Abstract
Strains of the phytopathogenic fungus Alternaria spp. have been found to contain a variety of double-stranded RNA (dsRNA) elements indicative of mycovirus infection. Here, we report the molecular characterization of a novel dsRNA mycovirus, Alternaria arborescens victorivirus 1 (AaVV1), from A. arborescens, the tomato pathotype of A. alternata. Using next-generation sequencing of dsRNA purified from an A. arborescens strain from the United States of America, we found that the AaVV1 genome is 5203 bp in length and contains two open reading frames (ORF1 and 2) that overlap at the tetranucleotide AUGA. Proteins encoded by ORF1 and ORF2 showed significant similarities to the coat protein (CP) and the RNA-dependent RNA polymerase (RdRp), respectively, of dsRNA mycoviruses of the genus Victorivirus. Pairwise comparisons and phylogenetic analysis of the deduced amino acid sequences of both CP and RdRp indicated that AaVV1 is a member of a distinct species of the genus Victorivirus in the family Totiviridae.
Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
Mycoviruses are widespread in a variety of fungal species [7]. An increasing number of mycoviruses have been reported in recent years, which have single-stranded RNA (ssRNA), double-stranded RNA (dsRNA) or single-stranded DNA (ssDNA) genomes. Of these, dsRNA genomes are the most prevalent. Mycoviruses with dsRNA genomes are classified into the seven families: Totiviridae, Partitiviridae, Chrysoviridae, Reoviridae, Endornaviridae, Megabirnaviridae and Quadriviridae [7]. Also, mycoviruses with linear positive-sense ssRNA genomes are currently classified into the five families: Alphaflexiviridae, Barnaviridae, Gammaflexiviridae, Hypoviridae and Narnaviridae. In order to screen for mycoviruses with different types of genomes, we can commonly employ dsRNA purification and subsequent electrophoresis, which can be applied to mycoviruses that possess a dsRNA genome as well as to those having a ssRNA genome, which produce a dsRNA intermediate during replication [15]. Although most mycoviruses cause no apparent symptoms on their host, some of them reduce the virulence of host-phytopathogenic fungi, which highlights the potential of mycoviruses as biological control agents against plant fungal pathogens [7].
Alternaria alternata is a ubiquitous fungus found in a variety of plants, causing leaf spot or other diseases. Many A. alternata strains have been found to harbor dsRNA elements of different sizes, which are indicative of mycovirus infections [9, 19]. Indeed, we previously found a mycovirus with four dsRNA segments, Alternaria alternata virus 1 (AaV1), from the non-pathogenic A. alternata strain EGS 35-193, which is closely related to Aspergillus mycovirus 341 and Aspergillus foetidus virus-fast (AfV-F) [2, 11]. Moreover, we have also found that a Japanese pear pathotype of A. alternata contains dsRNA elements responsible for phenotypic morphological changes in the host [6]. However, in most of these A. alternata strains, the nucleotide sequences and effects on host fungi of these dsRNAs remain unknown. Recently, mycoviruses have also been found in other members of the genus Alternaria. One example is Alternaria longipes dsRNA virus 1, found in A. longipes, which causes brown spot disease on tobacco plants. This virus contains a single dsRNA genome with two open reading frames (ORFs) and belongs to a new family of dsRNA mycoviurses [13]. Also, Alternaria brassicicola endornavirus 1 was found in A. brassicicola, which causes black spot disease in rapeseed [18].
Here, we report the complete genome sequence of a dsRNA virus, Alternaria arborescens victorivirus 1 (AaVV1), from A. arborescens (synonyms: A. alternata tomato pathotype, A. alternata f. sp. lycopersici) [1, 8, 16]. A single dsRNA segment of approximately 5.2 kbp has two ORFs putatively coding for the coat protein (CP) and RNA-dependent RNA polymerase (RdRp). Phylogenetic analysis of the RdRp indicated that AaVV1 is a member of a distinct virus species of the genus Victorivirus.
Provenance of the virus material
The A. arborescens strain used in this study was EGS 39-128 [6]. Fungi were initially grown on dextrose agar plates at 25 °C, and fungal mycelia from the plate were cultured in YG broth medium (0.5 % (w/v) yeast extract and 2 % glucose) with reciprocal shaking at 25 °C for 7 days. Total nucleic acid extraction and purification of dsRNA were performed as described previously [15]. The purity and size of the dsRNA were confirmed by electrophoresis in 1.0 % (w/v) agarose gels, which indicated that the strain EGS 39-128 contained an approximately 5.2-kbp dsRNA as described previously [6] (Fig. S1). Using an input of 27.5 ng of this dsRNA, library preparation for deep sequencing was carried out using an NEBNext® Ultra RNA Library Prep Kit for Illumina Version 2.0 (New England Biolabs, Ipswich, MA). Following measurement of the library quality and quantity on a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad CA), sequencing was performed on a MiSeq benchtop sequencer using a MiSeq Reagent Kit v2 (50 cycles) (Illumina, San Diego, CA) with 51 single-end reads. The raw sequence reads were formatted to FASTQ data using MiSeq Reporter v2.3 (Illumina), followed by read trimming and de novo assembly of contigs using CLC Genomics Workbench 6.5.1 (CLC bio, Aarhus, Denmark). Assembled sequences were compared to those in the GenBank database using BLASTP in the National Center for Biotechnology Information (NCBI). Multiple alignments and phylogenetic analysis of the amino acid sequences were perfomed using MEGA 6.0 [20].
Sequence properties
The 2,270,543 raw reads obtained by MiSeq sequencing were assembled into 2255 contigs, 28 of which had an average coverage of more than 100. One of these contigs, with an average coverage of 198, was 5206 bp in length, which corresponds well to the length of the dsRNA purified from EGS 39-128. A BLAST search of the 5206-bp contig revealed significant similarity to the complete genome sequence of Coniothyrium minitans RNA virus (CmRV), a dsRNA mycovirus that belongs to the genus Victorivirus (E-value, 0; greatest hit length, 2447) [4]. A whole-genome comparison between the 5206-bp contig and CmRV showed 66 % identity. The nucleotide sequence of this putative mycovirus, which we called “Alternaria arborescens victorivirus 1” (AaVV1), has been deposited in the GenBank/EMBL/DDBJ databases under accession number LC086813.
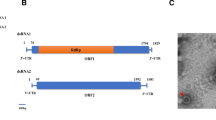
The 5’- and 3’-terminal genomic sequences of AaVV1 were determined using a SMARTer RACE cDNA Amplification Kit (Clontech, Mountain View, CA). As a result, we obtained almost identical terminal sequences by RACE, which indicated that the AaVV1 genome is 5203 bp in length with a GC content of 58.0 %. The GC content of the AaVV1 genome is similar to that of other victorivirus genomes, including CmRV (59.2 %), Magnaporthe oryzae virus 2 (MoV2) (61.9 %) and Epichloe festucae virus 1 (EfV1) (60.3 %). An ORF search revealed that the AaVV1 genome contains two ORFs in the same strand (Fig. 1a). ORF1 begins at an AUG codon (nt 327-329) and terminates with a UGA codon (nt 2655-2657). ORF2 begins at an AUG codon (nt 2654-2656) and terminates with a UAA codon (nt 5141-5143). ORF2 is in the -1 frame of ORF1, with the AUG codon of the ORF2 overlapping the UGA codon of ORF1 at the tetranucleotide AUGA at nt 2653-2656, which is a characteristic feature of many victoriviruses [10, 12, 14]. The 5’ and 3’ untranslated regions (UTR) were found to be 326 and 60 bases long, respectively. In the 5’-UTR, the octanucleotide sequence 5’-AGGGUUCC-3’, which is conserved in several victoriviruses, including CmRV and EfV1, was found at nt 264-271, 56 nt upstream of the AUG initiation codon of ORF1 [17].
Genome organization of Alternaria arborescens victorivirus 1 (AaVV1) and its phylogenetic relationships to selected viruses of the family Totiviridae. (a) Genome organization of AaVV1. (b) Phylogenetic tree of AaVV1 and other selected dsRNA viruses based on RdRp amino acid sequences, generated by the neighbor-joining method. A star indicates the position of AaVV1. Brackets indicate genera. Numbers proximal to nodes represent bootstrap support values for 1,000 replicates (only the bootstrap scores over 70 % are shown). The scale bar below the tree represents the phylogenetic distance. The following RdRp amino acid sequences were used to construct the tree, with their abbreviated names and accession numbers in parentheses: Epichloe festucae virus 1 (EfV1, AM261427), Coniothyrium minitans RNA virus (CmRV, AF527633), Saccharomyces cerevisiae virus L-A (ScV-L-A, J04692), Saccharomyces cerevisiae virus L-BC (ScV-L-BC, U01060), Gremmeniella abientina RNA virus L1 (GaRV-L1, AF337175), Leishmania RNA virus 1-1 (LRV1-1, M92355), Helicobasidium mompa totivirus 1-17 (HmTV1-17, AB085814), Helminthosporium victoriae virus 190S (Hv190SV, U41345), Sphaeropsis sapinea RNA virus 1 (SsRV1, AF038665), Magnaporthe oryzae virus 1 (MoV1, AB176964), Magnaporthe oryzae virus 2 (MoV2, AB300379), Trichomonas vaginalis virus 1 (TVV1, U08999), Ustilago maydis virus H1 (UmV-H1, U01059), Giardia lamblia virus (GLV, L13218), Botryotinia fuckeliana totiviurs 1 (BfTV1, AM491608), Beauveria bassiana RNA virus 1 (BbRV1, HE572591), and Eimeria brunetti RNA virus 1 (EbRV1, AF356189). Penicillium chrysogenum virus (PcV, AF296439) was used as a outgroup
ORF1 was predicted to encode a protein of 776 amino acids (aa) with a molecular mass of 80.9 kDa. A BLASTP search indicated that this protein has significant similarity to CPs of viruses of the family Totiviridae: 80 % identity to the CP of CmRV and 60-65 % identity to those of related victoriviruses, including MoV2. Notably, the CP of AaVV1 has a proline-rich region near its carboxy terminus, which is also prominent in CmRV, EfV1, Gremmeniella abientina RNA virus L1 (GaRV-L1), MoV2, and Helminthosporium victoriae virus 190S (Hv190SV) (Fig. S2).
The predicted translation product of the ORF2 of AaVV1 is 829 aa in length with a calculated molecular mass of 92.5 kDa. This protein was most closely related to the putative RdRp of CmRV, which has 70 % identity (E-value of 0 and query cover of 100 %). It also showed considerable sequence similarity to the RdRps of EfV1 (54 %) and MoV2 (49 %), suggesting that AaVV1 is closely related to dsRNA mycoviruses in the genus Victorivirus. After we had begun our analysis, a partial sequence of a victorivirus isolated from A. arborescens was reported with the accession number KJ013354, but we found that the RdRp amino acid sequence of AaVV1 and the protein sequence of KJ013354 shared only 40 % identity, indicating that they belong to distinct virus species. A conserved domain search revealed that the ORF2 product of AaVV1 possessed the eight conserved motifs of RdRps of dsRNA viruses (RdRP_4; pfam02123), which was confirmed by an amino acid sequence alignment of RdRps of selected victoriviruses [3] (Fig. S3).
A phylogenetic tree based on RdRp amino acid sequences of AaVV1 and related mycoviruses demonstrated that AaVV1 belongs to a well-supported clade containing viruses of the genus Victorivirus, especially in a subclade containing CmRV, EfV1, MoV2 and GaRV-L1 (Fig. 1b). A phylogenetic tree based on CP amino acid sequences had a similarity topology to the one based on RdRp (Fig. S4). As mentioned above, viruses in the subclade containing AaVV1 have a proline-rich region at the carboxy terminus of their CP, which supports the idea that they are evolutionary closely related. However, the topology of the tree based on both CP and RdRp amino acid sequences of victoriviruses does not match the host phylogeny, suggesting that horizontal transfer between different host fungi could contribute to the evolution of victoriviruses.
Moreover, RNA secondary predictions revealed that AaVV1 has an H-type pseudoknot structure and a strong stem-loop structure upstream and downstream, respectively, of the tetranucleotide AUGA motif located at the junction of ORF1 and ORF2 (Fig. S5). These unique RNA structures may be important for the translation of ORF2 of AaVV1, as has been suggested for other victoriviruses [5].
Overall, based on the results of the present study, we propose that AaVV1 should be considered a distinct member of the genus Victorivirus. Further studies are needed to determine whether AaVV1-related viruses infect other A. alternata strains and to evaluated their effect on the growth and virulence traits of the host fungus.
References
Akimitsu K, Tsuge T, Kodama M, Yamamoto M, Otani H (2014) Alternaria host-selective toxins: determinant factors of plant disease. J Gen Plant Pathol 80:109–122
Aoki N, Moriyama H, Kodama M, Arie T, Teraoka T, Fukuhara T (2009) A novel mycovirus associated with four double-stranded RNAs affects host fungal growth in Alternaria alternata. Virus Res 140:179–187
Bruenn JA (1993) A closely related group of RNA-dependent RNA polymerases from double-stranded RNA viruses. Nucleic Acids Res 21:5667–5669
Cheng J, Jiang D, Fu Y, Li G, Peng Y, Ghabrial SA (2003) Molecular characterization of a dsRNA totivirus infecting the sclerotial parasite Coniothyrium minitans. Virus Res 93:41–50
Chiba S, Lin YH, Kondo H, Kanematsu S, Suzuki N (2013) A novel victorivirus from a phytopathogenic fungus, Rosellinia necatrix, is infectious as particles and targeted by RNA silencing. J Virol 87:6727–6738
Fuke K, Takeshita K, Aoki N, Fukuhara T, Egusa M, Kodama M, Moriyama H (2011) The presence of double-stranded RNA in Alternaria alternata Japanese pear pathotype is associated with morphological changes. J Gen Plant Pathol 77:248–252
Ghabrial SA, Castón JR, Jiang D, Nibert ML, Suzuki N (2015) 50-plus years of fungal viruses. Virology 479–480:356–368
Gilchrist DG, Grogan RG (1976) Production and nature of a host-specific toxin from Alternaria alternata f. sp. lycopersici. Phytopathology 66:165–171
Hayashi N, Tsuge T, Kobayashi H, Nishimura S (1988) The presence of double-stranded RNAs in Alternaria alternata Japanese pear pathotype and their participation in AK-toxin productivity. Annu Phytopathol Soc Jpn 54:250–252
Huang S, Ghabrial SA (1996) Organization and expression of the double-stranded RNA genome of Helminthosporium victoriae 190S virus, a totivirus infecting a plant pathogenic filamentous fungus. Proc Natl Acad Sci USA 93:12541–12546
Kozlakidis Z, Herrero N, Ozkan S, Kanhayuwa L, Jamal A, Bhatti MF, Coutts RHA (2013) Sequence determination of a quadripartite dsRNA virus isolated from Aspergillus foetidus. Arch Virol 158:267–272
Li H, Havens WM, Nibert ML, Ghabrial SA (2015) An RNA cassette from Helminthosporium victoria virus 190S necessary and sufficient for stop/restart translation. Virology 474:131–143
Lin Y, Zhang H, Zhao C, Liu S, Guo L (2015) The complete genome sequence of a novel mycovirus from Alternaria longipes strain HN28. Arch Virol 160:577–580
Maejima K, Himeno M, Komatsu K, Kakizawa S, Yamaji Y, Hamamoto H, Namba S (2008) Complete nucleotide sequence of a new double-stranded RNA virus from the rice blast fungus, Magnaporthe oryzae. Arch Virol 153:389–391
Okada R, Kiyota E, Moriyama H, Fukuhara T, Natsuaki T (2015) A simple and rapid method to purify viral dsRNA from plant and fungal tissue. J Gen Plant Pathol 81:103–107
Peever TL, Su G, Carpenter-Boggs L, Timmer LW (2004) Molecular systematics of citrus-associated Alternaria species. Mycologia 96:110–134
Romo M, Leuchtmann A, García B, Zabalgogeazcoa I (2007) A totivirus infecting the mutualistic fungal endophyte Epichloë festucae. Virus Res 124:38–43
Shang HH, Zhong J, Zhang RJ, Chen CY, Gao BD, Zhu HJ (2015) Genome sequence of a novel endornavirus from the phytopathogenic fungus Alternaria brassicicola. Arch Virol 160:1827–1830
Shepherd HS (1988) Virus-like particles in tentoxin-producing strains of Alternaria alternata. J Virol 62:3888–3891
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Acknowledgments
We thank T. Peever (Washington State University, USA) for providing the A. arborescens strain.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was funded by a Grant-in-Aid for Scientific Research (C) (15K07838) from the Japan Society for the Promotion of Science.
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with animals or human participants performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
705_2016_2796_MOESM1_ESM.pptx
Fig. S1 Agarose gel electrophoresis of dsRNA purified from A. arborescens strain EGS 39-128. Ethidium bromide staining showed dsRNA elements with about 5.2 kb (PPTX 51 kb)
705_2016_2796_MOESM2_ESM.pptx
Fig. S2 Multiple amino acid sequence alignment of the carboxy terminus of the deduced CP encoded by AaVV1 and related victoriviruses (see the legend to Fig. 1 for viruses whose CP amino acid sequences were used for the alignment). The proline-rich region is shown as a black line under the alignment (PPTX 94467 kb)
705_2016_2796_MOESM3_ESM.pptx
Fig. S3 Multiple alignment of the deduced amino acid sequence of the RdRp encoded by AaVV1 and related victoriviruses (see the legend to Fig. 1 for viruses whose RdRp amino acid sequences were used for the alignment). The eight conserved motifs in RdRps of dsRNA viruses are indicated as numbers 1 to 8 [3]. Numbers at the beginning of the alignment represent the amino acid positions from the start of the predicted gene product (PPTX 8587 kb)
705_2016_2796_MOESM4_ESM.pptx
Fig. S4 Phylogenetic tree of AaVV1 and other selected dsRNA viruses based on CP amino acid sequences, generated by the neighbor-joining method. A star indicates the position of AaVV1. Brackets indicate genera. Numbers proximal to nodes represent bootstrap support values for 1,000 replicates (only the bootstrap scores over 70% are shown). The scale bar below the tree represents the phylogenetic distance. Viruses used to construct the tree and their abbreviated names are listed in the caption of Fig. 1b, except the accession number of segment 2 of PcV, which was AF296440 (PPTX 51 kb)
705_2016_2796_MOESM5_ESM.pptx
Fig. S5 Predicted secondary structures of the upstream and downstream regions of the tetranucleotide AUGA motif located at the junction of ORF1 and ORF2 of AaVV1. An H-type pseudoknot and a stem-loop were predicted using the HPKNOTTER (http://genome.cs.nthu.edu.tw/HPKNOTTER/) and RNAstructure program (http://rna.urmc.rochester.edu/RNAstructureWeb/), respectively. ∆G values, which represent the free energy, are indicated below each structure. Nucleotide numbers of the AaVV1 genome are also indicated (PPTX 46 kb)
Rights and permissions
About this article
Cite this article
Komatsu, K., Katayama, Y., Omatsu, T. et al. Genome sequence of a novel victorivirus identified in the phytopathogenic fungus Alternaria arborescens . Arch Virol 161, 1701–1704 (2016). https://doi.org/10.1007/s00705-016-2796-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-016-2796-9