Abstract
Xenochila is a monospecific genus that has not yet been included in molecular phylogenies. Based on morphology it has been aligned with Plagiochilaceae. A chloroplast DNA phylogeny places Xenochila within Jungermanniaceae in a robust sister relationship with the monospecific genus Delavayella. Though outwardly disparate in form, Delavayella and Xenochila share the presence of multicellular parenchymatous propagules forming singly from leaf margins of specialized gemmiparous shoots, a papillose gametophyte surface, nearly undifferentiated stem cells, absence of underleaves and bracteoles, weakly to distinctly bifid female bracts that are slightly larger than the leaves, and long cylindrical perianths with a dentate mouth. With the addition of Xenochila, Jungermanniaceae include the genera Delavayella, Eremonotus, Jungermannia, Liochlaena and Mesoptychia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Molecular phylogenetic studies greatly improved the classification of liverworts (Crandall-Stotler et al. 2009) and culminated in the recent publication of a global species checklist reflecting current knowledge on systematic relationships (Söderström et al. 2016). However, despite comprehensive efforts to build the liverwort tree of life (e.g., Shaw et al. 2015) it has not yet been possible to include all currently accepted genera in molecular studies.
One such example is the monospecific Asian genus Xenochila R.M.Schust. Schuster (1959) described Xenochila based on its septate rhizoids and ovoid propagules developing singly from marginal teeth of young leaves, and treated Xenochila as a member of Plagiochilaceae. However, Schuster’s concept of this family was broad enough to include also Leptoscyphus Mitt. (Lophocoleaceae, Hentschel et al. 2006); Mylia S.F.Gray (Myliaceae, Heinrichs et al. 2007) and Syzygiella Spruce (Adelanthaceae, Feldberg et al. 2010b). Although Schuster’s concept of Plagiochilaceae is now largely rejected, Xenochila remains accepted as a member of Plagiochilaceae (Inoue 1963; Crandall-Stotler et al. 2009; Singh et al. 2015; Söderström et al. 2016). In the dissenting opinion, Inoue (1963) pointed to morphological similarities of Xenochila with Jamesoniella (now considered a synonym of Syzygiella, Feldberg et al. 2010a) and Jungermannia L. This view was shared by Patzak et al. (2016) who considered the undifferentiated stem cells of Xenochila and the rhizoids restricted to ventral leaf bases untypical for Plagiochilaceae.
Here we test the different hypotheses by presenting a chloroplast DNA phylogeny of Jungermanniales.
Materials and methods
DNA extraction, PCR amplification and sequencing
Gametophytical plant tissue was isolated from a herbarium specimen of Xenochila integrifolia (Mitt.) Inoue (Hepaticae Japonicae Exsiccatae Ser. 15, 750, M) and used to extract genomic DNA with the Invisorb Spin Plant Mini Kit (Invitek, Berlin).
PCR reactions were carried out with 0.25 µL MyTaq DNA Polymerase (Bioline Reagents Ltd, UK), 10 µL reaction buffer, 1 µL of upstream primer (10 µM), 1 µL of downstream primer (10 µM) and 1.5 µL of template DNA. This mix was filled up with double distilled water to a total volume of 50 µL. Amplification of rps4 used the primers rps5-F (Nadot et al. 1994) and trnS-R (Taberlet et al. 1991). Temperature profile for rps4 was 92 °C for 10 min, followed by 35 cycles of 92 °C for 60 s, 52 °C for 60 s, and 72 °C for 150 s. A final extension step of 72 °C for 10 min completed the thermal cycle.
For amplification of rbcL a nested PCR was carried out. Temperature profile for both PCR reactions was 95 °C for 2 min, followed by 30 cycles of 95 °C for 60 s, 51 °C for 50 s, and 72 °C for 90 s. A final extension step of 72 °C for 10 min completed the thermal cycle. The primer pairs used for the nested rbcL PCR were rbcL1-Pl-F and rbcL-M1390-R (Wilson et al. 2004) for the first step using genomic DNA, and rbcL-M38F (Wilson et al. 2004) and rbcL-1379-Pl-R (Groth and Heinrichs 2005) in the second PCR seeded with 1.5 µL of the product from PCR 1.
Sequencing was carried out on an ABI 3730 capillary sequencer using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). Primers used for PCR amplification were also used for sequencing. The new sequences were assembled and edited in CodonCode Aligner 5.0.1 (CodonCode Corp., Dedham, MA, USA).
Phylogenetic analyses
First, the rbcL and rps4 sequences of Xenochila were compared with GenBank sequences using the BLASTN program (Altschul et al. 1990). The BLAST searches suggested an affiliation of Xenochila to Jungermanniales suborder Jungermanniineae. Based on the phylogenetic hypotheses of Heinrichs et al. (2007), Cailliau et al. (2013), Sun et al. (2014) and Shaw et al. (2015), sequences of Jungermanniales suborders Jungermanniineae, Cephaloziineae and Lophocoleineae were downloaded from Genbank. Two representatives of Jungermanniales suborder Perssoniellineae were chosen as outgroup (Table 1).
All sequences were aligned manually in Bioedit version 7.0.5.2 (Hall 1999). Lacking parts of sequences were coded as missing. Maximum likelihood (ML) inferrence was carried out using RAxML 8.1.2 (Stamatakis 2014; http://sco.h-its.org/exelixis/web/software/raxml/#documentation) as implemented in raxmlGUI 1.5b1 (Silvestro and Michalak 2012; http://sourceforge.net/projects/raxmlgui/). The best-fit models of evolution were selected in jModelTest 2 (Darriba et al. 2012) under the Akaike information criterion (Akaike 1973) following the suggestions given by Posada (2008). Initial analyses were carried out for the two chloroplast DNA datasets. Since there was no statistically supported (>70 % bootstrap value) contradiction (Mason-Gamer and Kellogg 1996), the two datasets were concatenated, resulting in an alignment of 1910 nucleotides (Online Resource 1). ML analyses of the concatenated datasets were conducted using the GTR model (Tavaré 1986) with a proportion of invariable characters (I) and among-site rate heterogeneity modeled as a discrete gamma distribution with four rate categories and its estimated parameters (Γ), and a rps4 and rbcL partition. Trees were generated by selecting ten independent runs and the multiparametric bootstrap option autoMRE resulting in 450 bootstrap replicates. ML bootstrap values (BV) of each node were visualized using FigTree 1.4 (http://beast.bio.ed.ac.uk/figtree). Clades with bootstrap values (BV) ≥70 % were considered to be well supported (Hillis and Bull 1993).
Morphological investigation
Specimens were studied using a Leica M50 dissection microscope and a Carl Zeiss AxioScope A1 compound microscope, the latter equipped with a Canon 60D digital camera. Incident and transmitted light were used simultaneously or separately. The presented illustrations (Fig. 2) are digitally stacked photomicrographic composites of moistened plants obtained with the software package HeliconFocus 6.5.1.
Results
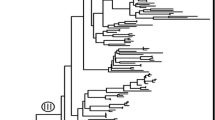
The ML phylogeny (Fig. 1) includes three major clades corresponding to Jungermanniales suborders Cephaloziineae (BV 78), Lophocoleineae (BV 97) and Jungermanniineae (BV 100). Xenochila integrifolia is placed in the Jungermanniineae lineage and sister to Delavayella serrata Steph. (BV 99); this clade forms a sister relationship with Liochlaena Nees (BV 100). The Delavayella Steph.-Liochlaena-Xenochila-clade is placed sister to a clade containing Eremonotus and Jungermannia (BV 97); these genera form a sister relationship with Mesoptychia (Lindb.) A.Evans (BV 97).
Discussion
Plagiochilaceae (Lophocoleineae) typically have reduced underleaves, scattered rhizoids, undivided leaves and laterally compressed, bilabiate perianths with a truncate mouth (Crandall-Stotler et al. 2009). Their classification is still in flux and several important changes have been proposed since the reviews of Söderström et al. (2015, 2016). These include the synonymy of the genera Acrochila R.M.Schust. and Plagiochilion S.Hatt. with Chiastocaulon Carl and the establishment of the genus Cryptoplagiochila S.Patzak, M.A.M.Renner & Heinrichs (Patzak et al. 2016) as well as a recircumscription of Dinckleria Trevis. (Renner et al. 2016). Our phylogeny (Fig. 1) leads to another change and provides convincing evidence that Xenochila is not related to Plagiochilaceae. It is a member of Jungermanniineae rather than Lophocoleineae and placed in the Delavayella-clade of Jungermanniaceae.
Numerous Jungermanniaceae concepts have been proposed since the initial description of this family by Reichenbach (1828). Jungermanniaceae were often considered to include nearly exclusively species with unlobed leaves (Amakawa 1959, 1960; Müller 1951–1958) yet Schuster (1970) extended the family to include former elements of Lophoziaceae, i.e., species with lobed leaves. This treatment rendered Jungermanniaceae one of the most speciose families of liverworts, but was not confirmed by molecular studies (Yatsentyuk et al. 2004; Heinrichs et al. 2005; He-Nygrén 2007). Schill et al. (2004) and He-Nygrén et al. (2006) were the first to demonstrate a close relationship of Jungermannia and Delavayella of the monogeneric Delavayellaceae. Based on phylogenetic analyses of rbcL sequences, Hentschel et al. (2007) excluded Liochlaena Nees and Solenostoma Mitt. from the synonymy of Jungermannia s.l. (Váňa 1973) and proposed to include Delavayella, Eremonotus Pearson, Jungermannia s.s., Leiocolea (Müll.Frib.) Buch, and Liochlaena in Jungermanniaceae. De Roo et al. (2007) showed that Mesoptychia is nested in Leiocolea and Váňa et al. (2012) consequently treated Leicolea as a synonym of Mesoptychia. The above genera have never been considered to form a monophyletic lineage representing Jungermanniaceae, however, Hentschel et al. (2007: 155) noted that they “share the exclusive presence of perianths, whereas related Jungermanniineae clades comprise genera with female involucres that are at least partly formed by stem tissue”. Shaw et al. (2015) considerably extended the taxon and marker sampling of Hentschel et al. (2007) and confirmed their treatment of Jungermanniaceae. They added further morphological characters supporting the recognition of the newly circumscribed Jungermanniaceae including “branching only from lateral merophytes, gynoecia and androecia only on leading stems, long emergent perianths that are contracted at the mouth, bistratose capsule walls, and in all taxa except M. sahlbergii (Lindb. & Arnell) A. Evans, calyptral development with little or no shoot involvement, no perigynial development, and no geocauly” (Shaw et al. 2015: 35). As a consequence of the new arrangement of Jungermanniaceae, this family includes species with either rounded or bifid leafs.
The position of Xenochila within Jungermanniaceae and its sister relationship with Delavayella have never seriously been considered although Inoue (1963) mentioned similarities with some papillose Jungermannia species. Schuster (1999) pointed to similarities in the method of asexual reproduction of Delavayella and Xenochila yet aimed at demonstrating morphological differences between Scapaniaceae and Delavayellaceae. Delavayella and Xenochila share the presence of multicellular parenchymatous green to red propagules forming singly from leaf margins on specialized gemmiparous shoots (Fig. 2c, e, h), a strongly papillose gametophyte surface (Fig. 2d, i), undifferentiated or weakly differentiated stem cells, absence of underleaves and bracteoles, weakly to distinctly bifid female bracts that are slightly larger than the leaves, and long cylindrical perianths with a dentate mouth (Inoue 1963; Schuster 2002; Singh Deo and Singh 2013). Small-leaved gemmiparous shoots are also known from the Jungermanniaceae elements Mesoptychia heterocolpos (Thed. ex Hartm.) L.Söderstr. & Váňa and Liochlaena subulata (A.Evans) Schljakov but these species produce only 1-2-celled gemmae on slightly bilobed, spoon-shaped (Mesoptychia) or unlobed (Liochlaena) leaves (Paton 1999). Xenochila also matches the other characters of Jungermanniaceae listed by Shaw et al. (2015) yet stands out by the presence of a somewhat plagiochiloid, laterally compressed perianth. Its sporophyte has not yet been described, but considering its expression in other Jungermanniaceae species it may comprise capsules with bistratose walls. The robust sister relationship and the morphological similarities brings into question the generic separation of Xenochila and Delavayella. We are aware that monospecific genera have limited information on the relationships of their members; however, need to consider important morphological differences of Xenochila and Delavayella. In particular and in contrast to all other Jungermanniaceae and nearly all other members of Jungermanniales, Delavayella has a ventral leaf margin forming a large water-sac (Fig. 2b). Its frontally rather than laterally compressed perianths (Schuster 1999) are also untypical for Jungermanniaceae. Delavayella is the only member of Jungermanniaceae with serrulate leaf margins, much more robust (Fig. 2a) than the “lax” Xenochila (Fig. 2e), and provided with diffusely distributed rather than fascicled rhizoids (Fig. 2g). In light of these differences we keep both genera.
Photomicrographs of Delavayella serrata (a–d) and Xenochila integrifolia (e–i). a Upper portion of gametophyte in dorsal view. b Leafs. Arrows point to water-sacs. c Portion of small-leaved gemmiparous shoot with a multicellular propagule. d, i Papillose leaf cells in surface view. e Upper portion of gametophyte with two lateral branches. Arrow points to gemmiparous shoot (propagules already detached). f Rhizoid covered with fungal hyphae. g Leaf and portion of stem tissue. Blue arrow points to insertion of dorsal leaf base; the vertically oriented cells derive from the outermost stem layer. Black arrow points to rhizoid bundle initiating from the ventral leaf base. h detached multicellular propagule (a–d from Nepal, Thodung, Poelt H22 (M); e, f from Hepaticae Japonicae Exsiccatae 750, Kitagawa Oct. 4, 1965 (M); g, h, i from Hepaticae Japonicae Exsiccatae 1100, Tatebe June 5, 1951 (M); scale bars a, b, g = 200 µm; c = 100 µm; d, f, h, i = 50 µm; e = 500 µm)
Schuster (1959) based Xenochila on its supposedly septate rhizoids; however, this description was based on Degenkolbe (1937) who mentioned the presence of septae in rhizoids of germinating propagules. Presence of septate rhizoids in Xenochila could not be confirmed by Inoue (1963), Yang and Hsu (1967), and Singh Deo and Singh (2013) and was also not observed by us. Septate rhizoids occur only in some members of the liverwort families Schistochilaceae, Lepidoziaceae and Lophocoleaceae (Heinrichs et al. 2015) and are—at least in part—induced by mycorrhizal fungi (Pressel et al. 2010). It is thinkable that fungal hyphae on the surface of Xenochila rhizoids (Fig. 2f) have misleadingly appeared to indicate septae.
Perspectives
Our study provided evidence for the affiliation of Xenochila to Jungermanniaceae rather than Plagiochilaceae, and indeed a robust sister relationship to Delavayella. Presence of rhizoid fascicles in the leaf axils of Xenochila and Syzygiella (Patzak et al. 2016) is the result of convergent evolution which is a major obstacle in reconstructing evolutionary relationships of liverworts based on morphology (Crandall-Stotler et al. 2005; Renner 2015). Only a single “plagiochiloid” genus (Söderström et al. 2016) has not yet been included in molecular phylogenies, Plagiochilidium Herzog. This genus includes only a single species and close relationships to either Syzygiella (Herzog 1931) or Xenochila (Inoue 1963) have been proposed. However, a more definite statement should be based on molecular phylogenetic evidence.
References
Akaike H (1973) Information theory as an extension of the maximum likelihood principle. In: Petrov BN, Csáki F (eds) Second international symposium on information theory. Akadémiai Kiadó, Budapest, pp 267–281
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Molec Biol 215:403–410
Amakawa T (1959) Family Jungermanniaceae of Japan. I. J Hattori Bot Lab 21:248–291
Amakawa T (1960) Family Jungermanniaceae of Japan. II. J Hattori Bot Lab 22:1–90
Cailliau A, Long DG, Price MJ, Perret M (2013) Phylogeny and systematic position of Mesoptychia (Lindb.) A. Evans. Pl Syst Evol 299:1243–1251
Crandall-Stotler B, Forrest LL, Stotler RE (2005) Evolutionary trends in the simple thalloid liverworts (Marchantiophyta, Jungermanniopsida subclass Metzgeriidae). Taxon 54:299–316
Crandall-Stotler B, Stotler RE, Long DG (2009) Phylogeny and classification of the Marchantiophyta. Edinburgh J Bot 66:155–198. doi:10.1017/S0960428609005393
Darriba D, Taboada GL, Doallo R, Posada D (2012) JModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772. doi:10.1038/nmeth.2109
De Roo RT, Hedderson TA, Söderström L (2007) Molecular insights into the phylogeny of the leafy liverwort family Lophoziaceae Cavers. Taxon 56:301–314
Degenkolbe W (1937) Brutorgane bei beblätterten Lebermoosen. Ann Bryol 10:43–96
Feldberg K, Váňa J, Hentschel J, Heinrichs J (2010a) Currently accepted species and new combinations in Jamesonielloideae (Adelanthaceae, Jungermanniales). Cryptog Bryol 31:141–146
Feldberg K, Váňa J, Long DG, Shaw AJ, Hentschel J, Heinrichs J (2010b) A phylogeny of Adelanthaceae (Jungermanniales, Marchantiophyta) based on nuclear and chloroplast DNA markers, with comments on classification, cryptic speciation and biogeography. Molec Phylogen Evol 55:293–304. doi:10.1016/j.ympev.2009.11.009
Groth H, Heinrichs J (2005) Maximum likelihood analyses of chloroplast gene rbcL sequences indicate relationships of Syzygiella (Jungermanniopsida) with Lophoziaceae rather than Plagiochilaceae. Cryptog Bryol 26:49–57
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Oxford University Press. Nucl Acids Symp Ser 41:95–98
Heinrichs J, Gradstein SR, Wilson R, Scheider H (2005) Towards a natural classification of liverworts (Marchantiophyta) based on the chloroplast gene rbcL. Cryptog Bryol 26:131–150
Heinrichs J, Hentschel J, Wilson R, Feldberg K, Schneider H (2007) Evolution of leafy liverworts (Jungermanniidae, Marchantiophyta): estimating divergence times from chloroplast DNA sequences using penalized likelihood with integrated fossil evidence. Taxon 56:31–44
Heinrichs J, Schmidt AR, Schäfer-Verwimp A, Gröhn C, Renner MAM (2015) The leafy liverwort Notoscyphus balticus sp. nov. (Jungermanniales) in Ecocene Baltic amber. Rev Palaeobot Palynol 217:39–44. doi:10.1016/j.revpalbo.2015.02.006
Hentschel J, Wilson R, Burghardt M, Zündorf HJ, Schneider H, Heinrichs J (2006) Reinstatement of Lophocoleaceae (Jungermanniopsida) based on chloroplast gene rbcL data: exploring the importance of female involucres for the systematics of Jungermanniales. Pl Syst Evol 258:211–226. doi:10.1007/s00606-006-0408-y
Hentschel J, Paton JA, Schneider H, Heinrichs J (2007) Acceptance of Liochlaena Nees and Solenostoma Mitt., the systematic position of Eremonotus Pearson and notes on Jungermannia L. s.l. (Jungermanniidae) based on chloroplast DNA sequence data. Pl Syst Evol 268:147–157. doi:10.007/s00606-007-0549-7
He-Nygrén X (2007) Multi-gene phylogeny supports single origin of jungermannioid perigynium. Ann Bot Fenn 44:450–462
He-Nygrén X, Juslén A, Ahonen I, Glenny D, Piippo S (2006) Illuminating the evolutionary history of liverworts. Cladistics 22:1–31
Herzog T (1931) Hepaticae. Mitt Inst Allg Bot Hamburg 7:182–216
Hillis DM, Bull JJ (1993) An empirical test of bootstrapping as a method for assessing the confidence in phylogenetic analysis. Syst Biol 42:182–192. doi:10.1093/sysbio/42.2.182
Inoue H (1963) Contribution to the knowledge of the Plagiochilaceae of Southeastern Asia IV. The genus Xenochila. Bull Natl Sci Mus, Tokyo 6:372–378
Mason-Gamer RJ, Kellogg EA (1996) Testing for phylogenetic conflict among molecular data sets in the tribe Triticeae (Gramineae). Syst Biol 45:524–545. doi:10.1093/sysbio/45.4.524
Müller K (1951–1958) Die Lebermoose Europas. In: Rabenhorst’s Kryptogamen-Flora von Deutschland, Österreich und der Schweiz, VI (1–2), 3rd edn. Geest & Portig, Leipzig
Nadot S, Bajot R, Lejeune B (1994) The chloroplast gene rps4 as a tool for the study of Poaceae phylogeny. Pl Syst Evol 191:27–38
Paton JA (1999) The liverwort flora of the British Isles. Harley Books, Colchester
Patzak SDF, Renner MAM, Schäfer-Verwimp A, Feldberg K, Heslewood MM, Peralta DF, Matos de Souza A, Schneider H, Heinrichs J (2016) A phylogeny of Lophocoleaceae-Plagiochilaceae-Brevianthaceae and a revised classification of Plagiochilaceae. Org Divers Evol. doi:10.1007/s13127-015-0258-y
Posada D (2008) jModelTest: phylogenetic model averaging. Molec Biol Evol 25:1253–1256
Pressel M, Bidartondo MI, Ligrone R, Duckett JG (2010) Fungal symbioses in bryophytes. New insights in the twenty first century. Phytotaxa 9:238–253. doi:10.11646/phytotaxa.9.1.13
Reichenbach HGL (1828) Botanik für Damen, Künstler und Freunde der Pflanzenwelt überhaupt, eine Anleitung zum Studium der Wissenschaft und zum Anlegen von Herbarien. Cnobloch, Leipzig
Renner MAM (2015) Lobule shape evolution in Radula (Jungermanniopsida): one rate fits all? Bot J Linn Soc 178:222–242. doi:10.1111/boj.12279
Renner MAM, Heslewood MM, Engel JJ, Glenny DS, von Konrat M, Patzak SDF, Schäfer-Verwimp A, Jamy M, Heinrichs J (2016) An integrative revision of Dinckleria (Plagiochilaceae: Jungermanniopsida). Austral Syst Bot
Schill DB, Long DG, Moeller M, Squirrel J (2004) Phylogenetic relationships between Lophoziaceae and Scapaniaceae based on chloroplast sequences. Monogr Syst Bot Missouri Bot Gard 98:141–149
Schuster RM (1959) A monograph of the Nearctic Plagiochilaceae, Part 1. Amer Midl Naturalist 62:1–166
Schuster RM (1970) Studies on Hepaticae, XVIII. The family Jungermanniaceae, s. lat.: a reclassification. Trans Brit Bryol Soc 6:86–107
Schuster RM (1999) Studies on Jungermannidae. IV. On Scapaniaceae, Blepharidophyllaceae and Delavayellaceae. J Bryol 21:123–132
Schuster RM (2002) Austral Hepaticae Part II. Nova Hedwigia Beih 119:1–606
Shaw B, Crandall-Stotler B, Váňa J, Stotler RE, von Konrat M, Engel JJ, Davis EC, Long DG, Sova P, Shaw AJ (2015) Phylogenetic relationships and morphological evolution in a major clade of leafy liverworts (phylum Marchantiophyta, order Jungermanniales): suborder Jungermanniineae. Syst Bot 40:27–45. doi:10.1600/036364415X686314
Silvestro D, Michalak I (2012) RaxmlGUI: a graphical front-end for RaxML. Org Divers Evol 12:335–337. doi:10.007/s13127-011-0056-0
Singh Deo S, Singh DK (2013) A note on the liverwort Delavayella serrata Steph. (Delavayellaceae) from Arunachal Pradesh, India. Indian J Forest 36:101–105
Singh DK, Majumdar S, Singh D (2015) Notes on scarcely collected Indian liverworts II. Xenochila integrifolia (Plagiochilaceae, Marchantiophyta). Indian J Forest 38:147–150
Söderström L, Hagborg A, von Konrat M (2015) Notes on Early Land Plants Today. 69. Circumscription of Plagiochilaceae (Marchantiophyta) with a preliminary infrageneric subdivision of Plagiochila. Phytotaxa 208:75–91. doi:10.11646/phytotaxa.208.1.8
Söderström L, Hagborg A, von Konrat M, Bartholomew-Began S, Bell D, Briscoe L, Brown E, Cargill DC, Cooper ED, Costa DP, Crandall-Stotler BJ, Dauphin G, Engel JJ, Feldberg K, Glenny D, Gradstein SR, He X, Heinrichs J, Hentschel J, Ilkiu-Borges AL, Katagiri T, Konstantinova NA, Larraín J, Long DG, Nebel M, Pócs T, Puche F, Reiner-Drehwald ME, Renner MAM, Sass-Gyarmati A, Schäfer-Verwimp A, Segarra-Moragues JG, Stotler RE, Sukkharak P, Thiers BM, Uribe J, Váňa J, Villarreal JC, Wigginton M, Zhang L, Zhu RL (2016) World checklist of hornworts and liverworts. PhytoKeys 59:1–828. doi:10.3897/phytokeys.@.6261
Stamatakis A (2014) RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi:10.1093/bioinformatics/btu033
Sun Y, He X, Glenny D (2014) Transantarctic disjunctions in Schistochilaceae (Marchantiophyta) explained by early extinction events, post-Gondwanan radiations and palaeoclimatic changes. Molec Phylogen Evol 76:189–201. doi:10.1016/j.ympev.2014.03.018
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of 3 noncoding regions of chloroplast DNA. Pl Molec Biol 17:1105–1109
Tavaré S (1986) Some probalistic and statistical problems on the analysis of DNA sequences. Lectures Math Life Sci 17:57–86
Váňa J (1973) Studien über die Jungermannioideae (Hepaticae) 1. Allgemeine Charakteristik. Folia Geobot Phytotax 8:181–208
Váňa J, Söderström L, Hagborg A, von Konrat M (2012) Notes on Early Land Plants Today. 8. New combinations and some lectotypifications in Mesoptychia. Phytotaxa 65:52–56
Wilson R, Gradstein SR, Heinrichs J, Groth H, Ilkiu-Borges AL, Hartmann FA (2004) Phylogeny of Lejeuneaceae: a cladistic analysis of chloroplast gene rbcL sequences and morphology with preliminary comments on the mitochondrial nad4-2 spacer region. Monogr Syst Bot Missouri Bot Gard 98:189–202
Yang BY, Hsu FM (1967) Studies on spore germination and gemmae development of Riccardia multifida (L.) S. F. Gray, Dumortiera hirsuta (Sw.) Reinw. Bl. et Nees, Xenochila integrifolia (Mitt.) Inoue and Marchantia polymorpha L. Taiwania 13:169–178
Yatsentyuk SP, Konstantinova NA, Ignatov MS, Hyvönen J, Troitsky AV (2004) On phylogeny of Lophoziaceae and related families (Hepaticae, Jungermanniales) based on trnL-F intron-spacer seqeunces of chloroplast data. Monogr Syst Bot Missouri Bot Gard 98:150–165
Acknowledgments
This study was supported by an LMU student research grant to SDFP, and an ABRS Reseach Grant (grant RFL213-14) to MAMR and the Royal Botanic Gardens and Domain Trust. We thank the directors and curators of GOET and M for the loan of specimens and Professor S. Renner (M) for the permission for destructive sampling.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare they have no conflict of interest.
Additional information
Handling editor: Karol Marhold.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Information on Electronic Supplementary Material
Information on Electronic Supplementary Material
Online Resource 1. Alignment of the concatenated rbcL and rps4 sequences.
Rights and permissions
About this article
Cite this article
Patzak, S.D.F., Váňa, J., Renner, M.A.M. et al. Transfer of the leafy liverwort Xenochila from Plagiochilaceae (Lophocoleineae) to Jungermanniaceae (Jungermanniineae). Plant Syst Evol 302, 891–899 (2016). https://doi.org/10.1007/s00606-016-1305-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-016-1305-7