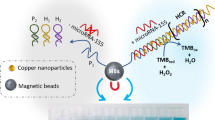
Abstract
An electrochemical strategy based on bimetallic nanozyme in collaboration with toehold-mediated DNA replacement effect is proposed for the sensitive determination of miRNA-21. The AuPt nanoparticles (AuPt NPs) are prepared as a catalytic beacon; it shows favorable peroxidase properties with a Michaelis contant (Km) of 0.072 mM for H2O2, which is capable of catalyzing H2O2 to induce an intense redox reaction, and causing a measurable electrochemical signal. To further enhance the strength of the signal response, a novel toehold-mediated DNA replacement strategy is employed. DNA strands with specific sequences are modified on electrodes and AuPt NPs, respectively. In the presence of miRNA-21, a cyclic substitution effect is subsequently activated via a specific toehold sequence and leads to a large accumulation of AuPt NPs on the electrodes. Subsequently, a strong signal depending on the amount of miRNA-21 is obtained after adding a small amount of H2O2. The analytical range of this determination method is from 0.1 pM to 1.0 nM, and the LOD is 84.1 fM. The spike recoveries for serum samples are 95.0 to 102.4% and the RSD values are 3.7 to 5.8%. The results suggests a promising application of the established method in clinical testing and disease diagnosis.
Graphical Abstract

Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
MicroRNA (miRNA) is recognized as a critical cancer marker [1, 2]. The miRNA-21 is strongly associated with colorectal cancer, breast cancer, lung cancer, and pancreatic cancer, as well as cervical cancer [3,4,5,6,7]. However, the miRNA-21 level in the human body is low. Hence, achieving a sensitive determination of miRNA-21 is important for clinical analysis and diagnosis of cancer. The usual ways to detect miRNA-21 are based on the Northern Blot and qRT-PCR [8, 9]. Despite the high sensitivity of these methods, they rely on complex equipment and require a rigorous experimental environment. This may restrict the further application of them for portable and on-site miRNA determination.
To achieve the portable determination, electrochemical techniques were wildly developed in recent years owing to the benefits like simple operation, effective response, and strong adaptability [10, 11]. However, there are two difficult requirements for the sensitive determination of miRNA-21, which includes a stabilized beacon and an enzyme-free signal amplification strategy. The general electrochemical beacons such as methylene blue, ferrocene, and thionine are easily influenced by modification status, interface condition, and solution pH, which may cause false negative or positive results. Accordingly, the nanozymes made by metal elementary substances or compounds and possess catalytic property are proposed [12]. For example, Xia et al. developed multiple Ag NPs–based electrochemical biosensors and strategies for the detection of miRNAs and other biomarkers [13, 14]. Additionally, metallic oxides like Fe3O4 NPs and CuO NPs, as well as some carbon-based nanomaterials, also possess favorable catalytic activity and strong stability, and can generate a measurable electrochemical signal with H2O2 [15,16,17,18]. Nonetheless, the catalysis levels of single metal nanozymes are constrained. For improved performance, bimetallic nanozymes with heterostructures or homostructures have been developed. The bimetallic nanozymes possess larger specific surface area and continuous ion exchange capability, resulting in a shorter interaction time and stronger electron transfer level [19,20,21], which provides a solid supporting for high-efficient catalysis.
As for signal amplification strategies, traditional methods based on the polymerase chain amplification technique were limited by enzymatic activity and the ion effect [22]. With the development of DNA self-assembling, multiple enzyme-free amplification ways were proposed like DNA train, DNA branch, and DNA polymer [23, 24]. However, the DNA strand cannot infinite assembling owing to the steric hindrance effect [25], and the unspecific DNA adsorption commonly causes an unreliable result. In comparison of this direct assembling, a new amplification way which depends on toehold-mediated DNA chain replacement was proposed. Toehold is a short oligonucleotides; it can accurately recognize targets, and orderly release the complementary sequence to perform a cyclical DNA assembling for signal amplification. Currently, the toehold technique provides a solid support for trace biomarker sensitive determination [26,27,28,29].
Herein, in consideration of catalytic ability, biocompatibility, and metallic property, Au and Pt are chosen to prepare AuPt nanoparticles (AuPt NPs) as catalytic beacon. A sensitive determination of miRNA-21 in serum has been established in synergy with a toehold-mediated DNA replacement strategy. Once the miRNA-21 is recognized by the toehold sequence, a continuous cyclic chain replacement can be performed, resulting in the accumulation of the AuPt NPs onto electrode surface. In the presence of H2O2, a measurable electrochemical signal is obtained via the catalytic interaction. Different from other electrochemical methods based on nanozymes, the proposed method utilizes a bimetallic nanozyme that interacts with the cyclic chain substitution effect to achieve selective and sensitive measurements.
Experimental section
Reagents and apparatus
L-ascorbic acid (AA), chloroauric acid (HAuCl4), chloroplatinic acid (H2PtCl6), and oligonucleotides (oligos) were obtained from Sangon Biotech Co. Ltd. (Shanghai, China); the oligos sequence listed in Table S1. Trimethylol aminomethane (Tirs), Tris (2-carboxyethyl) phosphine hydrochloride (TCEP), L-proline, 6-mercaptohexanol (MCH), 3,3′,5,5′-tetramethylbenzidine (TMB), 2,2′-azino-bis-(3-ethylbenzthiazoline- 6-sulfonic acid) (ABTS), and o-phenylenediamine (OPD) were purchased from Aladdin (Shanghai, China). Na2HPO4, NaH2PO4, K3Fe(CN)6, K4Fe(CN)6, and H2O2 were obtained from Guangzhou Chemical Reagent Factory (Guangzhou, China). The other reagents utilized in the experiments were analytical grades. The ultrapure water was obtained through Milli-Q® Integral (Milli-Q, France).
The oligos sequence is designed with an assist of ensemble website according to the complementary effect and toehold replacement requirements. The electrochemical workstation with a three-electrode system including a gold working electrode (3 mm in diameter), an Ag/AgCl reference electrode, and a platinum wire counter electrode belongs to Metrohm Autolab PGSTAT302N (Eco Chemie, Netherlands). This device is utilized to make electrochemical assays. The UV–Vis spectrum assays were performed by UV-2700 ultraviolet spectrophotometry (Shimadzu, Japan). The surface diffraction assays of the prepared nanozymes were performed by Ultima IV X-ray Powder Diffractometer (Rigaku, Japan) and nanoparticle size potential analyzer which belongs to Marvin Zetasizer Nano ZS90 (Malvern, Britain). In the assays, the 1.0 mg of AuPt NPs was dispersed into 1.0 mL H2O by ultraphonic for the XPS, the 1.0 μM H0, 1.0 mg/mL AuPt NPs, and 1.0 mg/mL of H0@AuPt NPs were prepared for Z-potential test respectively, and the 0.02 mg/mL of AuPt NPs was collected for the EDX test. The morphology and element distribution characterizations were performed by FEI Talos F200X field emission transmission electron microscope (FEI, USA) and Helios G4 CXTalos scanning electron microscope (Zeiss, Germany).
Preparation of AuPt NPs
The preparation of AuPt NPs is based on the procedure of the reported information [30]. The detailed information is shown in Supporting Information.
Enzymatic activity analysis of AuPt NPs
The enzymatic activity is assessed via the value of Km. The detailed information is shown in Supporting Information.
In addition, the level of colorimetric response was investigated by spectroscopic tests. Taking 1.5 mL of 0.2 M HAc-NaAc buffer (pH 3.5), 200 μL of 0.4 mM H2O2, 200 μL of 20 mM TMB, and 100 μL of 1 mg/mL AuPt NPs to mix and react at 37℃ for 5 min. UV–Vis absorption spectra in the wavelength range from 350 to 800 nm were recorded. Subsequently, the TMB was replaced by ABTS and OPD respectively to repeat the test.
Preparation of DNA@AuPt NPs
The 1.0 mg of AuPt NPs was dispersed into 1.0 mL of 0.1 M PBS (pH 7.4) by ultraphonic, and the 40 μL of 10 μM SH-DNA that activated by TCEP was added into 400 μL of the PBS containing AuPt NPs. The mixture was stirred at 4℃ for 30 min, and then centrifuged at 10,000 rpm for 5 min. The 20 mM Tris–HCl buffer (pH 7.4) contained 100 mM NaCl and 5 mM MgCl2 was utilized to wash the product two times. The final product was stored in 400 μL of 20 mM Tris–HCl buffer (pH 7.4) for further use.
Preparation of DNA sensor
Gold electrode (GE) was firstly immersed in piranha solution (30% H2O2:98% H2SO4 = 3:7/v:v) for 30 min, and rinsed by ultrapure water. It was then polished to a mirror-like surface by Al2O3 powder with 1.0, 0.3, and 0.05 µm diameter, respectively. Subsequently, an activation test was performed through a 10 cycle of scanning in 50 mM H2SO4 solution (− 0.2 ~ 1.6 V).
Taking 1 mL of 20 mM Tris–HCl buffer (pH 7.4) contained 100 mM NaCl and 5 mM MgCl2, 1 μM SH-DNA (H1), 1.5 μM complementary strand (H2), and 1.5 μM switch strand (H3) to make DNA hybridization at 37℃ for 3 h. The hybridized double-strand DNA (dsDNA) was then activated by TCEP. Subsequently, the 30 μL of the activated dsDNA was dropped onto the polished GE surface, and stored at 4℃ overnight. After washing by Tris–HCl buffer (pH 7.4), a 40 μL of 1.0 μM MCH was dropped onto the GE surface, and incubated at 4℃ for 30 min. After washing by the Tris–HCl buffer, the DNA sensor (GE/MCH/dsDNA) was formed.
Determination of miRNA-21
The 30 μL of 20 mM Tris–HCl buffer (pH 7.4) contained 100 mM NaCl and 5 mM MgCl2, as well as different concentrations of DNA@AuPt NPs and miRNA-21 was taken to the surface of the prepared DNA sensor to incubate at 37℃ for 3.5 h. After washing by 0.1 M PBS buffer (pH 7.4), the sensor is then immersed in 10 mL of the PBS buffer to perform “i-t” assays with the addition of H2O2. The initial potential is recorded at − 0.4 V, and after addition of 10 μL of 5 M H2O2, a gentle stirring step with 5 s is performed. Before the electrochemical measurements, the stirring is stopped.
Practical assays
The serum sample obtained from the Second People’s Hospital of Guangdong was filtered by a membrane with 0.2 μm diameter. Subsequently, they were extracted by a miRNA kit purchased from TianGen biochemical technology limited company. After the extraction, the miRNA contained in the serum sample was collected and transferred into the test tank with PBS buffer (pH 7.4). The electrochemical assays were then continued as before. In the spiking assays, different concentrations of miRNA-21 were respectively spiked in the serum sample. Similarly, they were filtered by a membrane with 0.2 μm diameter to perform the tests as before.
Results and discussion
Principle
The principle of the DNA sensor for the determination of miRNA is revealed in Scheme 1. Firstly, a SH-DNA (H1) is hybridized with a complementary strand (H2) and a switch strand (H3). Meanwhile, another SH-DNA (H0) is modified onto the AuPt NPs to form a beacon (H0@AuPt NPs). The H1 has performed a probe role which contained a toehold sequence, the H2 provides a supporting effect, and the H3 can avoid the combining of H0@AuPt NPs to the H1. Nonetheless, in the presence of miRNA-21, the H3 is released via toehold-mediated DNA chain replacement effect. The generated sequence change further led to a secondary replacement between the H0@AuPt NPs and the miRNA-21, and the released miRNA-21 is constrained to replace another H3. This results in a continuous cyclic replacement, which progressively accumulates plenty of H0@AuPt NPs on the electrode surface. While transferring the electrode into PBS buffer with H2O2, a redox was then catalyzed, which caused a measurable electrochemical response.
Characterization of AuPt NPs
To demonstrate the successful synthesis of AuPt NPs, TEM, SEM, and EDS are employed to investigate the morphology and constitution of the product. As the TEM graph displayed (Fig. 1A and B), the product presents a spherical structure with a diameter of ~ 50 nm. The following SEM and EDS graphs (Fig. 1C ~ G) indicated that the product contained Au and Pt elements with a uniformed distribution, and the atomic ratio of Au and Pt was 59:41. It is worth noting that Au and Pt coexist during the preparation procedure. The PtCl62− and AuCl4− precursors reduce to metallic Pt and Au atoms with the addition of AA, and fuse together to form AuPt nuclei. Afterwards, the AuPt nuclei gradually extended to larger nanoparticles by L-proline [30].
To further study the elemental composition and valence state of the AuPt NPs, a XPS assay is performed. In the full spectrum (Figure S1A), it contained Au and Pt, as well as C, N, and O elements which are from the sample substrate and air. In the high-resolution Au 4f XPS spectrum (Figure S1B), it shows two specific peaks at 86.68 eV and 83.04 eV; they belong to Au 4f 5/2 and Au 4f 7/2 in Au0 state, suggesting the Au precursor is reduced [31]. In the high-resolution Pt 4f XPS spectrum (Figure S1C), two pairs of peaks of Pt 4f 5/2 and Pt 4f 7/2 are observed. The peaks at 73.48 eV and 70.18 eV correspond to Pt0 state, indicating that Pt4+ is reduced. Another peak pair at 75.18 eV and 71.48 eV are derived from Pt2+, which are obviously lower than those of the Pt0, indicating the Pt0 is the dominant [32]. Besides, in comparison of the standard Pt peak, the peaks of the Pt 4f appeared displacement. This is because the electron interaction between Au and Pt can change the d-band structure of Pt, which is beneficial to performance and stability of the material [33, 34]. Therefore, the AuPt NPs are proved to be synthesized. Additionally, to verify the connection of AuPt NPs and DNA (H0), a Zeta electrical potential assay is made. Figure S1D shows that the Zeta potentials of the H0 and AuPt NPs are − 4.9 and − 27.8 mV. After modification, the Zeta potential is changed to − 32.8 mV, indicating the negatively charged DNA is covered on AuPt NPs to form the H0@AuPt NPs.
DNA sensor construction and performance test
Electrochemical impedance is utilized to illustrate the construction procedure of the DNA sensor. The sensors with different modification levels are respectively incubated into [Fe(CN)6]3−/4− solution to observe the signal. As Fig. 2A shows, little impedance signal is observed for bare electrode (line a), and the signal is orderly increased after the modifications of H1 ~ H3 (line b) and MCH (line c). It is worth noticing that the assembling of the electroactive material H0@AuPt NPs to H1 led to the increase of the impedance signal (line d). This is because the hybridized DNA strands between the AuPt NPs and electrode are not complete complementary; the mismatch bases led to a limited electron transfer. This phenomenon is proved by some reported literature [35, 36], whereas this cannot influence the signal response produced by the redox in the buffer. The sensor assembling procedure is additionally confirmed by the cyclic voltammetry assays in [Fe(CN)6]3−/4− solution (Figure S2). The CV signals (Fe(III)/Fe(II)) exhibited a reversed trend to that of the impedance, suggesting the modification is expected. Meanwhile, the SEM graphs (Figure S3) for the electrodes before and after the modifications further evidenced the construction of the DNA sensor.
(A) Nyquist plots of EIS to the [Fe(CN)6]3-/4- solution containing 0.1 M KCl (a) bare Au electrode, (b) Au electrode/H1/H2/H3, (c) Au electrode/H1/H2/H3/MCH, and (d) Au electrode/H1/H2/H3/MCH/H0@AuPt. (B) Lineweaver–Burk plot when the TMB concentration is fixed as 2.0 mM. (C) The ratio of current responses of the DNA sensor with toehold sequence to different concentrations of miRNA-21 (pM) (line a for 0, line b for 1.0, line c for 100). (D) The ratio of current responses of the DNA sensor with toehold sequence (line a) to 0 pM miRNA-21, as well as that without (line b) and with (line c) toehold sequence to 100 pM miRNA-21
The following assays focus on the signal response of the sensor. First, an i-t test is used to assess the sensor response in the absence of the target miRNA. As Figure S4A shows, a bare electrode (line a) displays little current response. However, after the assembling of the H0@AuPt NPs to the DNA sensor via toehold DNA replacement effect, it can result an obvious current signal (line b) with the addition of H2O2. While if the H2O2 is absent in the test solution, the current signal is little (line c) like that produced by the bare electrode. This indicates that the AuPt NPs has a satisfactory catalytic property for the electrochemical determination. Moreover, the colorimetric assays further indicate that AuPt NPs are capable of catalyzing H2O2 to generate ·OH, and results in the oxidation of TMB, ABTS, and OPD to appear color conversions (Figure S4B). In this case, the enzymatic activity is then investigated via the kinetic process and the corresponding Km. The assays are performed at 25℃, and the pH of the test buffer was controlled at 3.5. According to the obtained kinetic curve and Lineweaver–Burk plot (Fig. 2B), the Km value is calculated as 0.072 mM for H2O2, which is better than that of the horseradish peroxidase which is 3.7 mM [37, 38].
Encouraged by the above results, an assay for miRNA is performed. Figure 2C illustrates the i-t response of the DNA sensor containing toehold sequence to miRNA-21 at concentrations of 0, 1.0, and 100 pM. To perform the comparison more definitely, the responses are displayed by current ratio, which the level of the max current response is normalized as “1,” and the other current responses are shown according to the intensity ratio. The current responses to different concentrations of miRNA-21 exhibit measurable distinctions, suggesting a favorable determination feasibility of the DNA sensor for miRNA-21. In Fig. 2D, it indicates the responses of the DNA sensor with toehold sequence to 0 (line a) and 100 (line c) pM miRNA-21, as well as that without toehold sequence to 100 (line b) pM miRNA-21. The responses are displayed by current ratio like in Fig. 2C. The level of line b is within the fluctuation range of line a, suggesting the DNA sensor without toehold sequence is unable to recognize miRNA-21, and incapable to lead the assembling of H0@AuPt NPs. However, the response distinctions of the DNA sensor with toehold sequence to 0 and 100 pM miRNA-21 are obvious. This proves that the toehold sequence provides solid support for target recognition and signal amplification.
Determination of miRNA
To effectively measure the level of target miRNA, a condition optimization assay was initially performed. Firstly, the amount of the H0 modifying on AuPt NPs is investigated. As Fig. 3A shows, once the modified concentration arrived at 1 µM, the signal response is the largest. When the H0 amount continuously increased, the signal is declined owing to the steric-hinerance effect preventing further chain replacements. Similarly, the amount of the H1 modifying on the electrode is then assessed. As Fig. 3B shows, the 1 µM is the optimal usage. Subsequently, the incubation time at 37℃ is investigated. As Fig. 3C shows, when the time is controlled at 3.5 h, the signal is the most obvious, while the longer incubation can cause the deposition of H0@AuPt NPs, producing more background signal, and decreasing the specific signal distinction.
Under the optimal conditions, the electrochemical miRNA determinations are performed. As Figure S5 depicts, the relative current conversions ((I-I0)/I0) are gradually increase with the miRNA amount raising from 0.1 pM to 5 nM, and a linearity is plotted by the ((I-I0)/I0) versus the logarithmic concentrations (lgC) of 0.1 ~ 1000 pM (Fig. 4A). The fitted linear equation is (I-I0)/I0 = 0.2167lgC + 0.5147, the LOD is calculated as 84.1 fM (3σ/S), and the analytical range is confirmed from 0.1 pM to 1.0 nM. In comparison of other miRNA determination methods, this strategy has preferable benefits on the analytical range and LOD (Table S2). Subsequently, the selectivity of the strategy is evaluated. Four isogenous miRNAs of miRNA-141, miRNA-101, miRNA-122, miRNA-let7a are chosen as the interference; the concentrations of them are 100 pM, which is 100-fold to that of miRNA-21. As Fig. 4B reveals, solely miRNA-21 causes an obvious signal response, and the mixture containing the miRNA-21 and other four isogenous miRNA present a similar signal, which exhibits a satisfactory selectivity of this strategy. In addition, five prepared DNA sensors are respectively used to detect 1.0 pM miRNA-21, and the assays are repeated 3 times (n = 3). According to the results, the rates of the signal conversion of the electrodes are similar, and the RSD value from each electrode is under 3.0% (Fig. 4C). Moreover, the DNA sensors were then stored at 4℃ for 1, 2, 3, 4, and 5 days, and employed to perform the miRNA-21 assays. The results demonstrated the signals remaining at a similar level (Fig. 4D). Consequently, the favorable reproducibility and stability of the proposed strategy were verified.
(A) The working curve of i-t responses to target miRNA-21. (B) Investigation of the specificity of the sensor, the relative current change ratios responding to blank, miRNA-141, miRNA-101, miRNA-122, miRNA-let7a, miRNA-21, and their mixture. (C) Investigation of the repeatability of the sensor. (D) Investigation of the stability of the biosensor
To assess the practicability of the proposed eletcrochemical strategy, it is utilized to make the miRNA-21 determination in serum. The serum sample is filtered by a membrane (0.2 μm diameter) to dislodge impurities, and extracted by a commercial miRNA kit. The pre-treated serum sample is mixed with 20 mM Tris–HCl buffer (pH 7.4) to make measurement. Furthermore, different concentrations of miRNA-21 are respectively spiked into the original serum sample; after filtration and extraction, the product is then perform the measurements. As Table 1 displays, the miRNA-21 is not detected in the serum sample, and the spiked recoveries of the 1, 10, and 100 pM miRNA-21 are 95.0%, 101.5%, and 102.4% with the RSD values ranged from 3.7 to 5.8%. This indicated that the proposed electrochemical determination strategy owns a desirable application prospect.
Conclusion
This work proposed a sensitive electrochemical determination strategy for the miRNA-21 in serum. A bimetallic AuPt nanozyme possessing favorable peroxidase property is prepared as beacon, and a continuous toehold-mediated DNA replacement procedure is employed to make sensitive determination of miRNA-21. After addition of few H2O2, a measurable signal to miRNA-21 amount is obtained, the sensitive arrived at femtomole. By contrast to common miRNA-21 determination methods, this strategy has multiple benefits: achieving enzyme-free signal amplification and dispensing with high-cost instrument, as well as facilitating field detection. These indicate a promising application prospect in clinical testing and disease prediction.
References
Peng XX, Guo T, Lu H et al (2020) Nanostructuring synergetic base-stacking effect: an enhanced versatile sandwich sensor enables ultrasensitive detection of microRNAs in blood. ACS Sens 5:2514–2522. https://doi.org/10.1021/acssensors.0c00772
Huang X, Zhu X, Yu Y et al (2021) Dissecting miRNA signature in colorectal cancer progression and metastasis. Cancer Lett 501:66–82. https://doi.org/10.1016/j.canlet.2020.12.025
Asangani IA, Rasheed SAK, Nikolova DA et al (2008) MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 27:2128–2136. https://doi.org/10.1038/sj.onc.1210856
Frankel LB, Christoffersen NR, Jacobsen A et al (2008) Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem 283:1026–1033. https://doi.org/10.1074/jbc.m707224200
Wang ZX, Bian HB, Wang JR et al (2011) Prognostic significance of serum miRNA-21 expression in human non-small cell lung cancer. J Surg Oncol 104:847–851. https://doi.org/10.1002/jso.22008
Qu K, Zhang X, Lin T et al (2017) Circulating miRNA-21-5p as a diagnostic biomarker for pancreatic cancer: evidence from comprehensive miRNA expression profiling analysis and clinical validation. Sci Rep 7:1692. https://doi.org/10.1038/s41598-017-01904-z
Hasanzadeh M, Movahedi M, Rejali M et al (2019) The potential prognostic and therapeutic application of tissue and circulating microRNAs in cervical cancer. J Cell Physiol 234:1289–1294. https://doi.org/10.1002/jcp.27160
Huang Q, Mao Z, Li S et al (2014) A non-radioactive method for small RNA detection by northern blotting. Rice 7:26. https://doi.org/10.1186/s12284-014-0026-1
Tian H, Sun Y, Liu C et al (2016) Precise quantitation of microRNA in a single cell with droplet digital PCR based on ligation reaction. Anal Chem 88:11384–11389. https://doi.org/10.1021/acs.analchem.6b01225
Zhang JL, Yang HL, Liu WJ et al (2022) Rapid 16S rDNA electrochemical sensor for detection of bacteria based on the integration of target-triggered hairpin self-assembly and tripedal DNA walker amplification. Anal. Chim. Acta 1190:339266. https://doi.org/10.1016/j.aca.2021.339266
Hira SA, Nallal M, Rajendran K et al (2020) Ultrasensitive detection of hydrogen peroxide and dopamine using copolymer-grafted metal-organic framework based electrochemical sensor. Anal Chim Acta 1118:26–35. https://doi.org/10.1016/j.aca.2020.04.043
Robert A, Meunier B (2022) How to define a nanozyme. ACS Nano 16:6956–6959. https://doi.org/10.1021/acsnano.2c02966
Xia N, Wang X, Zhou BB et al (2016) Electrochemical detection of amyloid-β oligomers based on the signal amplification of a network of silver nanoparticles. ACS Appl Mater Interfaces 8:19303–19311. https://doi.org/10.1021/acsami.6b05423
Liu LL, Chang Y, Xia N et al (2017) Simple, sensitive and label-free electrochemical detection of microRNAs based on the in situ formation of silver nanoparticles aggregates for signal amplification. Biosens Bioelectron 94:235–242. https://doi.org/10.1016/j.bios.2017.02.041
Shi K, Dou B, Yang C et al (2015) DNA-fueled molecular machine enables enzyme-free target recycling amplification for electronic detection of microRNA from cancer cells with highly minimized background noise. Anal Chem 87:8578–8583. https://doi.org/10.1021/acs.analchem.5b02418
Wei H, Wang E (2013) Nanomaterials with enzyme-like characteristics (nanozymes): next- generation artificial enzymes. Chem Soc Rev 42:6060–6093. https://doi.org/10.1039/C3CS35486E
Wang HY, Xie AJ, Li SJ et al (2022) Three-dimensional g-C3N4/MWNTs/GO hybrid electrode as electro-chemical sensor for simultaneous determination of ascorbic acid, dopamine and uric acid. Anal Chim Acta 1211:339907. https://doi.org/10.1016/j.aca.2022.339907
Zhao RN, Feng Z, Zhao YN et al (2019) A sensitive electrochemical aptasensor for Mucin 1 detection based on catalytic hairpin assembly coupled with PtPd NPs peroxidase-like activity. Talanta 200:503–510. https://doi.org/10.1016/j.talanta.2019.03.012
Chen T, Xu J, Yang P et al (2019) Facile controlled synthesis of AuPd and AuPt bimetallic nanocrystals for enhanced electrocatalytic sensing. Sens Actuators B Chem 298:126724. https://doi.org/10.1016/j.snb.2019.126724
Liu J, Cao L, Huang W et al (2011) Preparation of AuPt alloy foam films and their superior electrocatalytic activity for the oxidation of formic acid. ACS Appl Mater Interfaces 3:3552–3558. https://doi.org/10.1021/am200782x
Liu Y, Xu Q, Zhang Y et al (2021) An electrochemical aptasensor based on AuPt alloy nanoparticles for ultrasensitive detection of amyloid-β oligomers. Talanta 231:122360. https://doi.org/10.1016/j.talanta.2021.122360
Özay B, McCalla ES (2021) A review of reaction enhancement strategies for isothermal nucleic acid amplification reactions 3:100033. https://doi.org/10.1016/j.snr.2021
Guo Q, Yu Y, Zhang H et al (2020) Electrochemical sensing of exosomal microRNA based on hybridization chain reaction signal amplification with reduced false-positive signals. Anal Chem 92:5302–5310. https://doi.org/10.1021/acs.analchem.9b05849
Wang F, Lu CH, Willner I (2014) From cascaded catalytic nucleic acids to enzyme-DNA nanostructures: controlling reactivity, sensing, logic operations, and assembly of complex structures. Chem Rev 114:2881–2941. https://doi.org/10.1021/cr400354z
Wang L, Zhang L, Yu Y et al (2021) DNA cyclic assembling control in an electrochemical strategy with MoS2@AuNPs for determination of kanamycin. Microchim Acta 188:264. https://doi.org/10.1007/s00604-021-04916-9
Wang S, Emery JN, Liu PA (2019) A novel synthetic toehold switch for microRNA detection in mammalian cells. ACS Synth Biol 8:1079–1088. https://doi.org/10.1021/acssynbio.8b00530
Miao P, Tang YG (2021) Cascade toehold-mediated strand displacement reaction for ultrasensitive detection of exosomal microRNA. CCS Chem 3:2331–2339. https://doi.org/10.31635/ccschem.020.202000458
Xu SH, Wang YL, Yao YY et al (2022) Toehold-mediated strand displacement coupled with single nanoparticle dark-field microscopy imaging for ultrasensitive biosensing. Nanoscale 14:3496–3503. https://doi.org/10.1039/d1nr08030j
Liang KL, Wang H, Li P et al (2020) Detection of microRNAs using toehold-initiated rolling circle amplification and fluorescence resonance energy transfer. Talanta 207:120285. https://doi.org/10.1016/j.talanta.2019.120285
Weng X, Liu Y, Xue Y et al (2017) L-Proline bio-inspired synthesis of AuPt nanocalliandras as sensing platform for label-free electrochemical immunoassay of carbohydrate antigen. Sens Actuators B Chem 250:61–68. https://doi.org/10.1016/j.snb.2017.04.156
Feng J, He LL, Fang R et al (2016) Bimetallic PtAu superlattice arrays: highly electroactive and durable catalyst for oxygen reduction and methanol oxidation reactions. J Power Sources 330:140–148. https://doi.org/10.1016/j.jpowsour.2016.08.094
Ren Y, Li C, Li B et al (2021) PtPd nanoframes derived from Pd@ PdPt core-shell rhombic dodecahedrals with excellent catalytic performance toward methanol oxidation. Inorg Chem Front 8:2280–2287. https://doi.org/10.1039/D1QI00081K
Khan IA, Sofian M, Badshah A et al (2020) Stable and efficient PtRu electrocatalysts supported on Zn-BTC MOF derived microporous carbon for formic acid fuel cells application. Front Chem 8:367. https://doi.org/10.3389/fchem.2020.00367
Fu X, Wan C, Zhang A et al (2020) Pt3Ag alloy wavy nanowires as highly effective electrocatalysts for ethanol oxidation reaction. Nano Res 13:1472–1478. https://doi.org/10.1007/s12274-020-2754-4
Nivedita B, Thu HH, Guillon FX et al (2019) Coupling electrochemical adsorption and long-range electron transfer: label-free DNA mismatch detection with ultramicroelectrode (UME). Electroanalysis 31:2232–2237. https://doi.org/10.1002/elan.201900357
Ferapontova EE (2019) Electron transfer in DNA at electrified interfaces. Chem Asian J 14:3773–3781. https://doi.org/10.1002/asia.201901024
Tan Z, Cao L, He X et al (2020) A label-free immunosensor for the sensitive detection of hepatitis B e antigen based on PdCu tripod functionalized porous graphene nanoenzymes. Bioelectrochem 133:107461. https://doi.org/10.1016/j.bioelechem.2020.107461
Xian Z, Zhang L, Yu Y et al (2021) Nanozyme based on CoFe2O4 modified with MoS2 for colorimetric determination of cysteine and glutathione. Microchim Acta 188:65. https://doi.org/10.1007/s00604-021-04702-7
Funding
This work is supported by the National Natural Science Foundation of China (Nos. 21575043, 52070080, 22004039, and 22206069), the Guangdong Basic and Applied Basic Research Foundation (No. 2020A1515110256), Science and Technology Projects Foundation (Basic and Applied Basic Research) in Guangzhou (Nos. 202102020043 and 202102080612), and Foundation of Department of Education of Guangdong Province (No. 2020KTSCX033).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Su, Z., Zhang, L., Yu, Y. et al. An electrochemical determination strategy for miRNA based on bimetallic nanozyme and toehold-mediated DNA replacement procedure. Microchim Acta 190, 149 (2023). https://doi.org/10.1007/s00604-023-05720-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-023-05720-3