Abstract
A novel lytic Raoultella phage, RP180, was isolated and characterized. The RP180 genome has 44,851 base pairs and contains 65 putative genes, 35 of them encoding proteins whose functions were predicted based on sequence similarity to known proteins. The RP180 genome possesses a gene synteny typical of members of the subfamily Guernseyvirinae. Phylogenetic analysis of the RP180 genome and similar phage genomes revealed that phage RP180 is the first member of the genus Kagunavirus, subfamily Guernseyvirinae, that is specific for Raoultella sp. The genome of RP180 encodes a putative protein with similarity to CRISPR-like Cas4 nucleases, which belong to the pfam12705/PDDEXK_1 family. Cas4-like proteins of this family have been shown to interfere with the bacterial host type II-C CRISPR-Cas system.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Raoultella spp. are gram-negative, aerobic, non-motile rods belonging to the family Enterobacteriaceae. Raoultella and Klebsiella are closely related genera, and Raoultella spp. were previously classified as members of the genus Klebsiella; however, in 2001, they were reclassified in the genus Raoultella [1]. Raoultella spp. are a rare source of infection in humans, although serious cases associated with sepsis, bacteremia, urinary tract infection, cholecystitis, immunodeficiency, and cirrhosis have been recorded [2,3,4,5]. Clinical isolates of the genus Raoultella can possess resistance to multiple antibiotics, and alternative therapeutics are therefore required [6]. One such alternative approach could be the use of lytic bacteriophages. However, nucleotide sequence data on bacteriophages specific for Raoultella spp. are limited to phage Ro1 [MG250486]. Here, we report the genome sequence and characteristics of the novel phage RP180, which infects Raoultella sp.
Materials and methods
Raoultella sp. strain CEMTC 2544 was obtained from the Collection of Extremophilic Microorganisms and Type Cultures of the ICBFM SB RAS and cultivated in Luria-Bertani (LB) medium and LB agar at 37 °C. Bacteriophage RP180 was isolated from a wastewater sample. Bacteriophage isolation and characterization of biological properties were performed as described previously [7].
RP180 genome library construction and sequencing were performed as described previously [7]. The genome was assembled de novo using SPAdes software of the Unipro UGENE software package [8]. SPAdes software was applied using default settings, and genome assembly resulted in one contig with an average coverage of 300. Putative open reading frames (ORFs) with a 150-bp minimum size were identified using Vector NTI [9] and UGENE software [8]. These ORFs were compared manually to sequences available in the GenBank database, using the BLASTx algorithm. The presence of tRNA genes was detected using tRNAscan-SE [10]. The Virulence Factor Database (VFDB) was used to search for genes encoding toxins [11]. MAFFT software was used to align genome sequences [12]. The CGView server [13] was used for comparative analysis of the genome of RP180 and those of phage K1G [GU196277], phage Jersey [KF148055], and phage FSL-SP031 [KC139518]. The overall identity of the aligned genome sequences was calculated using BioEdit [14]. A phylogenetic analysis of the RP180 genome and 20 related genomes was performed using PhyML3.0 [15].
Results and discussion
Phage RP180 formed clear plaques with a diameter of about 1─1.5 mm on a lawn of the host strain CEMTC 2544 (Supplementary Fig. 1S). Of the 94 Enterobacteria strains tested, RP180 could only infect Raoultella CEMTC 2544 (Supplementary Table 1S). One-step growth experiments revealed a latent period of 15 min with a burst size of ~40 phage particles per infected cell (Supplementary Fig. 2Sa). A multistep bacterial killing curve for phage RP180 showed that the number of live bacteria decreased dramatically in the three hours after infection and increased slowly thereafter (Supplementary Fig. 2Sb). These results indicate that phage RP180 is a virulent phage.
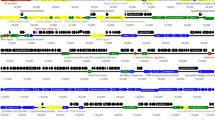
The RP180 genome is a double-stranded linear DNA molecule with a length of 44,851 bp. The GC content of the RP180 genome is 50.3%, while that of the host genome is 55.5%. The genome of RP180 contains 65 putative ORFs, 35 of which encode proteins whose function could be predicted due to their similarity to known proteins, whereas the other 30 were classified as hypothetical proteins. Genes encoding tRNAs, integrases, and toxins were not found (Fig. 1a). The complete genome sequence of Raoultella phage RP180 was deposited in the GenBank database of NCBI with the accession number MK737937.
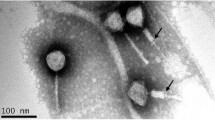
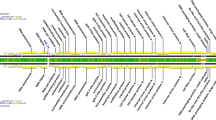
Electron microscopy revealed that RP180 has an icosahedral head with a diameter of 55-60 nm, which is connected to a non-contractile tail of approximately 120 nm in length, and a baseplate of 20 nm in width with spikes is located at the end of the tail (Fig. 1b). The morphology and size of the phage particle corresponds to those of Siphoviridae family members [16]. The RP180 genome exhibited the highest similarity to the genomes of the K1G-like phages, which belong to the subfamily Guernseyvirinae, family Siphoviridae. The overall nucleotide sequence identity of the RP180 genome to those of phages representing three genera of the subfamily Guernseyvirinae was 61% for Escherichia phage K1G (genus Kagunavirus), 49% for Salmonella phage Jersey (genus Jerseyvirus) [17], and 40.6% for Salmonella phage FSL-SP031 (genus Cornellvirus). In addition, the RP180 genome showed gene synteny typical of members of the subfamily Guernseyvirinae (Fig. 1a). Phylogenetic analysis of the RP180 genome and other similar phage genomes revealed that phage RP180 is a new member of the genus Kagunavirus (Fig. 2).
The RP180 genome was found to encode a CRISPR-like Cas4 nuclease belonging to the family pfam12705/PDDEXK_1. Cas4-like proteins have been shown to interfere with the host type II-C CRISPR-Cas system [18, 19]. Cas4-like nucleases have been found in phage infecting Campylobacter, Mycobacteria, Propionibacterium, Bacillus, Thermus and other bacteria; however, their exact functions are unknown [20].
In conclusion, a novel phage RP180 infecting Raoultella sp., belonging to the genus Kagunavirus, subfamily Guernseyvirinae was described. Almost all members of the subfamily Guernseyvirinae are Escherichia and Salmonella phages, with only a few exceptions: Xanthomonas phage f29-Xaj [KU595434] [21], Enterobacter phage phiEap-2 [KT287080] [22] and Serratia phage η [KC460990] [23]. Phage RP180 is one more phage of the subfamily Guernseyvirinae that does not infect Escherichia spp. or Salmonella spp.
References
Drancourt M, Bollet C, Carta A, Rousselier P (2001) Phylogenetic analyses of Klebsiella species delineate Klebsiella and Raoultella gen. nov., with description of Raoultella ornithinolytica comb. nov., Raoultella terrigena comb. nov. and Raoultella planticola comb. nov. Int J Syst Evol Microbiol 51(3):925–932
Ershadi A, Weiss E, Verduzco E et al (2014) Emerging pathogen: a case and review of Raoultella planticola. Infection 42(6):1043–1046
Fager C, Yurteri-Kaplan L (2018) Urinary tract infection with rare pathogen Raoultella planticola: a post-operative case and review. Urol Case Rep 22:76–79
Mehmood H, Pervin N, Ul Haq MI et al (2018) A rare case of Raoultella planticola urinary tract infection in a patient with immunoglobulin A nephropathy. J Investig Med High Impact Case Rep 6:1–3
Hajjar R, Schwenter F, Sebajang H et al (2018) Community-acquired infection to Raoultella ornithinolytica presenting as appendicitis and shock in a healthy individual. J Surg Case Rep 5:1–3
Chen DQ, Song JL, Tang HX et al (2014) Extensively drug-resistant Raoultella planticola carrying multiple resistance genes including blaNDM-1. JMM Case Rep 1(3):3–5
Morozova V, Kozlova Y, Shedko E et al (2016) Lytic bacteriophage PM16 specific for Proteus mirabilis: a novel member of the genus Phikmvvirus. Arch Virol 161(9):2457–2472
Okonechnikov K, Golosova O, Fursov M et al (2012) Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28(8):1166–1167
Lu G, Moriyama EN (2004) Vector NTI, a balanced all-in-one sequence analysis suite. Brief Bioinform 5(4):378–388
Lowe TM, Chan PP (2012) tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucl Acids Res 44:54–57
Liu B, Zheng D, Jin Q et al (2019) VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucl Acids Res 47(D1):D687–D692
Katoh K, Rozewicki J, Yamada KD (2017) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 18:1–7
Grant JR, Stothard P (2008) The CGView server: a comparative genomics tool for circular genomes. Nucl Acids Res 36:181–184
Phillips JL, Gnanakaran S (2015) BioEdit: an important software for molecular biology. Proteins Struct Funct Bioinform 83(1):46–65
Guindon S, Dufayard J, Lefort V et al (2010) New algorithms and methods to estimate maximim-likelihood phylogenies assessing the performance of PhyML 3.0. Syst Biol 59(3):1–37
Ackermann H-W (2009) Phage classification and characterization. In: Clokie MR, Kropinski AM (eds) Bacteriophages methods protocol, vol 1. ISOL character interaction. Humana Press, New York, pp 127–140
Anany H, Switt AIM, De Lappe N et al (2015) A proposed new bacteriophage subfamily: Jerseyvirinae. Arch Virol 160(4):1021–1033
Hooton SPT, Connerton IF (2015) Campylobacter jejuni acquire new host-derived CRISPR spacers when in association with bacteriophages harboring a CRISPR-like Cas4 protein. Front Microbiol 5:1–9
Hooton SPT, Brathwaite KJ, Connerton IF (2016) The bacteriophage carrier state of Campylobacter jejuni features changes in host non-coding RNAs and the acquisition of new host-derived CRISPR spacer sequences. Front Microbiol 7:1–8
Hudaiberdiev S, Shmakov S, Wolf YI et al (2017) Phylogenomics of Cas4 family nucleases. BMC Evol Biol 17(1):1–14
Retamales J, Vasquez I, Santos L et al (2016) Complete genome sequences of lytic bacteriophages of Xanthomonas arboricola pv. juglandis. Genome Announc 4(3):3–4
Li E, Wei X, Ma Y et al (2016) Isolation and characterization of a bacteriophage phiEap-2 infecting multidrug resistant Enterobacter aerogenes. Sci Rep 6(1):1–9
Denyes JM, Krell PJ, Manderville RA et al (2014) The genome and proteome of Serratia bacteriophage η which forms unstable lysogens. Virol J 11(1):1–8
Funding
This study was funded by RFBR Project No. 18-29-08015; Collection EMTC of ICBFM is supported by Russian State-Funded Budget Project ICBFM SB RAS # AAAA-A17-117020210027-9.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
There are no conflicts of interest.
Ethical approval
This article does not involve studies with human participants or animals.
Additional information
Handling Editor: Chan-Shing Lin.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fofanov, M.V., Morozova, V.V., Kozlova, Y.N. et al. Raoultella bacteriophage RP180, a new member of the genus Kagunavirus, subfamily Guernseyvirinae. Arch Virol 164, 2637–2640 (2019). https://doi.org/10.1007/s00705-019-04349-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-019-04349-z