Abstract
Attenuation of Gallid herpesvirus-2 (GaHV-2), the causative agent of Marek’s disease, can occur through serial passage of a virulent field isolate in avian embryo fibroblasts. In order to gain a better understanding of the genes involved in attenuation and associate observed changes in phenotype with specific genetic variations, the genomic DNA sequence of a single GaHV-2 virulent strain (648A) was determined at defined passage intervals. Biological characterization of these “interval-isolates” in chickens previously indicated that the ability to induce transient paralysis was lost by passages 40 and the ability to induce persistent neurological disease was lost after passage 80, coincident with the loss of neoplastic lesion formation. Deep sequencing of the interval-isolates allowed for a detailed cataloguing of the mutations that exist within a single passage population and the frequency with which a given mutation occurs across passages. Gross genetic alterations were identified in both novel and well-characterized genes and cis-acting regions involved in replication and cleavage/packaging. Deletions in genes encoding the virulence factors vLipase, vIL8, and RLORF4, as well as a deletion in the promoter of ICP4, appeared between passages 61 and 101. Three mutations in the virus-encoded telomerase which predominated in late passages were also identified. Overall, the frequency of mutations fluctuated greatly during serial passage and few genetic changes were absolute. This indicates that serial passage of GaHV-2 results in the generation of a collection of genomes with limited sequence heterogeneity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Marek’s disease (MD) is a lymphoproliferative disease of chickens responsible for great economic losses to the poultry industry [1, 2]. The causative agent of the disease is a Mardivirus, Gallid herpesvirus type 2 (GaHV-2), a member of the Alphaherpesvirinae subfamily in the family Herpesviridae [3]. GaHV-2 is the most oncogenic herpesvirus known and represents the first of four neoplastic diseases (including hepatocellular carcinoma: hepatitis B virus; cervical carcinoma: human papilloma virus and leukemia: feline leukemia virus) for which an effective vaccine has been developed [4–6]. Many GaHV-2 vaccines have been developed through extensive serial passage (35–100 passages) of virulent field isolates in avian embryo fibroblasts [7–10], but in vaccine trials all pale in comparison to the protection elicited by the avirulent strain CVI988 isolated from birds by Rispens et al. [11, 12]. To determine the genetic changes responsible for attenuation we have embarked on a study to identify the mutations that accumulate during extensive serial passage of the very virulent plus (vv+) strain of GaHV-2 known as 648A. Previously, the pathogenesis of regular passage levels (between 10 and 100) of this strain, propagated in avian embryo fibroblasts, was characterized in 15 × 7 cross-bred chickens [13]. These studies showed that the ability to induce transient paralysis (TP) was dramatically reduced between passages 30 and 40. However, persistent neurological disease (PND) was only reduced after an additional ten passages (p50) and disappeared between passages 80 and 90. The incidence of visceral tumors was also investigated and nearly 100 % of the chickens inoculated with passages 10 through 50 of 648A developed tumors. This frequency, however, dropped significantly between passages 50 and 60, and by passage 80 no tumors were observed in inoculated chickens for the period of the trial (12 weeks).
We have reported the genomic changes that occur during low (p11) and high (p101) in vitro passages of virulent plus strain 648A [14]. As a complementary study the sequentially passaged virus preparations between p11 and p101 were sequenced in a hope that a correlation between phenotype (e.g., PND, tumor formation, etc.) and genotype could be established. We have discovered that varying populations of polymorphisms (point mutations, deletions, and insertions) exist within the genomes at each passage interval. In addition to describing the mutations that occurs during serial-passage-induced attenuation of “interval-isolates”, this paper describes the fluctuations in the mutation frequencies for 14 single-nucleotide polymorphisms (SNPs) and details the nucleotide sequencing evidence for intra-strain diversity within populations of GaHV-2.
Methods
Virus and cells
Very virulent plus strain 648A of GaHV-2 isolated by Witter [15] was used in these experiments. Serial passage of this strain to generate virus stocks at passages 10, 20, 30, 40, 50, 60, 70, 80, 90, and 100 in line 0 chicken embryo fibroblast (CEF) culture was described previously by Gimeno and co-workers [13]. Secondary CEFs were used for a single round of virus propagation prior to nucleocapsid DNA isolation from passages 31, 41, 61, and 81 and were maintained in Dulbecco’s modified essential medium (DMEM) supplemented with 8 % fetal bovine serum as previously described [14].
Isolation of DNA
Nucleocapsid DNA for each passage was isolated when the cytopathic effect (CPE) reached >80 % with a protocol originally described by Sinzger et al. [16] and modified by Volkening and Spatz [17]. GaHV-2 infected cells were trypsinized, centrifuged, and suspended in 320 mM sucrose, 5 mM MgCl2, 10 mM TrisHCl (pH 7.5), and 1 % Triton X-100. Nuclei were pelleted by centrifugation at 1,300×g and suspended in 10 mM Tris HCl (pH 7.5), 2 mM MgCl2, and 10 % sucrose and mixed with an equal volume of 2× nuclease buffer [40 mM PIPES (pH 7.0), 7 % sucrose, 20 mM NaCl, 2 mM CaCl2, 10 mM 2-mercaptoethanol, and 200 μM PMSF]-containing 150 U of micrococcal nuclease and 1.0 μl of RNase A (20 mg/ml). Cellular and unpackaged viral nucleic acids were degraded by incubation at 37 °C. After stopping the nuclease reaction with EDTA, viral DNA was released from nucleocapsids by incubation at 50 °C in a buffer-containing 100 mM NaCl, 10 mM TrisHCl (pH 8.0), 25 mM EDTA (pH 8.0), 0.5 % SDS and 50 μg of proteinase K. The DNA was extracted with phenol/chloroform and precipitated with 6.5 % (w/v) PEG-8000 and 10 mM MgCl2 at room temperature for 20 min. After centrifugation at 16,000×g for 40 min, the pellets containing the high MW viral DNA were washed with 500 μl 70 % ethanol and re-centrifuged at 16,000×g for an additional 20 min. High MW DNA pellets were air-dried and dissolved in 1X TE buffer, pH 7.5.
DNA sequencing
Sequencing of 5.0 μg DNA from 648A passages 11, 31, 41, 61, 81, and 101 was carried out commercially on a Genome Sequencer 20 (GS20) System (454 Life Science Corporation). DNA from each passage was contained within its own section of the sequencing chip using an eight-lane gasket. Genomic nucleotide consensus sequences were assembled using the DS De Novo Assembler (454 Life Sciences, Branford, CT) and the Sequencher program (Gene Codes, Ann Arbor, MI). The average length of each read was 179 nucleotides. The final sequences on average represented a 47-fold coverage at each base pair. Problematic regions containing mononucleotide reiterations were sequenced from PCR products generated in reactions containing Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA) under standard PCR conditions. Sanger-based DNA sequencing of these PCR products was performed at the South Atlantic Area sequencing facility (Athens, GA) using the BigDye terminator cycle sequencing protocol and analyzed on a model ABI-3730 XL DNA Analyzer (Applied Biosystems, Foster City, CA). DNA sequences were maintained and analyzed using Lasergene (DNASTAR, Madison, WI), NCBI Entrez and other web-based tools. Origin of replication hairpins were analyzed using the mfold WebServer at the Rensselaer Polytechnic Institute, Troy, NY, USA [18]. Nucleotide sequence data reported in this paper have been submitted to GenBank and the accession numbers are as follows: 648Ap11-JQ806361; 648Ap31-JQ806362; 648Ap41-JQ809691; 648Ap61-JQ809692; 648Ap81-JQ820250, and 648Ap101-JQ836662.
Polymerase chain reaction (PCR) conditions
Deletions in the GaHV-2 genomes first identified through sequence analysis were verified using flanking oligonucleotides and PCR. A 632 base pair deletion within the ICP4 promoter (Md5 coordinates 151212–151845 and 166599–167232) was confirmed using 100 ng of nucleocapsid DNA from the six passages, one unit of Platinum Taq polymerase (Invitrogen, Carlsbad, CA) and the primers ir_94 (AACCGGATGGCGAACTTGC) and gap45-r (GAAATCAGGCGGGTTGTC) at 400 nM. The reaction contained 1× high fidelity buffer, 200 μM dNTP, 2 mM MgSO4, and 5 % dimethylsufoxide. The cycling conditions were as follows: templates were initially denatured at 94 °C for 2 min and then for 30 s at 94 °C at the beginning of each cycle. Oligonucleotides were allowed to anneal for 30 s at 60 °C and nucleotides were incorporated at 68 °C for 90 s. This cycle was repeated 34 additional times. PCR products were resolved by electrophoresis in a 0.6 % agarose gel with 1× TAE.
Results
DNA sequencing of attenuated passages
Since 454 pyrosequencing does not rely on cloned DNA, microgram quantities of nucleocapsid DNA were needed. To achieve this goal, between 8 and 16 flasks (75 cm2) of chick embryo fibroblasts were infected at an m.o.i. of ~1.0 with individual passage level stocks. The hypotonic lysis followed by micrococcal nuclease degradation provided a clear improvement over classical hypotonic lysis followed by sucrose gradient purification as reported previously [19]. Depending on the passage level and the number of cells infected, this protocol yielded 5–10 μg of DNA for most passages. The quality of the DNA was assessed both before and after PEG precipitation by separating restriction endonuclease-digested aliquots (500 ng) on 0.6 % agarose gels (data not shown).
Table 1 contains a summary of the pyrosequencing data for the 648A passages. The average read length was 179 nucleotides with an average of 45,663 reads per GaHV-2 genome. The number of reads excludes those that were avian specific (contaminating low MW host DNA). The fold-coverage varied over a wide range from 25 to 75 fold depending on the amount of contaminating avian DNA and the number of successful reads. This latter parameter is independent of the purity of the DNA and influenced by the chemistries involved in pyrosequencing. For example, p11 contained only 2.32 % contaminating DNA but only produced 43,601 GaHV-2-specific reads for a 44-fold coverage while p101-contained 20 % contaminating avian DNA but generated 84,144 GaHV-2-specific reads for a 73-fold coverage. Likewise p31 contained a lower percentage of contaminating DNA than p61 but sequenced poorly with only a 32.8 fold coverage relative to the 39.2 fold coverage obtained with p61. In general, however, DNA preparations with lower amounts of host DNA contamination yielded higher fold coverage thus making contig assembly easier.
Accumulation of mutations during serial passage
Deletions
Mutations were identified in each passage by comparison of DNA sequences to each other and the reference strain Md5 [20]. These mutations were catalogued as deletions, insertions, and single-nucleotide substitutions. Deletions were identified in the unique long, repeat long regions and the repeat short/unique short junctions (Fig. 1). As expected, deletions that occurred in genomes of early passages were fairly well maintained in later passages (Fig. 2). A 632 base pair deletion at the repeat short/unique short junction (Md5 coordinates 151,212–151,845 and 166,599–167,232) was identified in the genomes of passages 41 through 101. This deletion mapped 444 bp upstream of the ICP4 start codon within the 3′ terminus of the overlapping 10 Kb LAT [21, 22]. Five short-novel open reading frames (MDV084.5, 085, 085.3, 085.6 and 085.9) also mapped within the deleted region [23]. As shown in Fig. 1a, this deletion encompasses the Meq oncoprotein binding site [24] and MDV085.3 and alters the reading frames of the novel short ORFs MDV084.5, 085, 085.6, and 085.9.
Diagram of the locations of large deletions (>500 bp) in the repeat short, unique long, and repeat long regions of the 648A genome. Open reading frames are indicated by white arrowheads. The black boxes denoted the position of the deletions. (a) The 3′ terminus of the internal repeat short region showing the overlapping ICP4 and 10 Kb LAT genes and novel genes within the promoter of ICP4. The 632 bp deletion encompassing the Meq binding site (open box). b The 948 bp deletion at the 3′ termini of the overlapping LORF2 and V-lipase genes within the unique long region. c The 820 bp deletion within the terminal repeat long region encompassing vIL8, RLORF4, and RLORF5
Histograms showing the percentage of the 648A passages-containing deletions. a ICP4 promoters. b Lipase (MDV010). c vIL8, RLORF4, and overlapping RLORF5 genes. d diploid genes MDV05.5/075.91. e Origin of lytic replication and f A-like sequence involved in packaging. A 34 bp deletion was identified in all of the 648Ap101 populations. Black denotes the percentage of the population containing the full-length sequence and gray denotes the percentage of the population containing the deletion
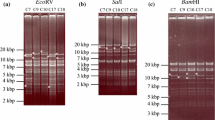
To investigate the proportion of genomes containing this deletion within each individual passage, flanking oligonucleotides were synthesized and used in Taq polymerase amplification reactions with DNA isolated from each interval-isolate. As shown in Fig. 3, a 1.1 Kb PCR product was generated in reactions containing DNA from early passages. As the passage number increased beyond 41, two PCR products (1.1 and 0.5 Kb) were generated. At passage 61, the predominating PCR product was the smaller 0.5 Kb product and by passage 101 only trace amounts of the larger 1.1 Kb (non-deleted) product were detected. The intensity of the PCR bands correlated well with the frequency of individual sequences calculated from aligned reads. Based on analysis of the sequencing data, the full-length sequences versus deleted sequences were present in the following proportions: p11–100/0 %: p31–100/0 %: p41–75/25 %: p61–6.7/93.3 %: p81–5.3/94.7 %: and p101–2.2/97.8 %.
Ethidium bromide-stained gel (0.6 % agarose) containing separated PCR products generated in reactions with primers that flank the 632 bp deletion found within the repeat short region of the 648A genomes. A 1,151 bp product was amplified in DNA extracted from early passages. The deletion, as evidenced by the appearance of the smaller 526 bp band, first appeared in DNA from p41 and predominated thereafter. The arrows indicate the positions of the 1.1 and 0.5 Kb products
Another large deletion (948 bp: Md5 coordinates 16,401–17,348) was identified within the UL region, at the MDV010 locus encoding the spliced virulence factor vLipase [25]. Based on analysis of the sequencing data, this deletion was found in 54 % of the p101 genomes. The deletion (Fig. 1b) removed 475 bp at the 3′ terminus of MDV010 and the resulting open reading frames codes for a fusion polypeptide of 609 aa. Two vLipase proteins (a full-length 756 aa protein and the COOH fusion form) are therefore predicted for the p101 genomic populations.
A less predominant 820 bp deletion mapping to the RL regions (MD5 coordinates 3,455–4,274 and 137,346–138,165) was identified in 3 % of the p81 sequences and 34 % of the p101 sequences. As shown in Fig. 1c this deletion affects the genes MDV003/MDV078 and MDV003.4/078.3 encoding the virulence factors vIL8 (RLORF2) [26–28] and RLORF4 [29] as well as the 5′ terminus of MDV003.6/078.2 encoding RLORF5.
A small deletion was identified in the novel gene MDV05.5/075.91 within the RL region of the p101 genome. A missing thymidine residue (after nucleotide 8,062 in the Md5 genome) is predicted to generate an 81 aa protein. A 99 aa protein is predicted from this gene within the genomes of earlier passages. The role of the gene in virulence is unknown.
In addition to deletions in protein coding regions, two cis-acting sites (the origin of replication and the a-like sequence) also contained deletions within the genomes of later serial passages. A TTA deletion in the origin of replication’s hairpin structure was identified within the genomes of passages 41 through 101. At passage 41, 9.5 % percent of the sequences contained the deletion. By passages 61 and 81, 73.9 and 77.3 %, respectively, contained the deletion. However, this deletion was not strictly maintained and by passage 101 only 64.2 % of the aligned reads contained the deletion, suggesting that genomes containing the deletion are not strongly favored. This is in agreement with predicted thermostability data indicating that the shortened origin of replication hairpin has a slightly higher ∆G value (−22.46 vs −21.88 kcal/mol). Although the difference in ∆G values is only 0.58 kcal/mol, the loop structure at the tip of the hairpin varies greatly (Fig. 4).
Computer generated representations and ∆G values of folded hairpin structures at the origin of lytic replication within the genomes of 648Ap11, 648Ap101, JM/102W, and RM-1. The latter three exhibit attenuated phenotypes. A three nucleotide deletion (arrow) is present in the 648A genomes from passages 81 and 101. The shaded boxes indicate the UL9 binding sites. A ten nucleotide deletion in the origin of lytic replication in the genomes of JM102/W (atatattata) and RM-1(tatattatat) maps to positions 30–40 on the 648Ap11 structure. The sequence of the origin of lytic replication of Md5 is the same as 648Ap11
Another region of sequence heterogeneity was identified in the a-like sequences [30]. A stretch of 89 nucleotides (Md5 coordinates 140,570–140,657; 177,787–177,874) between the pac-1 site and the telomeric repeats was deleted in the genomes of passage 61 though 101. The genome of passage 101 also contained a 34 bp deletion downstream from the 3′ DR1 sequence and a 22 bp sequence (GTCATGTAGAGGGTCATGCGCG) repeated 14 times upstream of the 60 bp repeats. Deletions in this region downstream from pac-2 have been reported in some attenuated GaHV-2 strains [30].
Insertions
Insertions accumulated less frequently than deletions in the serially passaged preparations. An insertion resulting in a reading frame shift was identified in the UL gene encoding the UL3 nuclear phosphoprotein (MDV015:Md5 coordinates 20,607–21,293). The mutation in this gene involved an additional adenosine residue in a stretch of mononucleotide reiterations (Md5 coordinates 21,039–21,046) that in most GaHV-2 genomes sequenced to date contain eight adenosine residues resulting in a 228 aa protein. Sixty-eight percent of the genomes in passage 81 contained an extra adenosine residue resulting in a frameshift that is predicted to generate a 149 aa protein. This mutation was selected against during additional passages and by p101 all of the reads contained the eight adenosine residues within this region of the UL3 gene. Interestingly, this gene was also mutated in the serially passaged virus 584Ap80 [9].
Variable thymidine reiterations of either seven (most common) or six residues were identified in UL5 gene encoding the DNA helicase–primase-associated protein). (MDV017:Md5 coordinates 22,669–25,245) of passages 11–101. The UL5 gene within the genomes of passages 11–81-contained seven thymidine residues (Md5 coordinates 24,772–24,779), and was predicted to generate a 858 aa protein. At passage 101 43 % of the UL5 genes contained seven thymidine residues while the majority contained a six thymidine stretch. It is predicted that genes with only six thymidines would generate a 29 aa peptide, but if a downstream start codon were used a 718 aa protein with a proper COOH terminus would be generated.
Insertions were also identified within the MDV002.6/078.5 diploid gene flanking the sequence CCCCCTTTTGCACGAAGAGT (Md5 coordinates 2,543–2,562 and 139,058–139,077). An additional cytosine residue at the 5′ end of the above sequence was present in the passage 81 genomes. This insertion would result is the production of a 51 aa protein, identical to the MDV002.6/078.5 protein of another attenuated strain (584Ap80) [9]. Two additional residues (cytosine and thymidine) at the 3′ end of the above sequence were identified within the genomes of passage 101. This would generate a 60 aa protein, in contrast to the 99 aa protein predicted from the translation of this gene within the genomes of the earlier passages as well as those of other sequenced GaHV-2 genomes annotated in GenBank.
Sequence variability was also identified in the MDV03.6/078.2 gene encoding RLORF5 (Md5 coordinates 4,154–4,327 and 13,7293–13,7466. Within the p101 genome, this gene contained extra guanosine and thymidine residues after nucleotide 4,301 (Md5 coordinates). This is predicted to generate a slightly larger protein (60 aa) than the RLORF5 homologues found within the genomes of earlier passages (57 aa). This ORF is not believed to be important in virulence since the region can be deleted without loss of virulence [29] and RLORF5 homologues in other sequenced genomes are predicted to encode proteins of varying lengths (57, 58, 59, 62, and 115 aa).
Single-nucleotide polymorphisms
With average fold coverage of 47, it was possible to identify a collection of SNPs that exist in differing proportions within a single-DNA preparation (Table 2). Fourteen SNPs were identified in the genomes of passages 11 through 101. The proportion of each SNPs within the genomes of each passage varied greatly. Only one G → A transition, within the promoter region of MDV006.1/075.1 encoding the hypothetical protein B68 [31, 32] transitioned completely by passage 101. The majority of the SNPs fluctuated to varying degrees but the proportion of the SNPs that predominated in the p11 genomes were largely inverted by passage 101. The changes were greatest for the SNPs found in MDV059 (UL46 encoding the tegument protein VP11/12), the untranslated 3′ end of MDV082/102, the Val1871 to Ala1871 polymorphism of MDV84/100 encoding the immediate early protein ICP4, and the thymidine to cytosine switch in the promoter of MDV086.1/097.9. One polymorphism affecting three overlapping genes (MDV008/073, 009, and 009.5/073.4) within the repeat long region was present in roughly half the genomes within each passage.
Discussion
Serial passage of virulent field isolates of GaHV-2 in cell culture from various avian and non-avian species for the purpose of attenuation has been reported over the last 40 years [35–40]. However, defining the mutations involved in attenuation has only recently been attempted through the generation of knockout mutants, primarily in unique Mardivirus genes, and through whole genome sequencing of virulent and attenuated strains [41–43]. This study addresses the accumulation of attenuating mutations by determining the nucleotide sequences at defined intervals of the serially passaged very virulent plus virus 648A. This collection was extensively characterized in MD susceptible chickens in an earlier report [13] and it is reported that acute TP was lost after passage 30, classical TP was lost after passage 40, and the ability to form tumors was lost after passage 70. It was the hope of this study to also match genetic mutations occurring at a specific passage level with the loss of a virulent phenotype. The most obvious and perhaps the easiest phenotype to correlate with a genotype was the loss of tumor formation which occurred after passage 70. Of the 140 genes encoded by GaHV-2, at least three genes encoding the oncoprotein Meq, the viral encoded RNA component of telomerase (vTR), and the large tegument protein UL36 have been identified as playing a role in tumor formation in chickens infected with GaHV-2 [44–46].
Analysis of the sequencing data for the Meq gene (MDV005/076) from each passage failed to identify a single mutation that differed among the passages. No mutations were identified in the ubiquitin-specific protease (USP) domain of UL36. Differences were noted in the 3′ hypervariable domain of UL36 [30], but nevertheless the genomes of each passage encoded a 3,327 aa UL36 protein. Mutations were, however, identified in the diploid gene encoding the viral encoded telomerase RNA subunit. Three mutations within the pseudoknot, CR4 and CR7 domains were identified within passages 81 and 101 genomes, although at low frequencies. Two of these mutations within the CR4 and CR7 domains are likely to affect stem structures based on the vTR model [33, 47] Kaufer et al. [48] have demonstrated that mutations of the template sequence of MDV-encoded vTR completely abrogated virus-induced tumor formation so it possible that the mutations we have identified may contribute to the lack of tumor formation as noted in the Gimeno study [13] after passage 70.
Based on the appearance of SNPs within the genomes of passages that failed to generate tumors and their generalized shift to nearly 100 % by passage 101 (Table 2), two candidates (UL46 and a non-coding sequence 3′ of RSORF1) may be important in tumor formation. UL46 encodes a tegument protein VP11/12 and it is highly unlikely that this well-conserved herpesviral protein plays a role in tumor formation. The point mutation downstream of RSORF1 is more attractive since it occurs in a region containing numerous microRNAs [49, 50]. However, it is far more likely that the 632 bp deletion identified within the promoter of ICP4 [51] plays a role in the lack of tumor formation by retarding the in vivo replication of the viral genome in passages p81 through p101 [52, 53]. A strong correlation between the ability of the virus to replicate and pathogenicity has been reported [54, 55] and the appearance of the deletion correlates well with the disappearance of tumor formation. However, since the deletion occurs in the region that Meq binds [24] and binding to the ICP4 promoter is thought to repress expression and maintain latency, it is counterintuitive to think that removal of a repressor-binding site would further suppress expression. This observation raises the possibility that the major role of Meq in GaHV-2 replication involves binding at the origin of replication [24].
Interestingly, mutations within the origin of replication were identified in the genomes of passages (p81 and p101) that reportedly were unable to generate tumors. These mutations may play a significant role in curtailing efficient replication by changing its tertiary structure to generate a bulky loop consisting of 8 mismatched adenosine and thymdine residues (Fig. 4) [56–59]. Similar positional mutations (Fig. 4 positions 30–40) were identified within the origins of the attenuated GaHV-2 strains RM-1 and JM/102W [34]. Studies to measure the replicative ability of passages 81 and 101 in chickens are needed since the preceding clinical study characterizing these interval passages only measured the in vivo replicative ability of 648A in passages 10–50 [13].
Other virulent phenotypes that disappeared after passage 50 include acute transient and classical TP. Acute TP, the hallmark of vv+ strains, was lost after passage 30, and classical TP was lost after passage 40. Only one mutation within the promoter of the diploid gene MDV006.6/075.1 and the 3′ untranslated region of hypothetical protein MDV007/074 appeared exclusively after passage 31 (Table 2). This guanosine to adenosine switch was present in roughly 50 % of the genomes in passage 41, completely transitioned to adenosine by passage 61 and was maintained through passage 101. It is therefore possible that this mutation was responsible for the lack of acute TP in chickens receiving passage 40 and later. Mutations in three other proteins (VHS, RLORF1, and ICP4) appeared in the p31 genomes and persisted through p101. The Ala359 Val mutation in the virus host shutoff ORF (UL41) appeared to fluctuate throughout later passages and it is unlikely to affect the tertiary structure of the VHS protein. However, the Arg180 Pro mutation which occurred at p31 in the Arg-rich protein (RLORF1) is more likely to affect its tertiary structure. Likewise, the Ser1641 Pro mutation in ICP4 which appeared at passage 31 and predominated throughout additional passages is also likely to affects its tertiary structure. It is therefore hypothesized that the loss of classical paralysis involved mutations in the Arg-rich protein RLORF1 and immediate early protein ICP4 which is essential for efficient replication.
One of the most interesting aspects of this study was the discovery that mutations (deletions, insertions, and SNPs) were present in differing proportions within individual passages. Polymorphisms within genes, whether synonymous or non-synonymous, tended to fluctuate throughout the passages (Table 2). Only one polymorphism (G → A) within the intergenic region between MDV006.6/075.1 and MDV007/074 transitioned completely during the course of serial passage. All others were in a state of dynamic equilibrium during passage.
The discovery that GaHV-2 exists as a collection of genomes with limited sequence heterogeneity has changed our views on the stability of the DNA genomes of herpesviruses. Recently it has been reported that other herpesviruses exist as a complex mixture of genome types [60, 61]. Mixed population of genome types or quasispecies has been described for RNA viruses since 1978 (RNA bacteriophage QB), [62] and later for most RNA viruses (e.g., HCV, HIV HBV, and influenza), but only within the last decade have viruses with small circular DNA genomes been reported to exist as quasispecies. Although most reports have involved geminiviruses and mosaic viruses that infect plants, some animal viruses within the family Circoviridae have also been reported to exist as a collection of genomes with great sequence heterogeneity [63, 64]. Our sequencing proof that an animal herpesvirus exhibits sequence heterogeneity within a single population has long been suspected by researchers in the field. Research by Witter [65] has demonstrated that upon back passage in vivo serially passaged attenuated viruses can regenerate virus “revertants” with virulent phenotype. Darwinian selection of minor virulent subpopulations was hypothesized to be the likely mechanism for the gain in virulence function. Silva et al. [66, 67] have shown that upon serial passage a region in the repeat long, known as the 132 bp repeat, is duplicated numerous times. When these viruses are back passaged in bird, this region reverts back to its low copy number. Preferential replication of subpopulations with high copy numbers of the 132 bp repeats also hinted at the existence of mixed genome populations [68].
The nucleotide sequencing of GaHV-2 genomes as bacterial artificial chromosomes (BACs) has also suggested the mixed nature of Marek’s disease virus genomes. Petherbridge et al. [69] have reported that recombinant BACs generated from the virulent RB-1B strain exhibit distinctly differing phenotypes in vitro and in vivo. These differences were shown not to be due to cloning artefacts but rather due to random cloning of genomes from a single-parental virus stock [70]. One MDV-1 BAC (designated RB-1B-5) with frameshift mutations in RLORF1, UL13, UL44 (gC), and US6 (gD) when reconstituted, was unable to infect horizontally. Interestingly the parental stock, which was horizontal transmission competent, contained a mixed population of genomes with wild type and mutant RLORF1, UL13, UL44 (gC), and US6 (gD), genes [70, 71]. Barrow and Venugopal [72] have also noted that two BAC clones of the virulent European strain C12/130, when reconstituted, exhibit distinctly different phenotypes in birds. Nucleotide sequencing of these BACs have identified minor differences, [73] further supporting the hypothesis that GaHV-2 exists as a collection of mixed genome populations.
Because of the nature of the 454 pyrosequencing technology, linking mutations on the same genome molecule is not easily accomplished. Since it is widely believed that attenuation is multigenic, genetic linkage is very important. To accomplish this, future comparative genomic studies are needed that analyze viruses that are reconstituted from bacterial artificial chromosomes with minimal in vitro propagation. After serial passage of these reconstituted viruses in cell culture it should be possible to recapture the recombinant BACs from Hirt’s extracts and stably propagate them in E. coli. Determination of the nucleotide sequences of theses GaHV-2 BACs prior to assessment of their level of virulence upon transfection of isolated chicken macrophages and injection back into the chicken should allow for the identification of mutations, linked on the same genome—that are involved in attenuation.
References
B.W. Calnek, Pathogenesis of Marek’s disease virus infection, in Current Topics in Microbiology and Immunology, ed. by K. Hirai (Springer, Berlin, 2001), p. 25
R.L. Witter, Marek’s disease virus vaccines-past, present and future (chicken vs. virus-a battle of the centuries), in Current progression Marek’s disease research. Proceedings of the 6th International Symposium on Marek’s disease, ed. by K.A. Schat et al. (American Association of Avian Pathologists, Kennett Square, 2001)
B. Roizman, A.E. Sears, in Fields Virology, ed by B.N. Fields, D.M. Knipe, P.M. Howley (Lippincott-Raven Press, New York, 1996), p. 2231
A.E. Churchill, L.N. Payne, R.C. Chubb, Marek’s disease immunization against Marek’s disease using a live attenuated virus. Lancet 1(7595), 610–611 (1969)
S.R. Pagliusi, M. Teresa Aguado, Efficacy and other milestones for human papillomavirus vaccine introduction. Vaccine 23(5), 569 (2004)
A.M. Prince, Prevention of liver cancer and cirrhosis by vaccines. Clin. Lab. Med. 16(2), 493–505 (1996)
E. Dudnikova et al., Factors influencing the attenuation of serotype 1 Marek’s disease virus by serial cell culture passage and evaluation of attenuated strains for protection and replication. Avian Dis. 53(1), 63–72 (2009)
R.C. Karpathy, G.A. Firth, G.A. Tannock, Derivation, safety and efficacy of a Marek’s disease vaccine developed from an Australian isolate of very virulent Marek’s disease virus. Aust. Vet. J. 80(1–2), 61–66 (2002)
S.J. Spatz et al., Clustering of mutations within the inverted repeat regions of a serially passaged attenuated Gallid herpesvirus type 2 strain. Virus Genes 37(1), 69–80 (2008)
R.L. Witter, K.S. Kreager, Serotype 1 viruses modified by backpassage or insertional mutagenesis: approaching the threshold of vaccine efficacy in Marek’s disease. Avian Dis. 48(4), 768–782 (2004)
R.L. Witter, L.F. Lee, A.M. Fadly, Characteristics of CVI988/Rispens and R2/23, two prototype vaccine strains of serotype 1 Marek’s disease virus. Avian Dis. 39(2), 269–284 (1995)
B.H. Rispens et al., Control of Marek’s disease in the Netherlands. I. Isolation of an avirulent Marek’s disease virus (strain CVI 988) and its use in laboratory vaccination trials. Avian Dis. 16(1), 108–125 (1972)
I. Gimeno et al., Differential attenuation of the induction by Marek’s disease virus of transient paralysis and persistent neurological disease: a model for pathogenesis studies. Avian Pathol. 30(4), 397–409 (2001)
S.J. Spatz, Accumulation of attenuating mutations in varying proportions within a high passage very virulent plus strain of Gallid herpesvirus type 2. Virus Res. 149(2), 135–142 (2010)
R.L. Witter, Increased virulence of Marek’s disease virus field isolates. Avian Dis. 41(1), 149–163 (1997)
C. Sinzger et al., A simple and rapid method for preparation of viral DNA from cell associated cytomegalovirus. J. Virol. Methods 81(1–2), 115–122 (1999)
J.D. Volkening, S.J. Spatz, Purification of DNA from the cell-associated herpesvirus Marek’s disease virus for 454 pyrosequencing using micrococcal nuclease digestion and polyethylene glycol precipitation. J. Virol. Methods 157(1), 55–61 (2009)
M. Zuker, Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31(13), 3406–3415 (2003)
S.J. Spatz, C.A. Rue, Sequence determination of a mildly virulent strain (CU-2) of Gallid herpesvirus type 2 using 454 pyrosequencing. Virus Genes 36, 478–489 (2008)
E.R. Tulman et al., The genome of a very virulent Marek’s disease virus. J. Virol. 74(17), 7980–7988 (2000)
D. Li et al., Further characterization of the latency-associated transcription unit of Marek’s disease virus. Arch. Virol. 143(2), 295–311 (1998)
R.W. Morgan et al., Marek’s disease virus latency, in Current Topics in Microbiology and Immunology, ed. by K. Harai (Springer, Berlin, 2001), p. 223
S.J. Spatz et al., Comparative full-length sequence analysis of oncogenic and vaccine (Rispens) strains of Marek’s disease virus. J. Gen. Virol. 88(Pt 4), 1080–1096 (2007)
A.M. Levy et al., Characterization of the chromosomal binding sites and dimerization partners of the viral oncoprotein Meq in Marek’s disease virus-transformed T cells. J. Virol. 77(23), 12841–12851 (2003)
J.P. Kamil et al., vLIP, a viral lipase homologue, is a virulence factor of Marek’s disease virus. J. Virol. 79(11), 6984–6996 (2005)
P.L. Cortes, C.J. Cardona, Pathogenesis of a Marek’s disease virus mutant lacking vIL-8 in resistant and susceptible chickens. Avian Dis. 48(1), 50–60 (2004)
X. Cui et al., A Marek’s disease virus vIL-8 deletion mutant has attenuated virulence and confers protection against challenge with a very virulent plus strain. Avian Dis. 49(2), 199–206 (2005)
M.S. Parcells et al., Marek’s disease virus (MDV) encodes an interleukin-8 homolog (vIL-8): characterization of the vIL-8 protein and a vIL-8 deletion mutant MDV. J. Virol. 75(11), 5159–5173 (2001)
K.W. Jarosinski et al., Attenuation of Marek’s disease virus by deletion of open reading frame RLORF4 but not RLORF5a. J. Virol. 79(18), 11647–11659 (2005)
S.J. Spatz, R.F. Silva, Sequence determination of variable regions within the genomes of gallid herpesvirus-2 pathotypes. Arch. Virol. 152(9), 1665–1678 (2007)
F. Peng et al., Isolation and characterization of cDNAs from BamHI-H gene family RNAs associated with the tumorigenicity of Marek’s disease virus. J. Virol. 66(12), 7389–7396 (1992)
L.J. Ross et al., Nucleotide sequence and characterization of the Marek’s disease virus homologue of glycoprotein B of herpes simplex virus. J. Gen. Virol. 70(Pt 7), 1789–1804 (1989)
L. Fragnet et al., The RNA subunit of telomerase is encoded by Marek’s disease virus. J. Virol. 77(10), 5985–5996 (2003)
S.J. Spatz, R.F. Silva, Polymorphisms in the repeat long regions of oncogenic and attenuated pathotypes of Marek’s disease virus 1. Virus Genes 35(1), 41–53 (2007)
A. Abujoub, P.M. Coussens, Development of a sustainable chick cell line infected with Marek’s disease virus. Virology 214(2), 541–549 (1995)
D. Jaikumar, K.M. Read, G.A. Tannock, Adaptation of Marek’s disease virus to the Vero continuous cell line. Vet. Microbiol. 79(1), 75–82 (2001)
X. Li, K.A. Schat, Quail cell lines supporting replication of Marek’s disease virus serotype 1 and 2 and herpesvirus of turkeys. Avian Dis. 48(4), 803–812 (2004)
V. Majerciak et al., Increased virulence of Marek’s disease virus type 1 vaccine strain CV1988 after adaptation to qt35 cells. Acta Virol. 45(2), 101–108 (2001)
T. Onoda et al., Propagation of herpes type virus isolated from chickens with Marek’s disease in Japanese quail embryo fibroblasts. Biken J 13(3), 219–228 (1970)
D. Schumacher et al., Generation of a permanent cell line that supports efficient growth of Marek’s disease virus (MDV) by constitutive expression of MDV glycoprotein E. J. Gen. Virol. 83(Pt 8), 1987–1992 (2002)
K. Osterrieder, J.-F. Vautherot, The genome content of Marek’s disease-like viruses, in Marek’s Disease: An Emerging Problem, ed. by F. Davison, V. Nair (Elsevier, Oxford, 2004)
N. Osterrieder et al., Marek’s disease virus: from miasma to model. Nat. Rev. Microbiol. 4(4), 283–294 (2006)
L. Petherbridge et al., Replication-competent bacterial artificial chromosomes of Marek’s disease virus: novel tools for generation of molecularly defined herpesvirus vaccines. J. Virol. 77(16), 8712–8718 (2003)
K. Jarosinski et al., A herpesvirus ubiquitin-specific protease is critical for efficient T cell lymphoma formation. Proc. Natl. Acad. Sci. USA 104(50), 20025–20030 (2007)
B. Lupiani et al., Marek’s disease virus-encoded Meq gene is involved in transformation of lymphocytes but is dispensable for replication. Proc. Natl. Acad. Sci. USA 101(32), 11815–11820 (2004)
S. Trapp et al., A virus-encoded telomerase RNA promotes malignant T cell lymphomagenesis. J. Exp. Med. 203(5), 1307 (2006)
L. Fragnet, E. Kut, D. Rasschaert, Comparative functional study of the viral telomerase RNA based on natural mutations. J. Biol. Chem. 280(25), 23502–23515 (2005)
B.B. Kaufer et al., Herpesvirus telomerase RNA(vTR)-dependent lymphoma formation does not require interaction of vTR with telomerase reverse transcriptase (TERT). PLoS Pathog. 6(8), e1001073 (2010)
J. Burnside et al., Marek’s disease virus encodes MicroRNAs that map to meq and the latency-associated transcript. J. Virol. 80(17), 8778–8786 (2006)
Y. Yao et al., MicroRNA profile of Marek’s disease virus-transformed T-cell line MSB-1: predominance of virus-encoded microRNAs. J. Virol. 82(8), 4007–4015 (2008)
A.S. Anderson, A. Francesconi, R.W. Morgan, Complete nucleotide sequence of the Marek’s disease virus ICP4 gene. Virology 189(2), 657–667 (1992)
P. O’Hare, G.S. Hayward, Comparison of upstream sequence requirements for positive and negative regulation of a herpes simplex virus immediate-early gene by three virus-encoded trans-acting factors. J. Virol. 61(1), 190–199 (1987)
P. O’Hare, G.S. Hayward, Evidence for a direct role for both the 175,000- and 110,000-molecular-weight immediate-early proteins of herpes simplex virus in the transactivation of delayed-early promoters. J. Virol. 53(3), 751–760 (1985)
I.M. Gimeno et al., Biocharacteristics shared by highly protective vaccines against Marek’s disease. Avian Pathol 33(1), 59–68 (2004)
R. Yunis, K.W. Jarosinski, K.A. Schat, Association between rate of viral genome replication and virulence of Marek’s disease herpesvirus strains. Virology 328(1), 142–150 (2004)
J.W. Balliet et al., Site-directed mutagenesis of large DNA palindromes: construction and in vitro characterization of herpes simplex virus type 1 mutants containing point mutations that eliminate the oriL or oriS initiation function. J. Virol. 79(20), 12783–12797 (2005)
J.W. Balliet, P.A. Schaffer, Point mutations in herpes simplex virus type 1 oriL, but not in oriS, reduce pathogenesis during acute infection of mice and impair reactivation from latency. J. Virol. 80(1), 440–450 (2006)
T.R. Hernandez et al., Mutations in a herpes simplex virus type 1 origin that inhibit interaction with origin-binding protein also inhibit DNA replication. J. Virol. 65(3), 1649–1652 (1991)
D.W. Martin et al., Analysis of the herpes simplex virus type 1 OriS sequence: mapping of functional domains. J. Virol. 65(8), 4359–4369 (1991)
N. Renzette et al., Extensive genome-wide variability of human cytomegalovirus in congenitally infected infants. PLoS Pathog. 7(5), e1001344 (2011)
M.L. Szpara et al., A wide extent of inter-strain diversity in virulent and vaccine strains of alphaherpesviruses. PLoS Pathog. 7(10), e1002282 (2011)
E. Domingo et al., Nucleotide sequence heterogeneity of an RNA phage population. Cell 13(4), 735–744 (1978)
T.F. Ng et al., Discovery of a novel single-stranded DNA virus from a sea turtle fibropapilloma by using viral metagenomics. J. Virol. 83(6), 2500–2509 (2009)
T. Nishizawa et al., Quasispecies of TT virus (TTV) with sequence divergence in hypervariable regions of the capsid protein in chronic TTV infection. J. Virol. 73(11), 9604–9608 (1999)
R.L. Witter, Attenuated revertant serotype 1 Marek’s disease viruses: safety and protective efficacy. Avian Dis. 35(4), 877–891 (1991)
R.F. Silva, S.M. Reddy, B. Lupiani, Expansion of a unique region in the Marek’s disease virus genome occurs concomitantly with attenuation but is not sufficient to cause attenuation. J. Virol. 78(2), 733–740 (2004)
R.F. Silva, R.L. Witter, Genomic expansion of Marek’s disease virus DNA is associated with serial in vitro passage. J. Virol. 54(3), 690–696 (1985)
M. Niikura, J.B. Dodgson, H.H. Cheng, Stability of Marek’s disease virus 132-bp repeats during serial in vitro passages. Arch. Virol. 151(7), 1431–1438 (2006)
L. Petherbridge et al., Oncogenicity of virulent Marek’s disease virus cloned as bacterial artificial chromosomes. J. Virol. 78(23), 13376–13380 (2004)
S.J. Spatz et al., Comparative sequence analysis of a highly oncogenic but horizontal spread-defective clone of Marek’s disease virus. Virus Genes 35(3), 753–766 (2007)
K.W. Jarosinski et al., Horizontal transmission of Marek’s disease virus requires US2, the UL13 protein kinase, and gC. J. Virol. 81(19), 10575–10587 (2007)
A. Barrow, K. Venugopal, Molecular characteristics of very virulent European MDV isolates. Acta Virol. 43(2–3), 90–93 (1999)
S.J. Spatz et al., Genotypic characterization of two bacterial artificial chromosome clones derived from a single DNA source of the very virulent gallid herpesvirus-2 strain C12/130. J. Gen. Virol. 92(Pt 7), 1500–1507 (2011)
Acknowledgments
The authors would like to thank Barbara Riegle of the Avian Disease and Oncology Laboratory for her assistance in the propagation of the virus strains.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Spatz, S.J., Volkening, J.D., Gimeno, I.M. et al. Dynamic equilibrium of Marek’s disease genomes during in vitro serial passage. Virus Genes 45, 526–536 (2012). https://doi.org/10.1007/s11262-012-0792-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-012-0792-z