase in beta vulgaris. Plant Physiol 24:1–15
Baisakh N, RamanaRao MV, Rajasekaran K, Subudhi P, Janda J, Galbraith D, Vanier C, Pereira A (2012) Enhanced salt stress tolerance of rice plants expressing a vacuolar H+-ATPase subunit c1 (SaVHAc1) gene from the halophyte grass Spartina alterniflora Loisel. Plant Biotechnol J 10:453–464
Balderhaar HJK, Arlt H, Ostrowicz C, Bröcker C, Sündermann F, Brandt R, Babst M, Ungermann C (2010) The Rab GTPase Ypt7 is linked to retromer-mediated receptor recycling and fusion at the yeast late endosome. J Cell Sci 123:4085–4094
Bassil E, Ohto MA, Esumi T, Tajima H, Zhu Z, Cagnac O, Belmonte M, Peleg Z, Yamaguchi T, Blumwald E (2011) The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. Plant Cell 23:224–239
Bassil E, Coku A, Blumwald E (2012) Cellular ion homeostasis: emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. J Exp Bot 63:5727–5740
Bolte S, Schiene K, Dietz KJ (2000) Characterization of a small GTP binding protein of the rab 5 family in Mesembryanthemum crystallinum with increased level of expression during early salt stress. Plant Mol Biol 42:923–936
Botha CEJ, Aoki N, Scofield GN, Liu L, Furbank RT, White RG (2008) A xylem sap retrieval pathway in rice leaf blades: evidence of a role for endocytosis? J Exp Bot 59:2945–2954
Bradford M (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Bruckert F, Laurent O, Satre M (2000) Rab7, a multifaceted GTP-binding protein regulating access to degradative compartments in eukaryotic cells. Protoplasma 210:108–116
Bucci C, Thomsen P, Nicoziani P, McCarthy J, van Deurs B (2000) Rab7: a key to lysosome biogenesis. Mol Biol Cell 11:467–480
Cheong MS, Yun DJ (2007) Salt-stress signaling. J Plant Biol 50:148–155
Cheung AY, de Vries SC (2008) Membrane trafficking: intracellular highways and country roads. Plant Physiol 147:1451–1453
Cruz de Carvalho MH (2008) Drought stress and reactive oxygen species: production, scavenging and signaling. Plant Signal Behav 3:156–165
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: version II. Plant Mol Biol Rep 1:19–21
Deunff EL, Davoine C, Dantec CL, Billard JP, Huault C (2004) Oxidative burst and expression of germin/oxo genes during wounding of ryegrass leaf blades: comparison with senescence of leaf sheaths. Plant J 38:421–431
Diaz-Vivancos P, Faize M, Barba-Espin G, Faize L, Petri C, Hernández JA, Burgos L (2013) Ectopic expression of cytosolic superoxide dismutase and ascorbate peroxidase leads to salt stress tolerance in transgenic plums. Plant Biotechnol J 11:976–985
Duan XG, Yang AF, Gao F, Zhang SL, Zhang JR (2007) Heterologous expression of vacuolar H(+)-PPase enhances the electrochemical gradient across the vacuolar membrane and improves tobacco cell salt tolerance. Protoplasma 232:87–95
Fedoroff N (2006) Redox regulatory mechanisms in cellular stress responses. Ann Bot 98:289–300
Garg AK, Kim JK, Owens TG, Ranwala AP, Choi YD, Kochian LV, Wu RJ (2002) Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc Natl Acad Sci U S A 99:15898–15903
George S, Parida A (2011) Over-expression of a Rab family GTPase from phreatophyte Prosopis juliflora confers tolerance to salt stress on transgenic tobacco. Mol Biol Rep 38:1669–1674
Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48:909–930
Hamaji K, Nagira M, Yoshida K, Ohnishi M, Oda Y, Uemura T, Goh T, Sato MH, Morita MT, Tasaka M, Hasezawa SI, Nakano A, Nishimura IH, Maeshima M, Fukaki H, Mimura T (2009) Dynamic aspects of ion accumulation by vesicle traffic under salt stress in Arabidopsis. Plant Cell Physiol 50:2023–2033
Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51:463–499
Jia Y, Yang X, Feng Y, Jilani G (2008) Differential response of root morphology to potassium deficient stress among rice genotypes varying in potassium efficiency. J Zhejiang Univ Sci B 9:427–434
Kader MA, Seidel T, Golldack D, Lindberg S (2006) Expressions of OsHKT1, OsHKT2, and OsVHA are differentially regulated under NaCl stress in salt-sensitive and salt-tolerant rice (Oryza sativa L.) cultivars. J Exp Bot 57:4257–4268
Kawano N, Ito O, Sakagami J (2009) Morphological and physiological responses of rice seedlings to complete submergence (flash flooding). Ann Bot 103:161–169
Kieffer F, Simon-Plas F, Maume BF, Blein JP (1997) Tobacco cells contain a protein, immunologically related to the neutrophil small G protein Rac2 and involved in elicitor induced oxidative burst. FEBS Lett 403:149–153
Levine A, Belenghi B, Damari-Weisler H, Granot D (2001) Vesicle associated membrane protein of Arabidopsis suppresses bax-induced apoptosis in yeast downstream of oxidative burst. J Biol Chem 276:46284–46289
Leyman B, Geelen D, Blatt MR (1999) Localization and control of expression of NtSyr1, a tobacco SNARE protein. Plant J 24:369–381
Limpens E, Ivanov S, van Esse W, Voets G, Fedorova E, Bisseling T (2009) Medicago N2-fixing symbiosomes acquire the endocytic identity marker Rab7 but delay the acquisition of vacuolar identity. Plant Cell 21:2811–2828
Liu F, Guo J, Bai P, Duan Y, Wang X, Cheng Y, Feng H, Huang L, Kang Z (2012) Wheat TaRab7 GTPase is part of the signaling pathway in responses to stripe rust and abiotic stimuli. PLoS ONE 7(5):e37146
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408
Mazel A, Leshem Y, Tiwari BS, Levine A (2004) Induction of salt and osmotic stress tolerance by overexpression of an intracellular vesicle trafficking protein AtRab7 (AtRabG3e). Plant Physiol 134:118–128
Mimura T, Kura-Hotta M, Tsujimura T, Ohnishi M, Miura M, Okazaki Y, Mimura M, Maeshima M, Washitani-Nemoto S (2003) Rapid increase of vacuolar volume in response to salt stress. Planta 216:397–402
Munns R (2002) Comparitive physiology of salt and water stress. Plant Cell Environ 25:239–250
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Ann Rev Plant Biol 59:651–681
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassay with tissue culture. Physiol Plant 15:473–476
Nahm MY, Kim SW, Yun D, Lee SY, Cho MJ, Bahk JD (2003) Molecular and biochemical analyses of OsRab7, a rice Rab7 homolog. Plant Cell Physiol 44:1341–1349
Niu X, Bressan RA, Hasegawa PM, Pardo JM (1995) Ion homeostasis in NaCl stress environments. Plant Physiol 109:735–742
O’Mahony PJ, Oliver MJ (1999) Characterization of a desiccation-responsive small GTP-binding protein (Rab2) from the desiccation-tolerant grass Sporobolus stapfianus. Plant Mol Biol 39:809–821
Park M, Lee H, Lee JS, Byun MO, Kim BG (2009) In planta measurements of Na+ using fluorescent dye corona green. J Plant Biol 52:298–302
Park JY, Kim YY, Martinoia E, Lee Y (2008) Long-distance transporters of inorganic nutrients in plants. J Plant Biol 51:240–247
Radomiljac JD, Whelan J, van der Merwe M (2013) Coordinating metabolite changes with our perception of plant abiotic stress responses: emerging views revealed by integrative-omic analyses. Metabolites 3:761–786
Randall SK, Crowell DN (1999) Protein isoprenylation in plants. Crit Rev Biochem Mol Biol 34:325–338
Rosales KR, Peralta ER, Wong SY, Edinger AL (2009) Rab7 activation by growth factor withdrawal contributes to the induction of apoptosis. Mol Biol Cell 20:2831–2840
Schimmoller F, Reizman H (1993) Involvement of Ypt7p, a small GTPase, in traffic from late endosome to the vacuole in yeast. J Cell Sci 106:823–830
Shi H, Lee BH, Wu SJ, Zhu JK (2003) Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat Biotechnol 21:81–85
Singh S, Ambastha V, Levine A, Sopory SK, Yadava PK, Tripathy BC, Tiwari BS (2015) Anhydrobiosis and programmed cell death in plants: commonalities and differences. Curr Plant Biol 2:12–20
Singla-Pareek SL, Reddy MK, Sopory SK (2003) Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proc Natl Acad Sci U S A 100:14672–14677
Singla-Pareek SL, Pareek A, Sopory SK (2007) Transgenic plants for dry and saline environments. In: Jenks MA et al (ed) Advances in molecular breeding toward drought and salt tolerant crops. Springer, pp 501-530
Stenmark H (2009) Rab GTPases as coordinators of vesicle traffic. Nat Rev 10:513–525
Sui JM, Li R, Fan QC, Song L, Zheng CH, Wang JS, Qiao LX, Yu SL (2013) Isolation and characterization of a stress responsive small GTP-binding protein AhRabG3b in peanut (Arachis hypogaea L.). Euphytica 189:161–172
Tripathy MK, Tyagi W, Goswami M, Kaul T, Singla-Pareek SL, Deswal R, Reddy MK, Sopory SK (2012) Characterization and functional validation of tobacco PLC delta for abiotic stress tolerance. Plant Mol Biol Rep 30:488–497
Verma D, Singla-Pareek SL, Rajagopal D, Reddy MK, Sopory SK (2007) Functional validation of a novel isoform of Na+/H+ antiporter from Pennisetum glaucum for enhancing salinity tolerance in rice. J Biosci 32:621–628
Wu SJ, Ding L, Zhu JK (1996) SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8:617–627
Yoshida S, Forno DA, Cock JH, Gomez KA (1972) Laboratory manual for physiological studies of rice. IRRI, Manila, Philippines
Zerial M, McBride H (2001) Rab proteins as membrane organizers. Nat Rev Mol Cell Biol 2:107–117
Zhang HX, Blumwald E (2001) Transgenic salt tolerant tomato plants accumulate salt in the foliage but not in the fruits. Nat Biotechnol 19:765–768
Acknowledgments
This work was supported by a grant from the Department of Biotechnology, New Delhi, Government of India to SKS. We are thankful to Mr. S. C. P. Sharma and Dr. G. Saini (Jawaharlal Nehru University, New Delhi, India) for their help during TEM study. We thank Prof. S. J. Roux (The University of Texas at Austin, Texas, USA) for his help in correcting the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: Bhumi Nath Tripathi
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
List of genes studied by real time PCR assay.
Fig. S1
Representative pictures to show relative salinity tolerance and morphology of wild-type and PgRab7 -transgenic plants. Transgenic plants and WT seedlings were grown under control (A and E), 100 mM NaCl (B and F), 175 mM NaCl (C and G), and 200 mM NaCl (D and H). Pictures showing (A, B, C, D) the number of seed germination (15-day-old seedlings), (E, F, G, H) shoot and root length (10-day-old seedlings), respectively, of WT and transgenic seedlings subjected to salt stress as mentioned above.
Rights and permissions
About this article
Cite this article
Tripathy, M.K., Tiwari, B.S., Reddy, M.K. et al. Ectopic expression of PgRab7 in rice plants (Oryza sativa L.) results in differential tolerance at the vegetative and seed setting stage during salinity and drought stress. Protoplasma 254, 109–124 (2017). https://doi.org/10.1007/s00709-015-0914-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-015-0914-2