Abstract
In this study, a novel virus designated Phomopsis vexans RNA virus 1 (PvRV1) was identified in a strain of Phomopsis vexans. The complete genomic nucleotide sequence was determined and analyzed. Sequence analysis indicated that PvRV1 is closely related to viruses in the genus Victorivirus of the family Totiviridae. Two open reading frames (ORF1 and 2) were found in the PvRV1 sequence, and these showed significant similarity to the capsid protein (CP) and RNA-dependent RNA polymerase (RdRp), respectively, of members of the family Totiviridae. The two ORFs were spaced 98 nt apart, which is unique to PvRV1 and different from the overlapping arrangement in most victoriviruses. The expression strategies of the CP and RdRp are discussed based on in silico RNA secondary structure analysis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phomopsis vexans, the causal agent of phomopsis blight of eggplant, is a very destructive disease and is considered to be the major constraint on productivity of eggplant in some areas of the world [1]. This pathogen is seed-borne both externally and internally and remains viable for about 14 months in soil debris and in the seeds of infected fruit [2, 3]. No satisfactory fungicide-based control is available for this disease. It is of considerable interest to develop biological control agents, possibly including mycoviruses, for phomopsis blight management. However, no virus has been reported to be a parasite of P. vexans to date.
Mycoviruses are widespread in filamentous fungi including phytopathogenic species [4, 5]. dsRNA mycoviruses are currently classified into six families: Chrysoviridae, Megabirnaviridae, Partitiviridae, Quadriviridae, Reoviridae and Totiviridae, whose members have 4, 2, 2, 4, 11 or 12, and 1 genome segment(s), respectively, all encapsidated in isometric particles [5, 6]. Mycoviruses are usually considered to lack an extracellular phase during their replication cycle and are transmitted only through intracellular mechanisms such as hyphal fusion or asexual sporulation [7]. Mycoviruses, in many cases, are cryptic, causing little or no visible abnormal symptoms in the host fungi. However, some of them have been shown to induce hypovirulence in their hosts and are considered potential biological control agents. Practical application of hypovirulence has been demonstrated by the successful use of Cryphonectria hypovirus 1 (CHV1) to control chestnut blight in Europe [8].
Most mycoviruses have either a double-stranded RNA (dsRNA) genome or a single-stranded RNA (ssRNA) genome that produces dsRNA replicative intermediates [9]. Because fungal cells lack large molecules of dsRNA, the isolation of dsRNA elements from mycelia serves as a step in RNA mycovirus diagnosis. In an attempt to investigate the presence of dsRNAs in P. vexans strain Pv-HN-1, we obtained three dsRNA bands with sizes of 12, 5 and 3 kbp, which were named dsRNA-L, dsRNA-M, and dsRNA-S, respectively. dsRNA-L might represent an endornavirus, based on the partial sequences obtained (data not show), and none of the dsRNA-S sequence has been determined until now. In this paper, we report the full nucleotide sequence of the 5-kbp dsRNA-M.
Provenance of the virus material
The P. vexans strain Pv-HN-1 was isolated from a diseased fruit from Hunan Province in China in 2012, and its identity was verified by the amplification and sequencing of the rDNA-ITS sequence (accession number, KP115580). The fungus was cultured in potato dextrose broth (PD) in a 27 °C shaker at 110 rpm for seven days for dsRNA multiplication. dsRNA was extracted and purified using CF-11 cellulose (Sigma, St. Louis, MO, USA) as described previously [9]. After being treated with RNase-free DNase I (Takara) and S1 nuclease (Takara) to eliminate contaminating ssRNA and DNA, the dsRNA samples were separated by agarose gel electrophoresis, and three bands, named as dsRNA-L, dsRNA-M and dsRNA-S, were obtained (Fig. 1a). Each of the three dsRNAs was purified and stored at -20 °C. dsRNA-M was selected for cDNA synthesis by reverse transcription (RT) and PCR amplification (RT-PCR). A cDNA library was constructed using random hexadeoxynucleotide primers (Takara). The regions not covered by the cDNA library clones were then filled by RT-PCR amplification using sequence-specific primers designed based on the sequences obtained. In order to clone the termini of the dsRNAs, an adaptor was ligated to the 3’ end of each strand with T4 RNA ligase (Fermentas) [10, 11], and terminal fragments were then amplified using a primer with reverse complementary sequence to the adapter and dsRNA-M-specific primers. All of the amplified cDNA products were cloned into the pMD18-T vector (Takara) and sequenced, with every base being determined independently at least three times. The resulting sequences were assembled and deposited in the GenBank database under the accession number KP090346. Sequence analysis and alignment were performed using DNAMAN and ClustalX [12], respectively. A phylogenetic tree was constructed using the neighbor-joining method in MEGA 4.0, with 1000 bootstrap re-samplings [13]. RNA pseudoknot prediction was carried out using DotKnot [14]. Potential RNA secondary structures and motifs were predicted using the Mfold (version 3.2) [15] and FSFinder [16] programs, respectively.
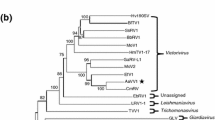
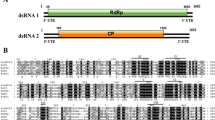
Agarose gel electrophoresis of dsRNAs extracted from the P. vexans strain Pv-HN-1 and the genomic organization and phylogenetic analysis of PvRV1. (a) dsRNA banding pattern of P. vexans strain Pv-HN-1 analyzed by 1 % agarose gel electrophoresis, with the size standard shown on the left. (b) Schematic representation of the genomic organization of the cloned dsRNA-M of the virus named PvRV1. (c) Phylograms based on the deduced amino acid sequences of the RdRps of PvRV1 and other members of the family Totiviridae. The phylogenic tree was constructed using the neighbor-joining method with 1000 bootstrap replicates in the program MEGA 4.0. The numbers near the branches indicate the percentage of bootstrap replicates supporting the branch. The names of the viruses used in the analysis and their respective GenBank accession numbers are as follows: Beauveria bassiana RNA virus 1 (CCC42235.1), Helicobasidium mompa No.17 dsRNA virus (NP_898833.1), Magnaporthe oryzae virus 1 (YP_122352.1), Tolypocladium cylindrosporum virus 1 (FR750562), Botryotinia fuckeliana totivirus 1 (AM491608), Helminthosporium victoriae virus 190S (U41345), Sphaeropsis sapinea RNA virus 2 (AF039080), Coniothyrium minitans RNA virus (AF527633), Epichloe festucae virus 1 (CAK02788.1), Magnaporthe oryzae virus 2 (AB300379), Gremmeniella abietina RNA virus L1 (NP_624332.2), Eimeria brunetti RNA virus 1 (NP_108651.1), Leishmania RNA virus 2–1 (U32108), Leishmania RNA virus 1-4 (NP_619653.1), Leishmania RNA virus 1–1 (M92355), Saccharomyces cerevisiae virus L-A (AAA50321.1), Saccharomyces cerevisiae virus L-BC (La) (NP_042581.1), Trichomonas vaginalis virus 2 (AAF29445.1), Trichomonas vaginalis virus 1 (ABC86751.1), Trichomonas vaginalis virus 3 (NP_659390.1), Giardia lamblia virus (NP_620070.1). Trichomonas vaginalis virus 4 (AED99798.1), Ustilago maydis virus H1 (NP_620728.1), Tuber aestivum virus 1 (ADQ54106.1), Ustilaginoidea virens RNA virus 1 (AGO04407.1), Ustilaginoidea virens RNA virus 2 (YP_007761589.1), Ustilaginoidea virens RNA virus 3 (YP_009004156)
Sequence properties
The complete genome sequence of dsRNA-M was determined to be 5076 nt in length, which was consistent with the estimated size of this dsRNA segment obtained by agarose gel electrophoresis (Fig. 1a). The dsRNA segment contained two non-overlapping ORFs (Fig. 1b): ORF1 (2301 nt), initiating at nt position 243 and terminating at a UAA codon at position 2543, and ORF2 (2385 nt), initiating at position 2642 and terminating at a UAA codon at position 5026. The molecular masses of the putative proteins encoded by ORF 1 and ORF 2 were calculated to be 80.0 kDa and 88.2 kDa, respectively. The untranslated regions (UTRs) at the 5’ and 3’ ends of the (+) sense strand were found to be 242 nt and 50 nt long, respectively.
A homology search with BLASTp showed that the deduced amino acid sequences of the two ORFs were very similar to that of the capsid protein (CP) and the RNA-dependent RNA polymerase (RdRp), respectively, of members of the family Totiviridae, particularly to that of Ustilaginoidea virens RNA virus 3 (UvRV3) (62 % aa sequence identity for the CP and 52 % for the RdRp), followed by Coniothyrium minitans RNA virus (CmRV) (64 % aa sequence identity for the CP and 46 % for the RdRp). The C-terminus of the putative CP encoded by ORF1 had a proline-rich region that is shared by many members in the family Totiviridae [17].
In addition, a conserved domain database (CDD) search and multiple protein alignments confirmed that the ORF2-encoded protein shared a conserved viral RdRp domain with eight conserved motifs characteristic of RdRps of dsRNA mycoviruses infecting lower eukaryotes [18] (Fig. S1). These results indicate that dsRNA-M comprises the genome of a novel mycovirus, which we designated as Phomopsis vexans RNA virus 1 (PvRV1).
Phylogenetic analysis was performed based on the deduced amino acid sequences of the RdRp in PvRV1 and other selected viruses in the family Totiviridae. The resulting neighbor-joining tree reflected the taxonomic structure of the family, with five clades corresponding to the genera Totivirus, Victorivirus, Giardiavirus, Leishmaniavirus, and Trichomonasvirus [19]. PvRV1 was clustered within the Victorivirus clade closer to Ustilaginoidea virens RNA virus 3 (Fig. 1c). A CP-based phylogenetic tree was also constructed, and it had a topology similar to that of the RdRp-based tree (Fig. S2), which further supported the taxonomic status of PvRV1.
It is of interest to note that the arrangement of ORF1 and ORF2 in the PvRV1 genome was unique and different from that of other victoriviruses (the two ORFs being separated by 98 nt instead of overlapping slightly). Magnaporthe oryzae virus 1 (MoV1), which contains only 2 nt between its two ORFs, is the other exception [17, 20, 21]. The existence of this intergenic region in the PvRV1 genome was confirmed by PCR amplification and cloning using specific primers spanning this region, ruling out the possibility of sequencing errors.
In victoriviruses, the RdRp is expressed as a separate protein by a coupled termination-reinitiation strategy. An upstream pseudoknot or long stem-loop structure located upstream of the stop-start region, together with the closely juxtaposed CP stop and RdRp start codons, are essential features of this mechanism [17, 21, 22]. Therefore, the distinct arrangement of its ORF might raise the question as to how the RdRp is expressed in PvRV1. According to our in silico analysis, a pseudoknot is present upstream of the stop codon of ORF1, with an estimated free energy of -22.72 kcal/mol. Its predicted structure is as follows (bold and italic lowercase letters indicated the base pairs that form stems): gcugggAACgcggc cccagcCCCgccgcUC (UAA). On the other hand, a slippery site, which would be required to induce ribosomal frameshifting, as observed in some totiviruses such as ScV-L-A and GLV [23, 24], was not found near the start codon of PvRV1 ORF2. The viruses that form the Victorivirus clade with PvRV1 in the RdRp-based tree all employ a stop/restart strategy to express their downstream ORF, consistent with the International Committee on Taxonomy of Viruses (ICTV) description of the genus Victorivirus.
It has been shown previously that the length of the spacer sequence between the upstream pseudoknot and the ORF1 stop codon is important for downstream ORF re-initiation, with spacers shorter than 6 nt or longer than 18 nt not supporting RdRp expression [21, 25]. Based on this, the PvRV1 RdRp may not be expressed effectively by coupled termination-reinitiation, since the spacer is only 2 nt long. However, two tandem stem-loop structures were detected within the intergenic region by Mfold at default settings (Fig. S3). The genome of another victorivirus, Magnaporthe oryzae virus 1 (MoV1), has only two nucleotides between the CP stop codon and the RdRp start codon but was shown to use the same stop/restart translation strategy. The common point for MoV1 and PvRV1 is the existence of a predicted pseudoknot [25]. Li et al. [21] provided evidence that HvV190S RdRp can initiate at a second AUG codon 90 nt downstream of the predicted pseudoknot and 80 nt downstream of the CP stop codon. Combing the information discussed above, we propose that viruses in the genus Victorivirus may not necessarily have overlapping ORFs, while still employing a coupled termination-reinitiation expression strategy. Further experiments to identify the polycistronic nature of the viral genomic RNA are required to confirm this hypothesis.
In summary, based on the predicted genome size, sequence similarities and phylogenetic analysis, we recommend the addition of this virus with a distinct ORF arrangement as a novel member of the genus Victorivirus in the family Totiviridae. As far as we know, this is the first report of a mycovirus found in the plant-pathogenic fungus Phomopsis vexans, the causal agent of phomopsis blight of eggplant.
References
Islam MM, Asaduzzaman M, Meah MB (2013) Molecular characterization of Phomopsis vexans isolates of eggplant of Bangladesh. J Sci Found 8(1–2):131–140
Singh RS (1992) Diseases of vegetable crops, 2nd edn. Oxford and IBH Publishing Company Pvt. Ltd., New Delhi, pp p119–p121
Kalda TS, Swarup V, Chowdhury B (1977) Resistance to Phomopsis blight in eggplant. Veg Sci 4(2):90–101
Ghabrial SA, Suzuki N (2009) Viruses of plant pathogenic fungi. Annu Rev Phytopathol 47:353–384
Pearson MN, Beever RE, Boine B, Arthur K (2009) Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol Plant Pathol 10:115–128
Kozlakidis Z, Herrero N, Ozkan S, Kanhayuwa L, Jamal A, Bhatti MF, Coutts RH (2013) Sequence determination of a quadripartite dsRNA virus isolated from Aspergillus foetidus. Arch Virol 158:267–272
Nuss DL (2011) Mycoviruses, RNA silencing, and viral RNA recombination. Adv Virus Res 80:25–48
Nuss DL (2005) Hypovirulence: mycoviruses at the fungal-plant interface. Nat Rev Microbiol 3:632–642
Morris TJ, Dodds JA (1979) Isolation and analysis of double stranded RNA from virus-infected plant and fungal tissue. Phytopathology 69:854–858
Chen L, Chen JS, Liu L, Yu X, Yu S, Fu TZ, Liu WH (2006) Complete nucleotide sequences and genome characterization of double-stranded RNA 1 and RNA 2 in the Raphanus sativus-root cv. Yidianhong [corrected]. Arch Virol 151:849–859
Zhong J, Lei XH, Zhu JZ, Song G, Zhang YD, Chen Y, Gao BD (2014) Detection and sequence analysis of two novel co-infecting double-strand RNA mycoviruses in Ustilaginoidea virens. Arch Virol 159:3063–3070. doi:10.1007/s00705-014-2144-x
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Sperschneider J, Datta A, Wise MJ (2011) Heuristic RNA pseudoknot prediction including intramolecular kissing hairpins. RNA 17:27–38
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Byun Y, Moon S, Han K (2007) A general computational model for predicting ribosomal frameshifts in genome sequences. Comput Biol Med 37:1796–1801
Ghabrial SA, Nibert ML (2009) Victorivirus, a new genus of fungal viruses in the family Totiviridae. Arch Virol 154:373–379
Bruenn JA (1993) A closely related group of RNA-dependent RNA polymerases from double-stranded RNA viruses. Nucleic Acids Res 21:5667–5669
Wickner RB, Ghabrial SA, Nibert ML, Patterson JL, Wang CC (2011) Family Totiviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowits EJ (eds) Virus taxonomy. Ninth report of the International Committee for the Taxonomy of Viruses. Elsevier/Academic Press Inc, New York, pp 639–650
Yokoi T, Yamashita S, Hibi T (2007) The nucleotide sequence and genome organization of Magnaporthe oryzae virus 1. Arch Virol 152:2265–2269
Li H, Havens WM, Nibert ML, Ghabrial SA (2011) RNA sequence determinants of a coupled termination–reinitiation strategy for downstream open reading frame translation in Helminthosporium victoriae virus 190S and other victoriviruses (Family Totiviridae). J Virol 85:7343–7352
Yie SW, Khalifa ME, Hahn T, Pearson MN (2013) Molecular characterization of a novel victorivirus from the entomopathogenic fungus Beauveria bassiana. Arch Virol 159:1321–1327
Ten Dam EB, Pleij CWA, Bosch L (1990) RNA pseudoknots: translational frameshifting and read-through on viral RNAs. Virus Genes 4:121–136
Wang AL, Yang HM, Shen KZ, Wang CC (1993) Giardiavirus double stranded RNA genome encodes a capsid polypeptide and a gag-pol-like fusion protein by a translation frameshift. Proc Nat Acad Sci USA 90:8595–8599
Li H, Havens WM, Nibert ML, Ghabrial SA (2015) An RNA cassette from Helminthosporium victoriae virus 190S necessary and sufficient for stop/restart translation. Virology 474:131–143
Acknowledgments
This study was funded by the National Key Technology R&D Program of China (2012BAD15B04-1).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, R.J., Zhong, J., Shang, H.H. et al. The complete nucleotide sequence and genomic organization of a novel victorivirus with two non-overlapping ORFs, identified in the plant-pathogenic fungus Phomopsis vexans . Arch Virol 160, 1805–1809 (2015). https://doi.org/10.1007/s00705-015-2420-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-015-2420-4