Abstract
Gallid herpesvirus 2 (GaHV-2) is the alphaherpesvirus responsible for Marek’s disease (MD), a T-cell lymphoma of chickens. The virulence of the GaHV-2 field strain is steadily increasing, but MD is still controlled by the CVI988/Rispens vaccine. We tried to determine distinguishing traits of the CVI988/Rispens vaccine by focusing on the 5′ end region of the latency-associated transcript (5′LAT). It includes a variable number of 60-bp tandem repeats depending on the GaHV-2 strain. By analyzing six batches of vaccine, we showed that CVI988/Rispens consisted of a population of 5′LAT molecular subtypes, all with deletions and lacking 60-bp tandem repeat motifs, with two major subtypes that probably constitute CVI988/Rispens markers. Serial passages in cell culture led to a substantial change in the frequency of CVI988/Rispens 5′LAT subtypes, with non-deleted subtypes harboring up to four 60-bp repeats emerging during the last few passages. Dynamic changes in the distribution of 5′LAT-deleted subtypes were also detected after infection of chickens. By contrast, the 5′LAT region of the oncogenic clonal RB-1B strain, which was investigated at every step from the isolation of the clonal bacmid RB-1B DNA to the isolation of the ovarian lymphoma cell line, consisted of non-deleted 5′LAT subtypes harboring at least two 60-bp repeats. Thus, vaccine and oncogenic GaHV-2 strains consist of specific populations of viral genomes that are constantly evolving in vivo and in vitro and providing potential markers for epidemiological surveys.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gallid herpesvirus type 2 (GaHV-2), also known as Marek’s disease (MD) virus serotype 1 (MDV-1), is an alphaherpesvirus that causes a rapid-onset T-cell lymphoma in chickens [1]. GaHV-2 belongs to genus Mardivirus of the subfamily Alphaherpesvirinae, together with two other closely related but non-oncogenic avian viruses, gallid herpesvirus type 3 (GaHV-3, MDV serotype 2), isolated from chickens, and serotype 3 herpesvirus of turkey (HVT) or meleagrid herpesvirus type 1 (MeHV-1). The highly contagious Marek’s disease has been controlled by worldwide vaccination since the 1970s, the initial MD vaccine being the first licensed commercially available vaccine against a neoplastic disease. Since the isolation and market release of the heterologous HVT vaccine [2], various strategies have been developed for the design of new and more effective vaccines [3, 4]. The CVI988/Rispens vaccine, isolated from a flock of viremic chickens without significant clinical signs of MD and then attenuated by serial passages in cell culture [5, 6], remains the gold standard MD vaccine [7]. As vaccinated chickens do not develop MD, but may still become infected with circulating strains of GaHV-2, vaccination seems to have played a major role in driving the evolution of GaHV-2 virulence, with waves of increase in field virus virulence being linked to the introduction of different generations of vaccines [7–9]. Consequently, GaHV-2 strains are now classified into virulent (v), very virulent (vv), and very virulent plus (vv+) pathotypes [8]. This increase in virulence over time strongly suggests that GaHV-2 strains display genomic variability that potentially contributes to their adaptability. GaHV-2 strains have been shown to consist of mixed viral populations, on the basis of molecular data from deep sequencing [10, 11] or BAC cloning [12, 13]. These findings are consistent with initial observations for human herpesvirus type 1 (HHV-1) [14] and subsequent reports for other members of the family Herpesviridae [15, 16]. The GaHV-2 genome is organized into two unique regions—the long (UL) and short (US) unique regions—encompassing core genes and flanked by the inverted internal and terminal long-region (IRL/TRL) and short-region (IRS/TRS) repeats, respectively [17]. Comparative genomics studies of attenuated GaHV-2 strains and strains with various levels of virulence have led to the identification of six major regions with highly variable sequences [18]. One of these regions is the a-like sequence region [19], positioned in a direct orientation at the terminus of each genome and in an inverted orientation at the L-S junction. The a region, which encodes several cis-acting elements, including conserved motifs such as the pac-1 and pac-2 sites directing the cleavage and packaging of unit-length genomes into the viral capsid in alphaherpesvirus genomes [20], has been shown to be highly variable in HHV-1 [21] and human herpesvirus type 6 (HHV-6) [22]. In addition to the conserved a-consensus motifs, the GaHV-2 a-like sequence contains two telomeric repeat islands, one of which is invariable, the other containing a highly variable number of repeats, depending on the virulence of the GaHV-2 strain [18, 23]. The 5′LAT (latency-associated transcript) region in the RS region [24, 25], just downstream from the a-like region, also encompasses a highly variable region consisting of either a variable number of 60-base-pair (bp) motifs tandemly repeated in virulent GaHV-2 strains, or lacking these motifs in attenuated and vaccine strains of GaHV-2 [18]. These polymorphic tandem 60-bp repeats carry no ORF but have been shown to control the expression of LAT transcripts [26] and intronic clustered microRNAs potentially involved in MD lymphomagenesis [27]. In their comparative sequence analysis, Spatz and Silva [18] showed that one of the three CVI988/Rispens isolates analyzed harbored two 60-bp repeats, whereas the other two had deletions of the 5′LAT region extending into the first intron of the LAT gene in both cases, with the deletion boundary at the 3′end preserving the entire microRNA cluster in one case and resulting in the deletion of mdv1-miR-M8 in the other [18]. CVI988/Rispens remains the most effective vaccine available for MD control worldwide. Thus, in this study, we focused on the 5′ LAT region, from nucleotide 142,484 to nucleotide 144,337 (EF523390.1), encompassing the 60-bp tandem repeat motifs, the TSS and the first exon of the LAT gene, and part of the first LAT intron, up to the 3′ end of the mdv1-miR-M8-M10 cluster [26]. We carried out a comparative study on the 5′ LAT region of CVI988/Rispens vaccines and that of the oncogenic RB1B strain cloned as a bacmid [28]. We identified distinguishing traits of CVI988/Rispens that might be useful for epidemiological surveys and for future development of vaccines.
Materials and methods
Ethics statement
All experiments were performed in accordance with the guidelines of the National Charter on the Ethics of Animal Experimentation. The protocols were approved by the Committee on the Ethics of Animal Experiments of Val de Loire (Numbers GD ST-04-06-A and GD 01-10B), according to directive 2010/63/UE. They were carried out in isolation units, under conditions of strict confinement and controlled access, at INRA (PFIE, 37380 Nouzilly, France).
Chickens
Four-week-old White Leghorn B13/B13 specific-pathogen-free (SPF) chickens lacking maternal antibodies against GaHV-2 and highly susceptible to MD were used. They were hatched and raised at INRA (PFIE, 37380 Nouzilly, France).
Vaccines and BAC-derived virus
Six batches of CVI988/Rispens vaccine were used. Four batches of monovalent vaccine consisting of CVI988 alone (MD9500, MD10300 and MD18800, provided by Pfizer Animal Health (AH), A290A provided by Intervet) and two batches of bivalent vaccine consisting of the CVI988/Rispens strain associated with MeHV-1 HVT (MC23900 and MC24000 provided by Pfizer AH) were used.
The oncogenic GaHV-2 strain RB-1B, cloned as a bacmid, was used in this study. BACRB1B-5 derived virus was recovered from the parental pRB1B-BAC5 as described below [28].
Cells, infection with CVI988/Rispens vaccine and reconstitution of BAC-derived virus
Chicken embryo fibroblasts (CEFs) were prepared from 11-day-old specific-pathogen-free (SPF) White Leghorn B13/B13 embryos raised at INRA (PFIE, 37380, Nouzilly, France) and used as secondary cultures [29]. CEFs were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 2.5 % fetal calf serum, 1.25 % chicken serum, 1 % penicillin/streptomycin, 1 % Fungizone and 2 % tryptose phosphate broth (TPB). Secondary CEF monolayer cultures were used to propagate CVI988/Rispens vaccine, which was stored in vials at -140 °C and then diluted 1/10, and CVI988/Rispens cell-associated vaccine from serial passages carried out every 3–4 days with 1/3 to 1/30 dilutions of the cell-associated CVI988/Rispens from the previous passage. CEF monolayer cultures were also used to reconstitute RB-1B BAC-derived virus by transfection with 1 μg of BAC DNA complexed with Lipofectamine® (Invitrogen®), in accordance with the manufacturer’s instructions.
GaHV-2 infection was monitored in indirect immunofluorescence assays in which the major capsid protein VP5 was labeled with anti-VP5 monoclonal antibody as described previously [30].
Vaccination of chickens with CVI988/Rispens
A group of chickens was vaccinated by intramuscular injection with 1000 PFU (plaque-forming units) of Pfizer AH CVI988/Rispens batch MD18800. PBLs (peripheral blood leucocytes) and FFs (feather follicles) were collected from three chickens at various time points after vaccination as described previously [31, 32].
Infection of chickens with BACRB1B-derived virus
A group of chickens was infected by intramuscular injection of 1000 PFU of BACRB1B-5 cell-associated virus. PBLs were collected at various time points after infection (pi) as described previously [31].
Building of 5′LAT DNA libraries
We investigated the 5′LAT region of GaHV-2 in more detail by establishing genomic libraries from different sources of CVI988/Rispens-vaccine-infected cells and at various stages of infection in vitro and in vivo with the BACRB1B oncogenic GaHV-2 strain. Genomic DNA was extracted from GaHV-2-infected CEFs or PBLs after incubation with 5 mg/ml proteinase K, 5 % sodium dodecyl sulfate and 100 mM Tris, pH 8, for 5 h to 24 h at 65 °C. DNA was then purified by phenol-chloroform extraction and precipitated in ethanol. The 5′LAT amplicons were obtained using the forward primer F-M448 (5′-GCTAGGGGTTCGACGAAAT-3′) and reverse primer R-M688 (5′-CCGGACCGAGAACACAGTGAT-3′) or R-M766 (5′-GATAGTTATATAAGCCGTTATATAG-3′). With the exception of the DNAs corresponding to BAC RB1B-5 samples, all 5′LAT DNAs were amplified by PCR (30 cycles of denaturation [94 °C for 1 min], annealing [55 °C for 1 min] and extension [72 °C for 2 min], followed by a final extension at 72 °C for 10 min) with the appropriate primers at a concentration of 100 nM (Table 1) in a final volume of 50 μl containing 1.5 mM MgCl2, 200 nM dNTPs and 0.01 units/μl ThermoPrime Taq™ polymerase in ReddyMix™ PCR Master Mix buffer (Thermo Scientific). For the BAC RB-1B5 samples, nested PCR was carried out using the same conditions, except that we used 0.01 units/μl GoTaq® Polymerase in Green GoTaq® Flexi reaction buffer (Promega). All PCR products were inserted into pGEM®-T Easy Vector (Promega) in accordance with the manufacturer’s instructions. Screening of the clones was carried out by PCR from bacterial colonies, using the forward primer M13 (−21) (5′-TGTAAAACGACGGCCAGT-3′) and the reverse primer M13 rev (29) (5′-CAGGAAACAGCTATGACC-3′). All of the clones were analyzed. Electrophoretic analysis was performed, sequencing was performed by GATC Biotech AG (Germany), and the corresponding sequences were aligned with the GaHV-2-RB1B genome sequence (EF523390.1). The number of 60-bp repeats was determined after PCR screening with F-M448 and R-M714 (5′-CGGATGCTGGAGCTGCCGCCAAACTTG-3′) primers, followed by electrophoretic analysis using a 1 % agarose gel performed with a ladder indicating the number of repeats. Directed PCRs with specific forward primers A366 (5′-TTCCGTAGTGTTCTCGTGACAC-3′) and A349 (5′-GGCCGCGAGAGGGTTAGAGGGCGT-3′) were designed and used to identify specific variants α and β. All of the CVI988/Rispens sequences were checked for the presence of the CVI988-characteristic SNP located at position 143,689 (EF523390.1).
Statistical analysis
Data were subjected to one-way analysis of variance with the R statistical package (R Project for Statistical Computing, www.r-project.org/). We then carried out a χ2 test to compare the distributions of 5′LAT molecular subtypes and a Student t-test to compare their frequencies. Comparisons were considered significant if P ≤ 0.05.
Results
CVI988 vaccines consist of sets of 5′LAT-deleted molecular subtypes
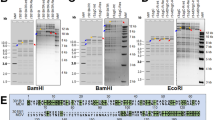
Going beyond the findings published by Spatz and Silva [18] showing that some CVI988/Rispens isolates have different 5′LAT regions, we carried out an extensive analysis of the 5′LAT region in six different batches of vaccine. We identified at least 29 different molecular subtypes among the 543 PCR product sequences obtained. All of these subtypes displayed deletions of part of the 5′LAT region, involving, in all cases, the 60-bp repeats and the TSS of the LAT gene (position 142,734 of the pRB-1B-5 genome; GenBank accession no. EF523390.1) [26] (Fig. 1, Table S1). With the exception of subtype N, all molecular subtypes shared the same 5′ boundary position (position 142544 of pRB-1B-5 genome; GenBank accession no. EF523390.1). By contrast, the 3′ boundary position differed between subtypes (Fig. 1, Table S1). Some deletions preserved the entire mdv1-miR-M8-M10 cluster (β subtype; A to G4i subtypes), whereas others extended into the mdv1-miR-M8-M10 cluster (α, δ, ε, H to L subtypes) or completely eliminated this cluster (γ, Li and M). Subtype N had a specified 5′ boundary position (142,607), resulting in the preservation of one 60-bp repeat motif and a 3′ boundary position (143,584) within mdv1-miR-M6-3p, affecting this 3′ arm and resulting in the deletion of the entire mdv1-miR-M8 and M6-5p cluster (Fig. 1, Table S1). The frequencies of the different subtypes in the various batches of CVI988 vaccine are reported in Table 2. Two groups of subtypes were distinguished. First, the prevailing subtypes α, β, γ, δ and ε were common to all vaccine batches. The frequencies of subtypes α, β, γ, δ, ε were similar for all batches of Pfizer AH vaccine tested, regardless of the production process, the CVI988 vaccine being present in isolation or associated with MeHV-1 HVT (Table 2). The α and β subtypes were always present at the highest frequencies, regardless of the vaccine producer. The α subtype was systematically more frequent (44 to 56 %) than the β subtype in Pfizer AH batches, whereas the β subtype was more frequent (45 %) than the α subtype in the Intervet A290A batch (Table 2). Unlike the α, β, γ, δ and ε subtypes, the minority subtypes A to N, accounting for 3 to 15 % of the subtypes per Pfizer AH batch and 17 % of the subtypes per Intervet A290A batch, were found only in certain batches. Their frequency differed significantly between Pfizer AH batches, and some subtypes were found in only one batch (Table 2). The distribution of the minority subtypes A to N may, therefore, constitute a hallmark of the CVI988 vaccine batch.
The 5′LAT molecular subtypes of CVI988/Rispens vaccines. (A) Schematic representation of the GaHV-2 genome (top), expanded below to show the details of the LAT locus between primers M448 and M766, with coordinates relative to the RB-1B genome (EF 523390). (B) Representation of the different molecular subtypes found in five batches of CVI988/Rispens vaccine from Pfizer AH and one from Intervet. Subtypes common to all batches of vaccine are named α to ε. The minority subtypes present only in some batches of vaccine, mostly at frequencies below 5 %, are named A to N. The letter “i” in lower case corresponds to subtypes detected only in the CVI988 batch from Intervet. Black arrows indicate microRNAs mdv1-miR-M8 to 10; black squares indicate sequences downstream from the telomeric region to the 60-bp repeat promoter region; white hatched boxes indicate 60-bp repeats; black diamonds indicate p53 response elements; the pale gray box indicates LAT gene exon 1; the gray box indicates LAT gene intron 1; the white boxes indicate microRNAs; the white triangles indicate the 5′ ends of molecular subtypes (α to ε, and A to N); the gray square indicates the transcription start site (TSS); black triangles indicate the 3′ boundaries of the molecular subtypes (α to ε, and A to N)
Dynamic changes in 5′LAT CVI988 subtypes occur in vitro and in vivo
The demonstration that the CVI988 vaccine consisted of a population of molecular subtypes with different 5′LAT regions led us to assess whether the frequencies of these subtypes were stable or varied during serial passages in cell culture, as currently used in vaccine production procedures. We infected secondary B13 CEFs with CVI988 MC18800, provided by Pfizer AH, and subjected the cells to 65 serial passages in culture. Every five passages, we performed a molecular analysis of the 5′LAT region as described previously, with the analysis of about 100 clones per passage sampled. We plotted the change in 5′LAT subtype frequency against the number of passages (Fig. 2). From passage 1 to 40, there was a gradual, continuous change in 5′LAT subtype distribution, characterized principally by a dynamic equilibrium involving mostly the α and β subtypes, with a continuous decrease in the frequency of the α subtype and a continuous increase in that of the β subtype (Fig. 2A). At passage 45, a breakdown of subtype evolution occurred, with an unusual pattern showing a sudden decrease in β subtype frequency and a substantial increase in the frequency of the minor N subtype (Fig. 2A). From passage 45 onwards, CVI988 infection was more difficult to manage; the frequency of the α subtype fell below 10 %, and that of the β subtype also decreased, in parallel with an increase in the frequency of the N subtype (Fig. 2A). At passage 65, a breakdown occurred, with an absence of detectable CVI988 infection (no labeling with VP5 antibody and no PCR product with the M448/M766 primers). However, we carried out additional serial passages after passage 65, and a few plaques typical of GaHV-2 infection were again observed from passage 66 onward. We also initiated a new series of passages from CVI988-infected frozen CEFs at passage 55 to prevent specific bottlenecks due to the loss of infection at passage 65 (Fig. 2B). Surprisingly, in both series, we observed the emergence of new CVI988 subtypes harboring complete 5′ LAT regions with one to four 60-bp repeats (Fig. 2). At passage 80, these complete CVI988 subtypes accounted for 25 and 15 % of all subtypes for the first and second series of passages, respectively.
Changes in 5′ LAT molecular subtype frequencies in CVI988 during serial passages in CEFs. The percentages of the different molecular subtypes are shown as a function of the number of passages, P0 to P80, with P0 representing the subtype distribution in the initial MC18800 batch. The α, β and N subtypes are represented individually, whereas the other subtypes are grouped together as “Others”. “Complete” subtypes correspond to the non-deleted 5′ LAT region with at least one 60-bp repeat. (A) First series, from P0 to P80. (B) Second series, initiated from frozen CVI988/Rispens-infected cells, from P55 to P80
We then investigated whether the distribution of the 5′LAT subtypes of the CVI988 vaccine also changed in vivo. We analyzed the pattern of change in 5′LAT subtypes at various time points after infection, in PBLs and FFs, both representative of GaHV-2 infection. PBLs and FFs were sampled from B13 chickens vaccinated with the same batch of vaccine used for serial passages, CVI988 MC18800. CVI988 infection rates were lower in PBLs and FFs than in CEFs, and nested PCR was therefore required for detection. We analyzed about 1000 clones and observed changes in the distribution of CVI988 subtypes in both tissues at various times pi. The data obtained for PBLs on day 7 pi showed an increase in α subtype frequency and a decrease in β subtype frequency with respect to the content of the vial of CVI988 used to inoculate chickens (Fig. 3). The overall proportions of α and β subtypes were similar on day 14 pi, but the frequencies of these two subtypes differed from those in the vial and those on day 7 pi (Fig. 3A). Remarkably, as observed from passage 45 after CEF infection, the frequency of the minor subtype N reached 69 % on day 29 pi. The data for FFs were similar to those for PBLs, but with a time lag, demonstrating an absence of tissue specificity in the changes in CVI988 subtypes (Fig. 3B). No subtype without deletions was observed in samples collected for this in vivo CVI988 infection survey.
Changes in 5′ LAT molecular subtype frequencies in the CVI988 vaccine after infection of chickens. Percentages of the different molecular subtypes in PBLs at 7, 14 and 29 dpi (A) and in FF (feather follicles) at 14 and 21 dpi (B), in chickens vaccinated with batch MC18800. The α, β and N subtypes are represented individually, whereas the other subtypes are grouped together as “Others”
The number of 60-bp repeats in the 5′ LAT region of the clonal BACRB-1B-5 strain varies during in vitro and in vivo infection
We assessed whether the population subtypes of the 5′LAT region and their dynamic evolution were specific to the CVI988/Rispens vaccines. We investigated the pattern of the 5′LAT region of an oncogenic clonal GaHV-2 strain by monitoring the 5′LAT subtypes found at each step from the isolation of the clonal pRB-1B-5 BAC DNA [28] to that of the BACRB-1B-5-induced 54-O ovarian lymphoma cell line (Fig. 4A). In contrast to our observations for the CVI988/Rispens vaccines, we detected no deletion in the 5′LAT region during the course of isolation of the 54-O ovarian GaHV-2 lymphoma cell line. However, we did observe changes in the number of 60-bp repeats during the isolation of the 54-O lymphoma cell line. As expected, the initial pRB-1B-5 BAC contained only three 60-bp repeats. Notably, a polymorphism with respect to the number of repeats, which ranged from two to nine, was detected by the third passage after the transfection of CEFs with pRB-1B-5 BAC DNA; the form with the highest frequency was that with three 60-bp repeats (Fig. 4). This polymorphism was also observed in an independent experiment in which the number of repeats ranged from two to seven, with the three 60-bp-repeat form having a frequency of 68 % (data not shown). During the course of in vivo infection, polymorphism in the number of repeats was observed in PBLs collected on days 7 and 14 pi from female chicken no. 54. Again, the form with the highest frequency was consistently that with three 60-bp repeats (Fig. 4). At the time of lymphoma development, the frequency of the three-60-bp repeat form in PBLs from female chicken no. 54 increased from 63 % on day 21 pi to 98 % on day 35 pi, corresponding to the selection of three 60-bp repeats only at this time (Fig. 4). On day 35 pi, a necropsy was carried out on chicken no. 54, which was found to have an ovarian lymphoma in which the frequency of the three-60-bp form was also 98 % (Fig. 4). We cultured the ovarian lymphoma cells [33] and determined the number of 60-bp repeats after four months of permanent culture. The frequency of the three-60-bp form had decreased to about 80 % by this time (Fig. 4).
Changes in the number of 60-bp repeats during infection with BACRB-1B-5. (A) Diagram of the successive steps from the transfection of CEFs with clonal pRB-1B-5 BAC DNA to isolation of the 54-O ovarian cell line, beginning with three CEF passages to reconstitute BACRB-1B-5-derived virus from CEFs (BACRB-1B-5 CEF (3)). Seven-week-old chickens were inoculated with 1000 PFU of BACRB-1B-5 CEF (3) virus. Blood samples were obtained from chicken C54 on days 7, 14, 21, 28 and 35 days postinfection (C54PBL7 to 35), and an ovarian lymphoma (C54COL) was collected 38 days postinfection (dpi) for the establishment of a cell line (54-O). (B) Percentages corresponding to the frequency of the forms with different numbers of 60-bp repeats at these successive steps are shown
Discussion
In this study, we characterized the genotypic composition of current commercial CVI988/Rispens vaccines and showed that CVI988/Rispens vaccine batches actually consist of a heterogeneous population of 5′LAT molecular subtypes (Table 2). This region was previously reported to vary with GaHV-2 pathotype, with oncogenic vv and vv+ strains having two to nine repeats, whereas most mildly virulent and vaccine strains have no 60-bp repeats, their deletions sharing the same 5′ end boundary and extending to various extents into the LAT gene, with variable 3′-end boundaries [18]. We extended these findings by identifying, in six commercial batches, 29 molecular subtypes in total, all displaying deletions of various lengths, but invariably encompassing the 60-bp repeats (Fig. 1). The two most frequently detected CVI988/Rispens molecular subtypes in the vaccine batches analyzed, which we named a and b, were previously found in the comparative analysis carried out by Spatz and Silva [18] on CVI988/Rispens Intervet and BP5 isolates [34]. The characteristics of the distribution of populations of 5′LAT molecular subtypes in the vaccine batches analyzed were developed further in the case of the CVI988/Rispens vaccine. Indeed, in addition to the α and β subtypes, three other molecular subtypes were detected in all vaccine batches. These five molecular subtypes common to all vaccine batches could thus be used as genetic markers of the CVI988/Rispens vaccine (Table 2). By contrast, the other molecular subtypes, referred to as minority subtypes and detected only in specific vaccine batches, may represent signatures of particular batches (Table 2). Genetic variability among CVI988/Rispens vaccine batches had been previously reported at the locus encoding viral telomerase RNA [35]. The identification of two sorts of molecular subtypes, one common to all CVI988/Rispens vaccine batches and one specific to particular batches, could be used to develop new tools for controlling vaccine production and for vaccine epidemiological surveys. Moreover, it would be informative to determine the genotypic composition of CVI988/Rispens clone C, which has been reported to be less protective than other CVI988/Rispens isolates [36–38]. We further assessed the effect of serial passages in CEFs, which are currently used in CVI988/Rispens vaccine production, on the molecular composition of the 5′LAT of the resulting vaccine. By comparison with the initial frequencies found in the vaccine batch MC18800, we observed a dynamic change in the frequencies of 5′LAT molecular subtypes with the number of serial passages, consistent with the currently observed bottleneck effect of cell culture on viral populations, and also consistent with the presence of quasispecies, which were first described for RNA viruses and have also recently been reported for HHV-1 [39] (Fig. 2). However, we again observed a dynamic pattern of change in 5′LAT molecular subtypes (Fig. 3) after infection of chickens with CVI988/Rispens. Our data extend previous findings indicating that GaHV-2 exists as a collection of mixed populations in vitro [10, 11] and definitively demonstrate the existence of mixed populations of GaHV-2 genomes in vivo, as first suggested by the reversion of attenuation upon back passages of the vaccine in vivo [40–42]. We investigated whether CVI988/Rispens vaccines consisted of specific of 5′LAT molecular subtypes. We therefore carried out a survey on an oncogenic GaHV-2 strain, using a clonal BACRB-1B strain reconstituted from a BAC and containing three 60-bp repeats in both the IRs and TRs regions. In contrast to the 5′LAT region of CVI988/Rispens, the 5′LAT region of BACRB-1B was always complete and contained at least two 60-bp repeats. In addition, a rapid amplification of the 60-bp repeat obtained with the clonal BAC occurred, generating a pattern of two to nine repeats after as few as three serial passages in CEFs following transfection with BAC DNA (Fig. 4), consistent with the rapid variations observed in nucleotide stretches [43]. We again observed a pattern in the number of 60-bp repeats in PBLs from chickens infected with BACRB-1B, with variability until day 21 pi, followed by selection at 35 days pi (Fig. 4), probably reflecting an evolutionary bottleneck phenomenon at the time of lymphomagenesis, consistent with a clonal origin of MD lymphomas [44–46]. The rapid amplification of the 60-bp repeat obtained with the clonal BAC strongly suggests that recombination events commonly occurring during replication of the DNA of viruses of the family Herpesviridae [47, 48] may be involved in generating the genetic diversity of the 5′LAT region in GaHV-2 strains. The location of the 60-bp repeats in the vicinity of the a-like sequence involved in the cleavage-packaging of the viral DNA genome [19, 20, 23] and displaying the pac2 signal 21 bp upstream from the 5′-end boundary of the 60-bp repeat deletion may have favored genetic variation in this area, resulting, in particular, in variability of the number of 60-bp repeats and their deletions, as shown for the expansion and contraction of closely located telomeric repeats [23]. The dynamic changes in the frequencies of the 5′LAT molecular subtypes of CVI988/Rispens with the number of serial passages in cell culture observed here may account for previous observations of differences in residual pathogenicity and protection between CVI988/Rispens clones isolated at different passages [36, 37]. Three main characteristics emerge from this dynamic pattern of change. First, the frequencies of the two major subtypes changed considerably, with a large increase in β subtype frequency accompanied by a decrease in the frequency of the a subtype (Fig. 2). This suggests that the commercial CVI988/Rispens vaccines provided by Fort Dodge/Pfizer Animal Health may have been produced after fewer passages than the Intervet vaccine. Second, we observed a considerable increase in the frequency of minority subtype N (Fig. 2), which also occurred in PBLs from chickens inoculated with the MC18800 vaccine (Fig. 3). The increase in the frequency of minority subtype N, therefore, did not result from a specific CEF culture bottleneck (Fig. 2 and 3), and its origin remains unclear. Further investigations are required to determine whether the frequencies of the other minority subtypes increased and whether the single 60-bp repeat present in subtype N played a role in the increase in the frequency of this subtype. Third, from passage 65, in both experimental series, we detected unexpected 5′LAT molecular subtypes without deletions, harboring one to four 60-bp repeats (Fig. 2). This observation was made on two occasions, suggesting that the detection of non-deleted 5′LAT molecular subtypes was not an incidental event. It seems likely that non-deleted 5′LAT molecular subtypes are present in CVI988/Rispens vaccine batches at a frequency too low to be picked up in our PCR and cloning protocols. This hypothesis is consistent with the presence of two 60-bp repeats in the 5′LAT regions of the CVI988/Rispens BAC clone [12]. The results presented here extend previous observations of changes in the CVI988/Rispens genome during serial passages, including (i) a deletion within the LAT gene, located downstream from the 5′LAT region analyzed in this study [49], and (ii) an increase in the number of head-to-tail copies of a 132-bp repeat located in the RL region, in the vicinity of the GaHV-2 origin of replication [50]. Non-deleted 5′LAT molecular subtypes may emerge at the same time as alterations in other CVI988/Rispens genomic regions, consistent with the occurrence of limited GaHV-2 genome sequence heterogeneity during serial passages [11]. Further studies are required to determine the precise effect of these non-deleted 5′LAT molecular subtypes on CVI988/Rispens attenuation and protection.
Our data provide potential genetic markers useful in monitoring vaccine production and in epidemiological vaccine surveys. Moreover, they suggest that the signature and frequency of molecular subtypes, with a high prevalence of 5′LAT subtypes harbouring at least two 60-bp repeats, determine the degree of pathogenicity of GaHV-2 strains. The introduction of the molecular subtypes most frequently detected in all CVI988/Rispens vaccines into a virulent GaHV-2 strain may be used for the development of new vaccines [51].
References
Osterrieder N, Kamil JP, Schumacher D, Tischer BK, Trapp S (2006) Marek’s disease virus: from miasma to model. Nat Rev Microbiol 4:283–294
Okazaki W, Purchase HG, Burmester BR (1970) Protection against Marek’s disease by vaccination with a herpesvirus of turkeys. Avian Dis 14:413–429
Calnek BW, Schat KA, Peckham MC, Fabricant J (1983) Field trials with a bivalent vaccine (HVT and SB-1) against Marek’s disease. Avian Dis 27:844–849
Witter RL, Lee LF (1984) Polyvalent Marek’s disease vaccines: safety, efficacy and protective synergism in chickens with maternal antibodies. Avian Pathol 13:75–92
Rispens BH, van Vloten H, Mastenbroek N, Maas HJ, Schat KA (1972) Control of Marek’s disease in the Netherlands. I. Isolation of an avirulent Marek’s disease virus (strain CVI 988) and its use in laboratory vaccination trials. Avian Dis 16:108–125
Rispens BH, van Vloten H, Mastenbroek N, Maas JL, Schat KA (1972) Control of Marek’s disease in the Netherlands. II. Field trials on vaccination with an avirulent strain (CVI 988) of Marek’s disease virus. Avian Dis 16:126–138
Gimeno IM (2008) Marek’s disease vaccines: a solution for today but a worry for tomorrow? Vaccine 26(Suppl 3):C31–C41
Witter RL (1997) Increased virulence of Marek’s disease virus field isolates. Avian Dis 41:149–163
Atkins KE, Read AF, Savill NJ, Renz KG, Islam AF, Walkden-Brown SW, Woolhouse ME (2013) Vaccination and reduced cohort duration can drive virulence evolution: Marek’s disease virus and industrialized agriculture. Evolution 67:851–860
Spatz SJ (2010) Accumulation of attenuating mutations in varying proportions within a high passage very virulent plus strain of Gallid herpesvirus type 2. Virus Res 149:135–142
Spatz SJ, Volkening JD, Gimeno IM, Heidari M, Witter RL (2012) Dynamic equilibrium of Marek’s disease genomes during in vitro serial passage. Virus Genes 45:526–536
Petherbridge L, Howes K, Baigent SJ, Sacco MA, Evans S, Osterrieder N, Nair V (2003) Replication-competent bacterial artificial chromosomes of Marek’s disease virus: novel tools for generation of molecularly defined herpesvirus vaccines. J Virol 77:8712–8718
Spatz SJ, Smith LP, Baigent SJ, Petherbridge L, Nair V (2011) Genotypic characterization of two bacterial artificial chromosome clones derived from a single DNA source of the very virulent gallid herpesvirus-2 strain C12/130. J Gen Virol 92:1500–1507
Kaerner HC, Schroder CH, Ott-Hartmann A, Kumel G, Kirchner H (1983) Genetic variability of herpes simplex virus: development of a pathogenic variant during passaging of a nonpathogenic herpes simplex virus type 1 virus strain in mouse brain. J Virol 46:83–93
Renzette N, Bhattacharjee B, Jensen JD, Gibson L, Kowalik TF (2011) Extensive genome-wide variability of human cytomegalovirus in congenitally infected infants. PLoS Pathog 7:e1001344
Szpara ML, Tafuri YR, Parsons L, Shamim SR, Verstrepen KJ, Legendre M, Enquist LW (2011) A wide extent of inter-strain diversity in virulent and vaccine strains of alphaherpesviruses. PLoS Pathog 7:e1002282
Tulman ER, Afonso CL, Lu Z, Zsak L, Rock DL, Kutish GF (2000) The genome of a very virulent Marek’s disease virus. J Virol 74:7980–7988
Spatz SJ, Silva RF (2007) Sequence determination of variable regions within the genomes of gallid herpesvirus-2 pathotypes. Arch Virol 152:1665–1678
Kishi M, Bradley G, Jessip J, Tanaka A, Nonoyama M (1991) Inverted repeat regions of Marek’s disease virus DNA possess a structure similar to that of the a sequence of herpes simplex virus DNA and contain host cell telomere sequences. J Virol 65:2791–2797
Roizman B, Knipe DM, Whitley RJ (2007) Herpes simplex virus. In: Knipe DM, Howley PM (eds) Fields virology. Lippincott Williams & Wilkins, Philadelphia, pp 2502–2601
Umene K, Oohashi S, Yoshida M, Fukumaki Y (2008) Diversity of the a sequence of herpes simplex virus type 1 developed during evolution. J Gen Virol 89:841–852
Deng H, Dewhurst S (1998) Functional identification and analysis of cis-acting sequences which mediate genome cleavage and packaging in human herpesvirus 6. J Virol 72:320–329
Volkening JD, Spatz SJ (2013) Identification and characterization of the genomic termini and cleavage/packaging signals of gallid herpesvirus type 2. Avian Dis 57:401–408
Cantello JL, Anderson AS, Morgan RW (1994) Identification of latency-associated transcripts that map antisense to the ICP4 homolog gene of Marek’s disease virus. J Virol 68:6280–6290
Li DS, Pastorek J, Zelnik V, Smith GD, Ross LJ (1994) Identification of novel transcripts complementary to the Marek’s disease virus homologue of the ICP4 gene of herpes simplex virus. J Gen Virol 75(Pt 7):1713–1722
Strassheim S, Stik G, Rasschaert D, Laurent S (2012) mdv1-miR-M7-5p, located in the newly identified first intron of the latency-associated transcript of Marek’s disease virus, targets the immediate-early genes ICP4 and ICP27. J Gen Virol 93:1731–1742
Stik G, Laurent S, Coupeau D, Coutaud B, Dambrine G, Rasschaert D, Muylkens B (2010) A p53-dependent promoter associated with polymorphic tandem repeats controls the expression of a viral transcript encoding clustered microRNAs. RNA 16:2263–2276
Petherbridge L, Brown AC, Baigent SJ, Howes K, Sacco MA, Osterrieder N, Nair VK (2004) Oncogenicity of virulent Marek’s disease virus cloned as bacterial artificial chromosomes. J Virol 78:13376–13380
Schat KA, Purchase HG (1998) Cell culture methods. In: Swayne DE, Glisson JR, Jackwood MW, Pearson JE, Reed WM (eds) A laboratory manual for the isolation and dentification of avian pathogens. Am Assoc Avian Pathol, Kennet Square, PA, pp 223–234
Amor S, Strassheim S, Dambrine G, Remy S, Rasschaert D, Laurent S (2011) ICP27 protein of Marek’s disease virus interacts with SR proteins and inhibits the splicing of cellular telomerase chTERT and viral vIL8 transcripts. J Gen Virol 92:1273–1278
Djeraba-AitLounis A, Soubieux D, Klapper W, Rasschaert D (2004) Induction of telomerase activity in avian lymphoblastoid cell line transformed by Marek’s disease virus, MDCC-MSB1. Vet Pathol 41:405–407
Baigent SJ, Petherbridge LJ, Howes K, Smith LP, Currie RJ, Nair VK (2005) Absolute quantitation of Marek’s disease virus genome copy number in chicken feather and lymphocyte samples using real-time PCR. J Virol Methods 123:53–64
Parcells MS, Lin SF, Dienglewicz RL, Majerciak V, Robinson DR, Chen HC, Wu Z, Dubyak GR, Brunovskis P, Hunt HD, Lee LF, Kung HJ (2001) Marek’s disease virus (MDV) encodes an interleukin-8 homolog (vIL-8): characterization of the vIL-8 protein and a vIL-8 deletion mutant MDV. J Virol 75:5159–5173
Witter RL, Kreager KS (2004) Serotype 1 viruses modified by backpassage or insertional mutagenesis: approaching the threshold of vaccine efficacy in Marek’s disease. Avian Dis 48:768–782
Debba-Pavard M, Le Galludec H, Dambrine G, Rasschaert D (2008) Variations in the H/ACA base sequence of viral telomerase RNA of isolates of CVI988 Rispens isolates. Arch Virol 153:1563–1568
de Boer GF, Groenendal JE, Boerrigter HM, Kok GL, Pol JM (1986) Protective efficacy of Marek’s disease virus (MDV) CVI-988 CEF65 clone C against challenge infection with three very virulent MDV strains. Avian Dis 30:276–283
Pol JM, Kok GL, Oei HL, de Boer GF (1986) Pathogenicity studies with plaque-purified preparations of Marek’s disease virus strain CVI-988. Avian Dis 30:271–275
Gimeno IM, Witter RL, Hunt HD, Reddy SM, Reed WM (2004) Biocharacteristics shared by highly protective vaccines against Marek’s disease. Avian Pathol 33:59–68
Jaramillo N, Domingo E, Munoz-Egea MC, Tabares E, Gadea I (2013) Evidence of Muller’s ratchet in herpes simplex virus type 1. J Gen Virol 94:366–375
Witter RL (1987) New serotype 2 and attenuated serotype 1 Marek’s disease vaccine viruses: comparative efficacy. Avian Dis 31:752–765
Witter RL (1991) Attenuated revertant serotype 1 Marek’s disease viruses: safety and protective efficacy. Avian Dis 35:877–891
Witter RL, Lee LF, Fadly AM (1995) Characteristics of CVI988/Rispens and R2/23, two prototype vaccine strains of serotype 1 Marek’s disease virus. Avian Dis 39:269–284
Niikura M, Kim T, Silva RF, Dodgson J, Cheng HH (2011) Virulent Marek’s disease virus generated from infectious bacterial artificial chromosome clones with complete DNA sequence and the implication of viral genetic homogeneity in pathogenesis. J Gen Virol 92:598–607
Delecluse HJ, Hammerschmidt W (1993) Status of Marek’s disease virus in established lymphoma cell lines: herpesvirus integration is common. J Virol 67:82–92
Delecluse HJ, Schuller S, Hammerschmidt W (1993) Latent Marek’s disease virus can be activated from its chromosomally integrated state in herpesvirus-transformed lymphoma cells. EMBO J 12:3277–3286
Mwangi WN, Smith LP, Baigent SJ, Beal RK, Nair V, Smith AL (2011) Clonal structure of rapid-onset MDV-driven CD4+ lymphomas and responding CD8+ T cells. PLoS Pathog 7:e1001337
Umene K (1999) Mechanism and application of genetic recombination in herpesviruses. Rev Med Virol 9:171–182
Thiry E, Meurens F, Muylkens B, McVoy M, Gogev S, Thiry J, Vanderplasschen A, Epstein A, Keil G, Schynts F (2005) Recombination in alphaherpesviruses. Rev Med Virol 15:89–103
van Iddekinge BJ, Stenzler L, Schat KA, Boerrigter H, Koch G (1999) Genome analysis of Marek’s disease virus strain CVI-988: effect of cell culture passage on the inverted repeat regions. Avian Dis 43:182–188
Bradley G, Lancz G, Tanaka A, Nonoyama M (1989) Loss of Marek’s disease virus tumorigenicity is associated with truncation of RNAs transcribed within BamHI-H. J Virol 63:4129–4135
Rasschaert D, Dambrine G, Labaille J, Lion A, Boissel E, Laurent S (2014) Deletion mutants of Gallidherpes virus and vaccinal compositions containing the same. n°PCT/IB2014/000244 (PCT/FR2014/050308)
Acknowledgments
We thank Nicolas Osterrieder (ZIBI, Berlin, Germany) for providing the BAC clone pRB-1B and bacterial strains for Red recombination. We thank Sylvie Laurent for helpful discussions. We thank Bruno Campone, Patrice Cousin, Jean Méry, Laurence Mérat and Edouard Guitton at PFIE, INRA, for taking excellent care of the animals. The study was supported by Zoetis Animal Health (www.zoetis.com) through a grant for the “Marek MIRES Collaboration Project”, the Fonds Européen de Développement Régional (FEDER) (www.europe-centre.eu ) through the EXMIR grant (no. 2835-34204) and the Région Centre (www.regioncentre.fr) MIRES (no. 200900038264). We thank Hervé Le Galludec (Zoetis Animal Health), who played a major role in the implementation and support of this project.
Conflict of interest
The authors declare that they have no competing interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
J. Labaille and A. Lion contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Labaille, J., Lion, A., Boissel, E. et al. Vaccine and oncogenic strains of gallid herpesvirus 2 contain specific subtype variations in the 5′ region of the latency-associated transcript that evolve in vitro and in vivo . Arch Virol 160, 161–171 (2015). https://doi.org/10.1007/s00705-014-2248-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-014-2248-3