Abstract
The northern Chilean Atacama Desert is among those regions on Earth where life exists at its dry limits. There is almost zero rainfall in its core zone, and the only source of water is a spatio-temporally complex fog system along the Pacific coast, which is reaching far into the hyperarid mainland. Hardly any vascular plants grow in these areas, and, thus, it is intriguing to be faced with a vegetation-type build-up by one single and highly specialized bromeliad species, Tillandsia landbeckii Phil., forming regular linear structures in a sloped landscape. We studied the genetic make-up of a population system extending an area of approximately 1500 km2 and demonstrated a fine-scale correlation of genetic diversity with spatial population structure and following an elevational gradient of approximately 150 m. Increase in genetic diversity is correlated with increased fitness as measured by flowering frequency, and evidence is provided that outbreeding is linked with a large-distance flying pollinator feeding occasionally as generalist on its flowers, but not using the plant as source for larvae feeding. Our data demonstrate that establishment of linear vegetation structure is in principle a process driven by clonal growth and propagation of ramets over short distances. However, optimal conditions (slope, elevation, fog occurrence) for linear growth pattern formation also increase sexual plant reproductive fitness, thus providing the reservoir for newly combined genetic variation and counteracting genetic uniformity. Our study highlights the Tillandsia vegetation, also called Tillandsia lomas, as unique and genetically diverse system, which is highly threatened by global climate change and disturbance of the coastal fog system.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The coastal zone of northern Chile and southern Peru ranging from approximately 18°S–30°S is home to unique ecosystems which depend on coastal fog. Here, marine low stratocumulus clouds are reaching far towards the elevated inland and act as the main source of water in an otherwise arid or even hyperarid environment. This region, the Atacama Desert, runs over 1300 km at elevations from sea level up to 3000 m altitude, and consists of very unique vegetation types (Luebert and Pliscoff 2006). The origin of the arid climate dates back to the mid-Miocene and was induced by the Andean uplift which blocked westward movement of humidity from the Amazon Basin (Houston and Hartley 2003; Rech et al. 2010). Hyperarid conditions were formed in the Atacama Deserts not earlier than during the Pleistocene (Hartley et al. 2005) and were intensified by an expansion of coastal upwelling in the southeast Pacific, the abrupt cooling of surface water temperatures along the coast of Ecuador and a global cooling trend (Ibaraki 1997; Zachos et al. 2001). Vegetation today occurs in disjunct patches, the so-called fog oases or lomas assumed to be the remnants of a continuous vegetation belt during the Pleistocene (Cereceda et al. 1999; Rauh 1958). These are made up of at least 300–400 vascular plant species in Chile (Schulz 2009)—many of them endemic and/or threatened (Schulz 2009)—and c. 850 species in Peruvian loma formations (Dillon et al. 2011). The fog oases constitute hot spots of biodiversity and important conservation areas (Zizka et al. 2009). In particular, phylogenetic diversity (PD) is high, because there are often only few species from a given genus or family (Heibl and Renner 2012). This may be considered as a signature for deep impact of evolutionary history (Scherson et al. 2017); and it has been shown, for instance, for genus Oxalis that there is no evidence for rapid adaptive radiation (Heibl and Renner 2012). However, little is known about spatio-temporal patterns of plant species (Heibl and Renner 2012), and vegetation dynamics throughout Pleistocene climate cycles (Luebert and Wen 2008).
In recent decades, the Chilean–Peruvian fog ecosystems have shown increasing signs of decline, which might be linked to abrupt mesoscale climate shifts since the mid-1970s (Schulz et al. 2010). However, the magnitude of the decline and the underlying causal ecologic-atmospheric relationships are yet to be investigated (Latorre et al. 2011). Reports are alarming and indicate high mortality within the coastal lomas, but currently the extent and quality of the dieback as well as the effects of changes in the fog dynamics on the degradation of the lomas are not understood (Muñoz-Schick et al. 2001; Pinto et al. 2006; Schulz et al. 2010). Similarly, topographic features, such as saddlebacks, depressions, fog-corridors, aspect, and elevation above sea level, may play an important role by affecting the local geographic distribution, frequency and water content of fog (Osses et al. 2007), with direct effect on the distribution of coastal ecosystems (Cereceda et al. 2008; Larraín et al. 2002; Latorre et al. 2011; Pinto et al. 2006; Westbeld et al. 2009).
The northern Chilean Atacama Desert in particular is among those regions where life exists at its dry limits. In major areas, there is hardly any vascular plant growing, and, thus, it is intriguing to be faced with a vegetation-type build-up by one single and highly specialized bromeliad species, Tillandsia landbeckii Phil., forming regular linear structures in a sloped desert landscape. In Chile, there are few terrestrial Tillandsia species growing on bare sand (T. landbeckii, T. marconae Till & Vitek, rarely T. virescens Ruiz & Pavon). Since they are lacking a typical root system, they depend on sloped dunes (coppice dunes) to stabilize plant growth. Moreover, lacking a system of water uptake via root, these “grey tillandsias” harvest water with highly specialized trichomes utilizing incoming fog during the night. This adaptation to extreme water uptake efficiency is accompanied by obligate CAM (Crassulacean Acid Metabolism) photosynthesis representing another efficient way to cope with extreme environments and open stomata during the night only (Haslam et al. 2003).
Linear or banded pattern formation of vegetation in arid climates has been analysed in the past, but is still lacking a deeper understanding (Deblauwe et al. 2012). Studies from various arid regions all over the world developed models and elaborated on parameters to describe the observed patterns, such as wavelength of banding patterns, slope or precipitation (Deblauwe et al. 2012; Tlidi et al. 2018). Such descriptive models have also been generated for T. landbeckii (Borthagaray et al. 2010) and Tillandsia latifolia Meyen in Peru (Aponte and Flores 2013; Hesse 2012, 2014).
However, these models do not allow to draw conclusions about important population characteristics such as the relative contributions of vegetative or generative reproduction and dispersal to population dynamics, either as seeds or ramets, which may have severe consequences on population fitness often positively correlated with genetic diversity (Takahashi et al. 2018). In principal substantial genetic variation in T. landbeckii populations, which may be essential to survive in a hyperarid environment for thousands of years (Latorre et al. 2011), can be maintained by both strategies: either a large number of genotypes is propagated clonally, or geneflow via pollen and subsequent seed dispersal is maintaining and continuously creating genetic diversity. The formation of regular linear banding patterns might favour the hypothesis of clonal population growth, whereas the extreme habitats within the entire distribution range may require continuous local adaptation, which is dependent on geneflow and sexual reproduction.
Therefore, elaborating on a comprehensive understanding of tillandsia lomas and their contemporary population dynamics needs to consider genetic information, which also reflects the species evolutionary history. We screened genetic variation in Tillandsia landbeckii within the centre of its distribution in the northern Chilean Atacama Desert covering a metapopulation system that spans approximately 1500 km2. Within this area, we selected a representative population (Cerro Oyarbide) with optimal linear growth patterning to study the fine-scale distribution of genetic diversity in the context of the entire metapopulation system. Using this data, we aim to test the two competing hypotheses—whether population structure in tillandsia lomas is dominated by clonal growth indicated by genetically identical individuals leading to linear vegetation patterns or by geneflow and seed dispersal indicated by a spatial distribution of genetic variation.
We (1) study genetic connectivity among populations in the best-developed metapopulation system, (2) characterize fine-scaled genetic diversity at a representative study site (Oyarbide) to unravel modes and spatial structure of reproduction and dispersal, (3) and provide first evidence for fitness parameters important for long-term plant population survival within a (possibly) rapidly changing and fragile environment, and thereby (4) contribute to our understanding of linear vegetation pattern formation in this unique vegetation type.
Materials and methods
Study sites and plant material
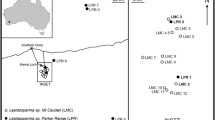
Satellite-based remote-sensing detection of Tillandsia landbeckii populations was performed (Wolf et al. 2016). The entire study area is part of the hyperarid Atacama Desert in the Tarapacá region in northern Chile near Iquique (Fig. 1). The analysed area extends over a 90 × 30 km coastal stripe at approximately 20°S (Wolf et al. 2016), and vegetation cover analyses were based on 30 × 30 m grids (Wolf et al. 2016). These data were used to navigate through the desert and sample individuals along a transect from North to South grouped into populations no. 1–7. In the centre of this population system, a large Tillandsia loma field (Oyarbide, pop. 7) was identified with a perfectly linear vegetation banding patters running orthogonally to the sloped terrain (Fig. 2). This study field has a total size of approximately 6 km2 and is our focal population in this study. In order to place the population in a genetic context, randomly selected individuals from additional six populations over the entire region were collected. These individuals (65 in total) served as a reference for subsequent genetic analysis (genepools and genetic parameters). To collect samples within this study field, we first overlaid the region with a georeferenced grid of 100 × 100 m. This grid was aligned to the linear arrangement of the vegetation. Coordinates were extracted for all grid centres and used as way-points to navigate and collect samples. Samples were ideally collected with the exact coordinates (centre of the grid). If this was not possible (either no Tillandsia at all present at coordinates or only dead plants), the nearest neighbouring Tillandsia was collected and the coordinates for the new respective sampling site within the given grid cell were adjusted accordingly before moving to the next way-point/sampling point. The Oyarbide sampling field (grid system) was structured with rows (A-O, from inland to coast; following linear vegetation) and 23 grids each resulting in a final coverage of 3.5 km2. In total, we collected 264 individuals, with 199 individuals from the described Oyarbide grid field. Collection of plant material was conducted in November 2016 under optimal field growing conditions.
Distribution of analysed populations and genetic results. a Study area in northern Chile (Map data: Google Earth, Landsat/Copernicus; accessed 20 February 2019). b Output from Barrier analysis. Numbers indicate populations. Barriers to geneflow are drawn as red lines. c Zoom into the Oyarbide study field (pop. 7) and the respective output from Barrier analysis with rows A–O indicated. d Genetic assignment revealed by Structure analysis based on optimal K = 2. Population numbers and rows (pop. 7) are indicated
Genetic diversity and nuclear AFLP analysis
AFLP profiles (Meudt and Clarke 2007; Vos et al. 1995) were generated for the entire data set with 264 individuals from all seven population areas (Online Resource 1). Leaf material was taken in the field and immediately stored in silica gel for fast drying. In the laboratory, material was homogenized by grinding with a pistil on liquid nitrogen. The Invisorb Spin Plant Mini kit was used following the manufacturer’s instructions, including the optional step of RNA digestion. The following modifications were applied to the CTAB protocol (Doyle and Doyle 1987): DNA pellets were washed twice with 70% ethanol and then dissolved in 100 μl TE-buffer (10 mM Tris–HCl, 1 mM EDTA, pH 7.5). Quality (high molecular weight) and quantity of DNA were checked prior to subsequent analytical steps. DNA quality and fragment length were visually checked on 1% agarose gels, and concentration was assessed via fluorescence spectroscopy using a high-sensitivity, double-stranded DNA-specific dye with the Qubit® dsDNA HS Assay (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Digestion of diluted genomic DNA and ligation of dsDNA adaptors was performed simultaneously by endonucleases EcoRI HF and MseI and T4 DNA Ligase (New England Biolabs GmbH, Frankfurt am Main, Germany). PCRs were carried out with AmpliTaq DNA Polymerase in AmpliTag buffer II for PCR step 1 and with AmpliTaq Gold and AmpliTaq Gold Buffer (Abi/Life Technologies, Darmstadt, Germany) for PCR step 2 (selective PCR). Oligonucleotides and fluorescent-labelled oligonucleotides (with fluorophores FAM, Hex and Atto550) were obtained from biomers.net GmbH Ulm, Germany. During an initial screening for variability, a set of six combinations of selective primers was chosen for the study: (A) EcoRI + ACA(FAM)/MseI + CAT, (B) EcoRI + AAC(Hex)/MseI + CAT, (C) EcoRI + AGC (Atto550)/MseI + CAT), (D) EcoRI + ACA(FAM)/MseI + CTC, (E) EcoRI + AAC(Hex)/MseI + CTC and (F) EcoRI + AGC (Atto550)/MseI + CAT). A detailed protocol of the laboratory routines including reagent concentrations and thermocycling conditions is described in Tewes et al. (2017). Prior to fragment detection, amplicons were poolplexed and purified by ultrafiltration (NucleoFast 96 PCR plate ultrafiltration kit, Macherery-Nagel, Düren, Germany). Fragment detection was performed by GATC Biotech AG (Konstanz, Germany). Size calling and manual genotype calling were performed with GeneMarker 1.95 (SoftGenetics LLC, State College, USA). Scored data were exported as a binary data table for further analysis. To assess the genotyping error, we included replicate samples from duplicated DNA extractions in our setup. In total, we generated 20 replicate genotypes for 10 individuals plus several negative (water) controls. Samples were assigned randomly to an experimental block (96-well reaction plate). Experimental and genotyping errors were further analysed using a variance criterion testing plate-specific effect. For that, associations of all pairs of samples were expressed by a dissimilarity coefficient with Euclidean properties (Legendre and Legendre 2012) using Jaccard’s dissimilarity coefficient (Jaccard 1901). Principle coordinate analysis (PCoA) (Anderson and Willis 2003) and Eigen decomposition of the resulting association matrix were performed with the function pcoa R package “ape” (Paradis et al. 2004). The variance of the occurrence of any marker among plates was calculated, and all markers showing a variance bigger than 0.025 were removed. The final rate of genotyping error was determined over all pairs of replicates (Bonin et al. 2004).
Genetic assignment of accessions was inferred under the non-admixture model implemented in STRUCTURE 2.3.4. (Pritchard et al. 2000) using the correlated allele frequencies and the recessive alleles options (Falush et al. 2007). Burn-in period was set to 150,000 MCMC steps, and data collection was carried out over another 250,000 steps. Thirty replicate simulations were run for each value of K (the number of ancestral clusters assumed by STRUCTURE) ranging from 1 to 10. Individual ancestry was averaged across all replicate simulations for each value of K using CLUMPP Version 1.1.2 (Jakobsson and Rosenberg 2007). A formal determination of the optimal value of K was carried out using Evanno’s Mean Delta K (Evanno et al. 2005). Analysis of molecular variance (AMOVA) and estimation of molecular diversity indices were performed with Arlequin v. 3.5.2.2 (Excoffier and Lischer 2010).
Possible genetic barriers in a geographically defined landscape were analysed using a Monmonier’s maximum difference algorithm as implemented with the software BARRIER version 2.2 (Manni et al. 2004). This program calculates zones where abrupt changes in genetic variations occur and visualizes them on a geographic map presenting the sample’s locations. These abrupt changes occur when the IBD (isolation–by–distance) model does not apply to a population due to barriers of gene flow. Basically, the IBD model states that genetic differentiation between individuals uniformly increases with a geographic distance due to a decline in gene flow at increasing distances (Manel et al. 2003). If isolating factors prevent gene flow, irregularities appear in the pattern, represented as barriers in the map. Barrier analyses were conducted (i) with the total data set (264 individuals), and (ii) with a reduced set of samples from the grid field (Oyarbide) (199 individuals).
Screening of flowering phenotypes
Estimation of flower frequency at Oyarbide was performed on a 2 m2 plot scale with the respective georeferenced and sampled individual in its centre. Flowering categories were 0 = 0–6 flowers per 100 cm2, 1 = 7–12 flowers per 100 cm2, and 2 =>12 flowers per 100 cm2. Final screening of flowering frequency was performed during a field campaign in November 2018. In order to obtain a better idea about flowering time and phenology, we selected 10 permanent plots (10x10 m) and marked 500 randomly chosen and non-flowering shoots. All of them were visited during March, May, August and November 2018 to score transition to flowering.
Results
Genetic data obtained from AFLP analysis indicate little genetic structure and limited barriers to geneflow in the metapopulation system
The AFLP analysis of core (pop. 7, Oyarbide field) and peripheric populations (pops. 1–6) included 264 individuals. Of 287 semi-automatically scored loci, 131 loci were kept after rigorous quality control following a method introduced earlier (Koch et al. 2017); 106 of these 131 loci were found to be polymorphic. The combined experimental and genotyping error rate (Bonin et al. 2004) in the final data set was below 0.5%. AFLP data indicated little genetic structure within the entire study region of about 1500 km2. The optimal number of genetic clusters (K) was estimated to be 2 (Fig. 1d) with highest DeltaK at K = 2 (Table 1).
Individuals assigned to both genetic clusters were also detected in the core population and study field at Oyarbide (Fig. 1c and Fig. 2a, c, population no. 7 with rows A–O, inland to coast direction). However, genetic clusters are partitioned geographically with cluster 1 (grey) confined mostly to upper rows A–E and cluster 2 (blue) confined to lower rows F–O. Neighbouring populations (1–6) largely follow this pattern, and AFLP data indicate genetic coherence.
Analysis of barriers to geneflow between core and peripheric populations was congruent (Fig. 1B, C), and no major barriers to geneflow between the different populations were detected. Within the entire distribution area, significant barriers were found only between population 1 and 2 (Fig. 1B), which were also characterized by Structure with two different genetic assignments (Fig. 1D). Within the core population at Oyarbide (Fig. 1C), barriers to geneflow following linear vegetation were detected in most parts of the study site when analysing this population alone. Linear patterns are most pronounced in the upper rows compared to rows at lower elevation which are less sloped (see Fig. 2C). This is in agreement with Structure results indicating a transition zone from genepool 1 (grey) to genepool 2 (blue) at row E/F. Following Structure results, the Oyarbide field can be divided into row A–E and F–O and the mean number of fragments can be compared as an indicator of genetic diversity. Data in both classes followed a normal distribution, and a two-tailed Mann–Whitney U test was applied. The mean number of AFLP fragments among individuals (n = 199) was significantly different at p < 0.001: mean A–E = 118.6 (SD 16.3) compared to F–O = 104.3 (SD 10.0), with 13% more alleles in the upper part of the field.
Further genetic diversity statistics of all populations using Arlequin (Table 2) indicated that 32.4% of the genetic variation is distributed among populations, whereas the majority of variation (67.6%) is found within populations. The overall fixation index (FST) was 0.32. Within the Oyarbide population, summary statistics also showed a strong difference of θ (S) and θ (π) between rows A–E and F–O and higher genetic diversity in rows A–E.
Mantel tests of genetic and geographic distance indicate linear genetic population structure in distinct parts of a population
A Mantel test for distance classes over the entire Oyarbide field showed significant correlation between genetic distances (Jaccard index) and geographic distances (p < 0.01) for distance classes from 100 to 400 m only. A less significant negative correlation (p < 0.05) was observed for distance classes from 900 to 1200 m (Fig. 3). The positive correlation may coincide with local clusters of genetically descendent individuals considering the grid size of 100 m. Negative Mantel’s r in distance classes below 700 m indicated the separation of the entire field in two genepools (upper rows A–E and lower rows F–O). Under the assumption that there is increased clonal dispersal along a given line, there should be no correlation of genetic and spatial distances. In order to test whether any correlation from the total data set at Oyarbide is sub-structured within given rows, these Mantel tests were conducted for individual rows A–O. None of the tests showed any correlation between genetic and geographic distances (Online Resource 2), indicating that limited genetic variation is distributed within rows and that individuals are more similar to each other, possibly representing a more pronounced clonal structure. These results are consistent with BARRIER analyses that also demonstrated a linear arrangement of genetic diversity. Because of the rigorous filtering of potential genotyping error, we may not be able to distinguish between clonal variants (identical genotypes) and genetically very similar individuals. Therefore, we followed the above-described results to further elaborate on the fine-scale structure and distribution of genetic variation. Using Arlequin, we first extracted genetically identical pairs and groups of individuals from the data matrix, and second, we identified individuals in direct proximity (neighbouring 100 × 100 m grid), and finally, we tested whether identical and neighbouring individuals were spatially arranged following vegetation banding patterns (horizontally) or whether they were following an up/downward direction. Along rows A–E, only 3% of all individuals had AFLP haplotypes identical to any other individual within these rows, in contrast to rows F–O with 36% of identical haplotypes. Those identical AFLP haplotypes along rows F–O were largely shared across rows (90%) and not within rows (10%) [total pairwise hits n = 782]. The number of identical AFLP types was gradually increasing from 13% in row F to a maximum of 67% in row K and decreasing from 47% in row L to 22% in row O. This might indicate more extensive dispersal of clonal variants within the lower part of the Oyarbide populations represented by rows F–O. A significantly negative Tajima’s D was found for the same area, signifying an excess of low-frequency polymorphisms relative to expectation and thereby potentially indicating population size expansion.
Plant reproductive fitness correlates with genetic diversity
Mean flowering frequency was determined in the core population at Oyarbide within plots from which samples screened for genetic variation were collected (n = 199 plots). Flower frequency grouped into ranks (0, 1, 2) did not violate an assumption of a normal distribution of data. Mean flowering frequency along rows A–E (rank = 1.03, SD 0.43) was significantly higher (p < 0.05) than along rows F–O (rank = 0.86, SD 0.48) (Wilcoxon-Mann–Whitney U test). Thus, the differences between “flowering frequency” phenotypes were not only associated with a distinction between genepool 1 (grey) and genepool 2 (blue) (Fig. 2D), but also coincided with mean number of AFLP fragments/gene diversity (Table 2) indicating most likely a higher level of heterozygosity in rows A–E. During flowering frequency inspection and other field campaigns (November 2016, January–February 2017, October 2017, March, May, August, November–December 2018), we also searched for putative pollinators. We identified adults of the seasonally occurring hawkmoth Hyles annei (= Sphinx annei Guérin-Méneville) on our field collection trip in October in the given region indicating the early start of seasonal flowering. Phenological data revealed a transition rate towards flowering of 9.5% (from November to March), 5% (from March to May), 5% (from May to August), and 2.2% (from August to November) totalling to a yearly chance of 21.5% for any randomly chosen shoot (n = 500) to transition towards flowering.
Discussion
An important hypothesis to be tested was that whether Tillandsia landbeckii lomas in our study area represent a metapopulation system without significant barriers to geneflow, thus enabling us to make firm conclusions from genetic fine-scale analysis at core Oyarbide study site. The presented data reflect historical and contemporary population dynamics. The historical dimension spanning the history of many generations and the sum of any dispersal event did not reveal any obvious and significant spatial pattern across the entire study area (peripheral and core population) (Fig. 1), indicating that the entire region can be considered a metapopulation system with substantial migration of genetic migrants over time. Our main study site at Oyarbide in the centre of the metapopulation is a genetically representative population allowing to present conclusive results. The fine-scale analysis of the core population at Oyarbide mirrors substantial contemporary population dynamics and geneflow. Therefore, our genetic data provide a clue for the understanding of the establishment of the linear arranged tillandsia lomas in coastal fog systems. There are several approaches to explain the genesis of linear structures in T. landbeckii lomas. A very strict correlation between increasing total vegetation cover and increasing elevational occurrence has been described between 950 and 1025 m a.s.l. (Rundel et al. 1997), and it was suggested that a dynamic pattern of colonial development of Tillandsia landbeckii starts from a single point of origin. The radiative growth of each lens-like clonal structure should then be accompanied by a dieback of original and older parts of the clonal patch. However, it is still unclear how this results in linear and regular patterns running orthogonally to the slope of the terrain (Rundel et al. 1997). Here, it should be considered that several of those expanding rings interfere with each other and are then forced to form a more continuous line (see Fig. 2). In our analysis at lower elevations (less-sloped terrain), spatial distances between rows are larger compared to higher elevations at the same population. This has been modelled and explained by a correlated nonlinear function of decreasing fog shadow with increasing slope (Borthagaray et al. 2010). In a similar way, the model predicted an increase in regular patterning (linearity) with increased vegetation density (Borthagaray et al. 2010) and vice versa. The model, however, did not integrate two factors—sand movement and growth behaviour—which are both important for the system to be maintained. The rootless tillandsias are only able to grow in a sympodial and strictly “forward manner”. Considering a sloped terrain and facing incoming fog from downhill, plants have to grow “downhill”. At the same time, plants lack their stabilizing substrate. The wind system transporting fog during the night, however, also allows the transport of sand in an “uphill” direction, thereby stabilizing the growing plants which act as sand trap. The dynamics and speed of this process is neither understood nor modelled and is awaiting detailed analyses (Fig. 4). Tillandsia landbeckii lomas are only distributed between approximately 18 and 21.5°S with a maximum of total vegetation cover between 20 and 20.5°S (Pinto et al. 2006), where the Oyarbide study field is located. A more detailed analysis of a geographically defined niche has divided topological constraints of T. landbeckii growth (Wolf et al. 2016) and provided respective optimal median values and range from lower to upper quartile (elevation: 1090 m a.s.l. [1000–1160 m], slope: 8° [5°–12°], aspect: 220°, facing the southwest [160°–270°], distance from coast: 15.5 km [11–21 km].
Factors and variables determining terrestrial growth of Tillandsia landbeckii. Wind brings in fog (water source from the Pacific) and transports sand as stabilizing substrate modulating Tillandsia growth down the sloped landscape. The plant itself serves as a sand trap, and over decades and centuries plants will be over-sanded repeatedly leaving fossil carbon bands (Latorre et al. 2011). For further details, refer to the main text
Our genetic data are supporting several aspects of the above-described growth traits and support the hypothesis that linearity of vegetation patterns along an altitudinal gradient is correlated with spatial distribution of genetic variation. Genetic diversity is highest at the upper part of our focal population (Oyarbide field), where the terrain is steeper, rows are closer together and the overall “linearity” of the entire system is maximized. This is exactly what BARRIER analyses indicate (Fig. 1). Furthermore, higher genetic diversity can be attributed to increased heterozygosity of individual plants but also total genetic diversity is increased (Table 2), which may favour the idea that the linear system is the result of U-shaped and interfering growth of different individuals (Rundel et al. 1997). In contrast, at lower elevations (and less-sloped areas) vegetation is patchy and distance between rows is much larger accompanied by suboptimal growth conditions, which leaves ample space for vegetative (clonal) remnants acting as colonizers. Hence, we do not see horizontal dispersal as the preferred direction. Rather, plants migrate in any direction with downhill movement even preferred. The gradually changing dispersal and growth pattern along an altitudinal gradient result in decreased genetic variation and genetically less linearly arranged vegetation systems at lower elevations at our study field.
Accordingly, genetic data indicate local and effective outcrossing as suggested by pollinator visits. The various populations showed surprisingly high levels of genetic diversity and expected heterozygosity (Table 1). This is best explained by outbreeding and an effective pollination system. Although different mating systems ranging from SI (self-incompatible) to SC (self-compatible) and autonomous self-pollination have been described for more than 80 species of Tillandsia (Orozco-Ibarrola et al. 2015), most relatives of T. landbeckii from subgenus Diaphoranthema have been classified as possessing polyembryonic seeds (rare in Bromeliaceae) and an autogamous breeding system with a few number of species having cleistogamous flowers (Bianchi and Vesprini 2013; Donadio et al. 2015; Gardner 1986). In T. landbeckii, seeds are not polyembryonic, and endosperm is present in mature seeds (Donadio et al. 2015). In conclusion, we may have to consider a SC system acting in T. landbeckii, allowing self-pollination. However, nectar production is triggering insect-mediated pollination resulting in varying outcrossing rates. The dynamics of breeding systems involving T. landbeckii are also reflected by hybridization between different species, and, thereby, also documenting the evolutionary consequences of outbreeding even across species: the northern limit of T. landbeckii distribution building respective mono-specific coastal lomas in Chile is also the southern limit of lomas built up by T. purpurea Ruiz & Pavon, which are mostly found in Peru. In the respective contact zone, T. marconae has been found (Zizka and Munoz-Schick 1993). There is some evidence that T. marconae is a hybrid between both species, T. landbeckii and T. purpurea, with varying degrees of backcrossing in one or the other species (AFLP data and DNA sequence data from agt1, microanatomical studies; Koch M.A. et al., Zizka G. et al. unpublished). The presence of the putative pollinator Hyles annei in our study fields with a peak during October considered as the end of the flowering season and which is feeding on Tillandsia flowers can be seen as an indicator for seasonal outbreeding, thereby maintaining substantial levels of heterozygosity within populations. The flowers have a narrow perianth tube considered as adaptation to the arid environment to insulate the nectar and thereby reduce evaporation. Long, narrow tubes in combination with yellowish, whitish flower colour combined with pleasant odours suggest shingophily (Leins and Erbar 2010). There are no other nectar feeding resources around across many kilometres, and therefore Tillandsia landbeckii serves as the only feeding source for the hawkmoth. Hyles annei is a widely distributed species from Chile, Bolivia, western Peru and Argentina and is thought to feed as larvae on many plant species and, therefore, is considered a generalist. The large insect with up to 60–70 mm in length can travel large distances, and the strong and inland-directed winds might support long-distance migration. We were not able to monitor the species during the rest of the year. Details on natural feeding plants of larvae are largely unknown, but a polyphagous behaviour has been shown (Hundsdörfer et al. 2019); however, we did not observe feeding larvae on T. landbeckii nor damage on the leaves possibly caused by feeding larvae.
From an evolutionary perspective, it may make sense that an environmentally highly specialized plant species with high risk of local extinction but an ample capacity to distribute over larger distances by vegetative means (e.g. wind-mediated dispersal of plant fragments) is not depending on specialized pollinators. Such a pollinator would suffer even harder from stochasticity of population collapse in a landscape with no alternative feeding resources. The reasoning for higher number of flowers in the upper parts of the Oyarbide remains open. Higher flower number may either be directly linked to better environmental conditions, in particular due to fog occurrence supplying sufficient water and thereby increasing plant fitness (growth and thereby flowering), or increased flowering frequency is genotype dependent and increased heterozygosity counteracts negative fitness effects otherwise acting via inbreeding, which may result in increased drift and genetic depletion. Alternatively, both effects act hand in hand and affect each other in a feed-back loop. Thereby, the upper part of the population can operate as a long-term genetic source for successful colonization either downwards or towards different regions, which is an open question to be addressed in future.
In summary, our data demonstrate that establishment of linear vegetation structure is primarily a process driven by clonal growth and propagation of ramets over short distances. Optimal conditions (slope, elevation, fog occurrence) for linear growth pattern formation, however, also increase sexual plant reproductive fitness, thus providing the reservoir for newly combined genetic variation and thereby counteracting genetic uniformity. Under less favourable conditions with less dense vegetation and disturbed linear vegetation patterns, clonal propagation and spread is prevailing in any direction. Since sexual reproductive fitness is also highly reduced at lower elevation at Oyarbide, total genetic variation is lower compared to optimal sites at higher elevation with optimal growth conditions. It is remarkable to see that maintaining sexual reproductive fitness plays a key role in population dynamic processes while growing at the dry limits, most likely also on long-term evolutionary time scales.
The herein described vegetation type is threatened by global climate change. The rapid decline of total size of the Tillandsia landbeckii lomas during the past 50 years is alarming. Unfortunately, we largely lack information on past evolutionary dynamics of distribution ranges for plants from the hyperarid Atacama Desert, where episodes of high aridity were interrupted by intervals of increased rainfall (Latorre et al. 2005) to make any comparisons with past climate change. First evidence for an evolutionary north–south gradient of declining genetic diversity was detected in Nolana (Ossa et al. 2013). This supports that populations survived in northern arid sites during wetter and colder episodes of the glacial cycles, and present-day distribution is the result of the last southward expansion of the coastal desert vegetation. Accordingly, fossil evidence for T. landbeckii lomas in our study area can be traced back for 3290 years (study site pop. 12), 3500 years (study site pop. 15) or 3310 years (study site pop. 16) (Latorre et al. 2011). This indicates long-term stable vegetation for thousands of years after the Last Glaciation Maximum. Currently, many Tillandsia fields, which are easily accessible and close to main roads, largely consist of dead vegetation lines. However, aside general observations of plant dieback along the coastal desert of northern Chile during the second half of the past century (Schulz et al. 2010), there is only one systematically conducted survey focusing on T. landbeckii (Osses et al. 2007). This study compared the situation close to our Oyarbide study site from 1955 with 1997 (Osses et al. 2007; Schulz et al. 2010) and postulated a total loss of living Tillandsia landbeckii lomas of about 35%. Generally, information about range contraction of Tillandsia lomas is scarce, because of the difficulties to access and monitor the landscape and because of generally missing documentation of biodiversity change in these areas. Distribution surveys and loma vegetation observation differed significantly among studies (Cereceda et al. 1999; Pinto et al. 2006; Rundel et al. 1997; Wolf et al. 2016) with significant increases in reliability and accuracy following the establishment of remote sensing-based technologies (satellite- and drone-based imaging). Although there is a bias because of differing methods to estimate vegetation cover, the negative trend in coverage by Tillandsia lomas is obvious. The reason, however, is unclear, and direct human impact such as recreational activities (off-road motorcycling) may have an impact. The recent literature has reported high mortality within the coastal lomas, but at present we understand neither the regional extent nor how widespread such mortality is, nor the effect of topography (aspect, elevation) and changes in fog dynamics on loma degradation (Muñoz-Schick et al. 2001; Schulz et al. 2010). However, there is evidence that loma vegetation and its decline are mostly linked to changes in the intensity of fog fluxes (Pinto et al. 2006), and we know from isotope analysis from living and fossil plant material that there were past fog changes (Latorre et al. 2011). It has been shown that regional-wide El Niño Southern Oscillation (ENSO) has a potential causal link to fog along the Atacama (Del Rio et al. 2018). Since El Niño events are in the process of becoming more intense due to global warming and climate change (Cai et al. 2014), the fog system might change accordingly. Within our project, we have set up a long-term monitoring scheme (monitoring total vegetation and biomass, screening individual and population growth, monitoring relevant climate parameters and fog precipitation along various transects in the Atacama, collecting cloud cover data and fog occurrence) to develop a respective geoecological niche model which will help to predict the fate of Tillandsia lomas. It has been suggested (Rundel et al. 1997) that Tillandsia lomas should deserve special attention as bioindicators of climatic changes. Our analyses were conducted in this light, and our monitoring system was set up to refine future analysis, thereby further strengthening the role of Tillandsia lomas as bioindicators in a changing world.
Data availability
The data sets generated and analysed during the current study are available from the corresponding author on reasonable request. Results from Mantel tests and accession data are presented in Online Resource 1.
References
Anderson MJ, Willis TJ (2003) Canonical analysis of principal coordinates: a useful method of constrained ordination for ecology. Ecology 84:511–525. https://doi.org/10.1890/0012-9658(2003)084%5b0511:CAOPCA%5d2.0.CO;2
Aponte H, Flores J (2013) Density and spatial distribution of Tillandsia latifolia on the tillandsial lomas of Piedra Campana (Lima, Perú). Ecol Aplicada 12:35–43
Bianchi MB, Vesprini JL (2013) Contrasting breeding systems in six species of Tillandsia L. (Bromeliaceae) from woody areas of Santa Fe Province, Argentinia. Pl Biosyst 148:956–964. https://doi.org/10.1080/11263504.2013.806965
Bonin A, Bellemain E, Bronken Eidesen P, Pompanon F, Brochmann C, Taberlet P (2004) How to track and assess genotyping errors in population genetics studies. Molec Ecol 13:3261–3273. https://doi.org/10.1111/j.1365-294X.2004.02346.x
Borthagaray AI, Fuentes ME, Marquet PA (2010) Vegetation pattern formation in a fog-dependent ecosystem. J Theor Biol 265:18–26. https://doi.org/10.1016/j.jtbi.2010.04.020
Cai W, Borlace S, Lengaigne M, Van Rensch P, Collins M, Vecchi G, Timmermann A, Santoso A, MvPhaden MJ, Wu L, England MH, Wang G, Guilyardi E, Jin FF (2014) Increasing frequency of extreme El Niño events due to greenhouse warming. Nat Clim Change 4:111–116. https://doi.org/10.1038/nclimate2100
Cereceda P, Larrain H, Lázaro P, Osses P, Schemenauer RS, Fuentes L (1999) Campos de Tillandsias y niebla en el desierto de Atacama. Revista Geogr Norte Grande 26:37–52
Cereceda P, Larrain H, Osses P, Farías M, Egaña I (2008) The climate of the coast and fog zone in the Atacama Desert of Tarapacá Region, Chile. Atmos Res 301:301–311. https://doi.org/10.1016/j.atmosres.2007.11.011
Deblauwe V, Couteron P, Bogaert J, Barbier N (2012) Determinants and dynamics of banded pattern migration in arid climates. Ecol Monogr 82:3–21. https://doi.org/10.1890/11-0362.1
Del Rio C, Garcia JL, Osses P, Zanetta N, Lambert F, Rivera D, Siegmund A, Wolf N, Cereceda P, Larraín H, Lobos F (2018) ENSO influence on coastal fog-water yield in the Atacama Desert, Chile. Aerosol Air Qual Res 18:127–144. https://doi.org/10.4209/aaqr.2017.01.0022
Dillon MO, Gonzáles SL, Cruz MZ, Asencio PL, Silvestre VQ (2011) Floristic checklist of the Peruvian lomas formation. Arnaldoa 18:7–32
Donadio S, Pozner R, Giussani LM (2015) Phylogenetic relationships within Tillandsia subgenus Diaphoranthema (Bromeliaceae, Tillandsioidea) based on a comprehensive morphological dataset. Pl Syst Evol 301:387–410. https://doi.org/10.1007/s00606-014-1081-1
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem Bull 19:11–15
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molec Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molec Ecol Res 10:564–567. https://doi.org/10.1111/j.1755-0998.2010.02847.x
Falush D, Stephans M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molec Ecol Notes 7:574–578. https://doi.org/10.1111/j.1471-8286.2007.01758.x
Gardner CS (1986) Inferences about pollination in Tillandsia (Bromeliaceae). Selbyana 9:76–87
Hartley AJ, Chong G, Houston J, Mather AE (2005) 150 million years of climatic stability: evidence from the Atacama Desert, northern Chile. J Geol Soc London 162:421–424. https://doi.org/10.1144/0016-764904-071
Haslam R, Borland A, Maxwell K, Griffiths H (2003) Physiological responses of the CAM epiphyte Tillandsia usneoides L. (Bromeliaceae) to variations in light and water supply. J Pl Physiol 160:627–634. https://doi.org/10.1078/0176-1617-00970
Heibl C, Renner SS (2012) Distribution models and a dated phylogeny for Chilean Oxalis species reveal occupation of new habitats by different lineages, not rapid adaptive radiation. Syst Biol 61:823–834. https://doi.org/10.1093/sysbio/sys034
Hesse R (2012) Spatial distribution of and topographic controls on Tillandsia fog vegetation in coastal southern Peru: remote sensing and modelling. J Arid Environm 78:33–40. https://doi.org/10.1016/j.jaridenv.2011.11.006
Hesse R (2014) Three-dimensional vegetation structure of Tillandsia latifolia on a coppice dune. J Arid Environm 109:23–30. https://doi.org/10.1016/j.jaridenv.2014.05.001
Houston J, Hartley AJ (2003) The central Andean west-slope rainshadow and its potential contribution to the origin of hyper-aridity in the Atacama Desert. Int J Climatol 23:1453–1464. https://doi.org/10.1002/joc.938
Hundsdörfer AK, Buchwalder K, O´Neill MAO, Dobler S (2019) Chemical ecology traits in an adaptive radiation: TPA-sensitivity and detoxification in Hyles and Hippotion (Sphingidae, Lepidoptera) larvae. Chemoecol 29:35–47
Ibaraki M (1997) Closing of the Central American Seaway and Neogene coastal upwelling along the Pacific coast of South America. Tectonophysics 281:99–104. https://doi.org/10.1016/S0040-1951(97)00161-3
Jaccard P (1901) Étude comparative de la distribution florale dans une portion des Alpes et des Jura. Bull Soc Vaud Sci Nat 37:547–579
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806. https://doi.org/10.1093/bioinformatics/btm233
Koch MA, Michling F, Walther A, Huang XC, Tewes L, Müller C (2017) Early-Mid Pleistocene genetic differentiation and range expansions as exemplified by invasive Eurasian Bunias orientalis (Brassicaceae) indicates the Caucasus as key region. Sci Rep 7:16764. https://doi.org/10.1038/s41598-017-17085-8
Larraín H, Velásquez F, Cereceda P, Espejo R, Pinto R, Osses P, Schemenauer RS (2002) Fog measurements at the site “Falda Verde” north of Chañaral compared with other fog stations of Chile. Atmos Res 273:273–284
Latorre C, Betancourt JL, Rech JA, Quade J, Holmgren C, Placzek C, Maldonado A, Vuille M, Rylander KA (2005) Late Quaternary history of the Atacama Desert. In: Smith M, Hesse P (eds) 23°S: archaeology and environmental history of the southern deserts. National Museum of Australia Press, Canberra, pp 73–90
Latorre C, Gonzáles AL, Quade J, Farina JM, Pinto R, Marquet PA (2011) Establishment and formation of fog-dependent Tillandsia landbeckii dunes in the Atacama Desert: Evidence from radiocarbon and stable isotopes. J Geophys Res 116:G03033. https://doi.org/10.1029/2010JG001521
Legendre P, Legendre L (2012) Numerical ecology. Elsevier, Amsterdam
Leins P, Erbar C (2010) Flower and fruit: morphology, ontogeny, phylogeny, function and ecology. Schweizerbart, Stuttgart
Luebert F, Pliscoff P (2006) Sinopsis bioclimática y vegetacional de Chile. Editorial Universitaria Santiago de Chile, Santiago de Chile
Luebert F, Wen J (2008) Phylogenetic analysis and evolutionary diversification of Heliotropium Sect. Cochranea (Heliotropiaceae) in the Atacama Desert. Syst Bot 33:390–402. https://doi.org/10.1600/036364408784571635
Manel S, Schwartz MK, Luikart G, Taberlet P (2003) Landscape genetics: combining landscape ecology and population genetics. Trends Ecol Evol 18:189–197. https://doi.org/10.1016/S0169-5347(03)00008-9
Manni F, Guérard E, Heyer E (2004) Geographic patterns of (genetic, morphologic, linguistic) variation: how barriers can be detected by “Monmonier’s algorithm”. Human Biol 76:173–190
Meudt HM, Clarke AC (2007) Almost forgotten or latest practice? AFLP applications, analyses and advances. Trends Pl Sci 12:106–117. https://doi.org/10.1016/j.tplants.2007.02.001
Muñoz-Schick M, Pinto R, Mesa A (2001) Oasis de Neblina en los Cerros Costeros del sur de Iquique, Región de Tarapacá, Chile, durante el evento El Niño 1997–1998. Revista Chilena Hist Geogr 74:389–405
Orozco-Ibarrola OA, Flores-Hernández PS, Victoriano-Romero E, Corona-López AM, Flores-Palacios A (2015) Are breeding system and florivory associated with the abundance of Tillandsia species (Bromeliaceae). Bot J Linn Soc 177:50–65. https://doi.org/10.1111/boj.12225
Ossa PG, Perez F, Armesto JJ (2013) Phylogeography of two closely related species of Nolana from the coastal Atacama Desert of Chile: post glacial population expansion in response to climate fluctuations. J Biogeogr 40:2191–2203. https://doi.org/10.1111/jbi.12152
Osses P, Cereceda P, Larrain H, Astaburuaga JP (2007) Distribution and geographical factors of the Tillandsia fields in the coastal mountain range of the Tarapacá Region, Chile. In: Biggs A, Cereceda C (eds) Proceedings of fourth international conference on fog, fog collection and dew, La Serena, Chile. La Serena, pp 93–96
Paradis E, Claude J, Strimmer K (2004) APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290. https://doi.org/10.1093/bioinformatics/btg412
Pinto R, Barria I, Marquet PA (2006) Geographical distribution of Tillandsia lomas in the Atacama Desert, northern Chile. J Arid Environm 65:543–552. https://doi.org/10.1016/j.jaridenv.2005.08.015
Pritchard JK, Stephens M, Donelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rauh W (1958) Beitrag zur Kenntnis der peruanischen Kakteenvegetation. Sitzungsber. Heidelberger Akad Wiss Math-Naturwiss Kl 395:1–543
Rech JA, Currie BS, Shullenberger ED, Dunagan SP, Jordan TE, Blanco N, Tomlinson AJ, Rowe H, Houston J (2010) Evidence for the development of the Andean rain shadow from a Neogene isotopic record in the Atacama Desert, Chile. Earth Planet Sci Lett 292:371–382. https://doi.org/10.1016/j.epsl.2010.02.004
Rundel PW, Palma B, Dillon MO, Sharifi MR, Boonpragob K (1997) Tillandsia landbeckii in the coastal Atacama Desert of northern Chile. Revista Chilena Hist Nat 70:341–349
Scherson RA, Thornhill AH, Urbin-Casanova R, Frayman WA, Pliscoff PA, Mishler BD (2017) Spatial phylogenetics of the vascular flora of Chile. Molec Phylogen Evol 112:88–95. https://doi.org/10.1016/j.ympev.2017.04.021
Schulz N (2009) Loma-Formationen der Küsten-Atacama/Nordchile unter besonderer Berücksichtigung rezenter Vegetations- und Klimaveränderungen. PhD Thesis, Geographical Institute, Erlangen-Nürnberg University, Nürnberg
Schulz N, Aceituno P, Richter M (2010) Phytogeographic divisions, climate change and plant dieback along the coastal desert of northern Chile. Erdkunde 65:169–187. https://doi.org/10.3112/erdkunde.2011.02.05
Takahashi Y, Tanaka R, Yamamoto D, Noriyuki S, Kawata M (2018) Balanced genetic diversity improves population fitness. Proc Roy Soc B 285:20172045. https://doi.org/10.1098/rspb.2017.2045
Tewes LJ, Michling F, Koch MA, Müller C (2017) Intracontinental plant invader shows matching genetic and chemical profiles and might benefit from high defence variation within populations. J Ecol 106:714–726. https://doi.org/10.1111/1365-2745.12869
Tlidi M, Clerc MG, Escaff D, Couteron P, Messaoudi M, Khaffou M, Makhoute A (2018) Observation and modelling of vegetation spirals and arcs in isotropic environmental conditions: dissipative structures in arid landscapes. Phil Trans Roy Soc A 376:20180026. https://doi.org/10.1098/rsta.2018.0026
Vos P, Hogers R, Bleeker M, Reijans M, Van de Lee T, Hornes M, Friters A, Pot J, Paleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucl Acids Res 23:4407–4414. https://doi.org/10.1093/nar/23.21.4407
Westbeld A, Klemm O, Grießbaum F, Sträter E, Larrain H, Osses P, Cereceda P (2009) Fog deposition to a Tillandsia carpet in the Atacama Desert. Ann Geophys 27:3571–3576. https://doi.org/10.5194/angeo-27-3571-2009
Wolf N, Siegmund A, Del Rio C, Osses P, Garcia JL (2016) Remote sensing-based detection and spatial pattern analysis for geo-ecological niche modelling of Tillandsia spp. in the Atacama Desert, Chile. Int Arch Photogramm Remote Sensing 41:B2. https://doi.org/10.5194/isprsarchives-XLI-B2-251-2016
Zachos J, Pagani M, Sloan L, Thomas E, Billups K (2001) Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 292:686–693
Zizka G, Munoz-Schick M (1993) Tillandsia-marconae W. Till & Vitek, a bromeliad species new to Chile. Bol Mus Nac Hist Nat Chile 44:11–17. https://doi.org/10.1126/science.1059412
Zizka G, Schmidt M, Schulte K, Nonoa P, Pinto R, König K (2009) Chilean Bromeliaceae: diversity, distribution and evaluation of conservation status. Biodivers Conservation 18:2449–2471. https://doi.org/10.1007/s10531-009-9601-y
Acknowledgements
This study was supported by HCE (Heidelberg Centre for the Environment) enabling funds to M.A.K., and by BMBF within the ERANet-LAC program of the EU to A. S. and M.A.K. J.L.G, C.D.R. and P.O. were supported by ELAC2015/T01-0872 and Centro UC Desierto de Atacama. We thank Peter Sack, Lisa Kretz and Florian Michling for technical laboratory support in Heidelberg. Nils Wolf assisted with GIS-based data processing and field collections.
Author information
Authors and Affiliations
Contributions
M.A.K. conceived the project and the experiments. D.K. and M.A.K. conducted the experiment(s), M.A.K., D.K., E.A. and C.K. analysed the results. A.S., C.D.R., P.O., J.L.G., G.Z. and M.V.M. assisted with sample plant material. M.A.K. drafted the manuscript. All authors contributed to the final manuscript draft.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they do have no conflict of interest.
Additional information
Handling Editor: Karol Marhold.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Information on Electronic Supplementary Material
Information on Electronic Supplementary Material
Online Resource 1. Information on accessions and individuals studied.
Online Resource 2. Information on the results from Mantel tests.
Rights and permissions
About this article
Cite this article
Koch, M.A., Kleinpeter, D., Auer, E. et al. Living at the dry limits: ecological genetics of Tillandsia landbeckii lomas in the Chilean Atacama Desert. Plant Syst Evol 305, 1041–1053 (2019). https://doi.org/10.1007/s00606-019-01623-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-019-01623-0