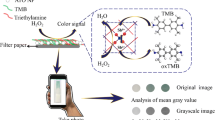
Abstract
A new nano-structured platform for fluorescent analysis using PQQ-dependent glucose dehydrogenase (PQQ-GDH) was developed, particularly using a smartphone for transduction and quantification of optical signals. The PQQ-GDH enzyme was immobilized on SiO2 nanoparticles deposited on glass microfiber filter paper, providing a high load of the biocatalytic enzyme. The platform was tested and optimized for glucose determination using a wild type of the PQQ-GDH enzyme. The analysis was based on the fluorescence generated by the reduced form of phenazine methosulfate produced stoichiometrically to the glucose concentration. The fluorescent signals were generated at separate analytical spots on the paper support under wavelength (365 nm) UV excitation. The images of the analytical spots, dependent on the glucose concentration, were obtained using a photo camera of a standard smartphone. Then, the images were processed and quantified using software installed in a smartphone. The developed biocatalytic platform is the first step to assembling a large variety of biosensors using the same platform functionalized with artificial allosteric chimeric PQQ-dependent glucose dehydrogenase activated with different analytes. The future combination of the artificial enzymes, the presently developed analytical platform, and signal processing with a smartphone will lead to novel point-of-care and end-user biosensors applicable to virtually all possible analytes.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Two major approaches are traditionally used for analysis of biochemicals: (i) bioanalytical assays (frequently used for the analysis of biocatalytic enzyme species) [1] and (ii) biosensors (broadly defined as instruments for biochemical analysis) [2]. The difference between the assays and biosensors is mostly technical. In bioanalytical assays, most of analytical steps are performed in solutions with processing steps requiring highly trained personnel, and then the solutions are analyzed using different instruments (optical, electrochemical/electronic, mass spectroscopy, etc.). In biosensors, the biochemical recognition and analysis are performed at a chemically modified interface. More specifically, in biosensors, there is integration of the bioanalytical biochemical material with a physical transduction interface. This integration allows miniaturization of the device and cancels the need of sophisticated instrumentation used in the case of assays. The interface modified with bioanalytical substances and used for the detection of analyte species can operate differently depending on the applied transduction modes, which might be optical (measuring changes in optical absorbance, fluorescence, surface plasmon resonance, etc.), electrochemical (measuring currents or potentials in amperometric or potentiometric sensors), and electronic (using, for example, field effect transistors). All these devices can operate continuously, automatically in fields without expensive and sophisticated instrumentation and with minimally trained personnel or even completely without any personnel (automatically with a wirelessly transmitted outcome). Sometimes, classification of the bioanalytical tools as the assays or biosensors can be not very clear. For example, immunosensing performed as an ELISA technique proceeds at a chemically modified interface [3], but the signal transduction requires an external device, and some processing steps performed by technical staff or automatically operating machines are still needed. Therefore, this analytical procedure is more related to an assay rather than a biosensor. Regardless of the exact mode of realization, there is a modern tendency to simplify the analytical procedures and signal transduction instrumentation and then moving from sophisticated machinery operated by a highly trained staff to point-of-care and end-user sites when the analytical procedure does not require any technical experience. This is applicable to biomedical analysis, environmental monitoring, and military and homeland security applications. One of the currently actively researched solutions is based on the use of commonly available electronics for transduction of biochemically generated signals. Such electronics may not be specially designed for bioanalytical applications, but it could be adapted for this use. The best example of it are smartphones used as electronic transducers in bioanalytical assays/biosensors. The smartphones with minimum adaptation might be integrated with various biosensors [4, 5, 6, 7, 8, 9, 10] (see additional references in the Supporting Information) including glucose biosensors [11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22]) and with almost zero changes they can be used for processing optical signals (color changes or light emission) when a digital photo camera is only needed. A very simple app installed in the smartphone might be needed for processing the obtained bioanalytical images. The main challenges for smartphone-adapted optical biosensors are the development of new technologies for creation of sensor plates, usage of new efficient and specific probes for the signal processing with smartphone camera, and adaptation of the available software for the analytical result calculation.
The present communication demonstrates a very simple and robust analytical procedure and instrumentation where biocatalytically generated optical signals are transduced by a smartphone to quantitatively read outputs. Our final goal is the development of this procedure as a standard tool for analysis of almost infinite library of analytes using artificial chimeric enzymes based on PQQ-dependent glucose dehydrogenase activated by various biomolecule signals [23, 24, 25, 26]. However, the present communication aims at a much more modest goal where the analyte is only glucose, and the enzyme is represented with a wide type of the PQQ-dependent glucose dehydrogenase [27]. This study can be considered as a preliminary step, a proof of the concept, prior to the use of the artificial chimeric enzymes. This study does not pretend to compete with well-developed glucose-sensing technology but provides experimental background for the next step when different analytes will be processed with the artificial enzymes but using the same interface as one developed with the wide type of the enzyme reported here.
Experimental
Chemicals and materials
PQQ-dependent glucose dehydrogenase (E.C. 1.1.1.118; PQQ-GDH) from nonspecified microorganisms was obtained from Toyobo Inc. Human serum from human male AB plasma, pyrroloquinoline quinone (PQQ), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES buffer), (3-aminopropyl)triethoxysilane (APTES), D-( +)-glucose, phenazine methosulfate (PMS), dichlorophenolindophenol (DCPIP), glutaric dialdehyde, and other standard organic and inorganic reagents were purchased from Acros Organics and Millipore Sigma (formerly Sigma-Aldrich). SiO2 nanoparticles (SiO2-NPs) (ca. 200 nm diameter) were purchased from Fiber Optics Center Inc. All commercial organic/inorganic chemicals, solvents, and materials were used without further purification. All experiments were carried out in ultrapure water (18.2 MΩ∙cm; Barnstead NANOpure Diamond) at room temperature (22 ± 2 °C).
Glass microfiber filter paper 0.5 micron, 90 mm, was obtained from Maine Manufacturing LLC, glass micro slides were purchased from VWR, and double-sided mounting tape produced by Scotch was purchased from Walmart. Glucose Assay Kit (Fluorometric) for 500 assays was purchased from Cell Biolabs, Inc. (San Diego, USA).
Instrumentation
A Shimadzu UV‐2450 UV–Vis spectrophotometer with 2.5-mL poly(methyl methacrylate) (PMMA) cuvettes was used for optical density measurements. Fluorescence measurements were performed using a fluorescent spectrophotometer (Varian, Cary Eclipse). A Mettler Toledo S20 SevenEasy pH meter was used to measure pH of solutions. Fluorescent images were obtained with Leica TCS SP5 II Tandem Scanning Confocal and Multiphoton Microscope. Field emission scanning electron microscopy (SEM) was used to characterize SiO2-NP-modified microfiber glass paper. The SEM imaging was conducted using FESEM JEOL-7400 electron microscope (JEOL USA, Inc.). Revlon Essentials Lightweight Ionic Hair Dryer (model RV474) was used for drying the plate with cool air. UVP Inc. UVSL-25 Mineralight Lamp with wavelength (365 nm) was used for fluorescence visualization. The Hioki 3664 optical power meter was used for measuring the wavelength and optical power of UVSL-25 Mineralight Lamp (Fig. S1 in the Supporting Information (SI)). A Samsung Galaxy S20 + (2020) smartphone was used for taking pictures of the sensors’ spots; settings were fixed as ISO 400 and speed 1/15 s.
Preparation of sensor’s plate
A sensor’s interface was constructed using a glass microscope slide as a support. A VWR glass microscope slide (75 mm × 25 mm × 1.2 mm) was cleaned with ethanol and distilled water prior to its modification. A piece of fiberglass (Maine Manufacturing LLC) was placed on a double-sided tape with a smoother side facing up, and then stuck together. A hole-punch tool (office puncher) was used to create fiberglass/tape circles with a diameter of 0.25 inches (6.35 mm). The circles (spots) were then put onto the glass microscope slide at even intervals to allow for using a multichannel pipette (Fig. S2 in the SI).
Functionalization of SiO2-NPs and immobilization of PQQ-GDH
SiO2-NPs (100 mg) were placed in a plastic tube (Eppendorf) containing 1 mL water with pH adjusted to ca. 9–10, and the suspension was sonicated for 60 min and then centrifuged at 1,000 rpm for 10 min at room temperature. Then, the water was replaced with ethanol and centrifuged 3 more times, changing ethanol each time. APTES (97%, 30 µL) was added to 970 µL of ethanol, and the resulting solution was added to the SiO2-NP suspension in ethanol. The mixture was vortexed and left in a shaker (mild shaking) for 3 h. The reacting mixture was centrifuged for 10 min at 10,000 rpm (20 °C), and the liquid supernatant was changed to a 25 mM HEPES buffer, pH 7.4, 1 mL (repeated 3 times changing the buffer each time). The suspension of the silanized SiO2-NPs was mixed with glutaric dialdehyde (0.04% v/v in a 25 mM HEPES buffer, pH 7.4) and slowly shaken with an orbital shaker for 1 h.
PQQ-GDH (20 μL solution, 0.4 mg·mL−1 with activity of 320 U·mL−1) in 25 mM HEPES buffer, pH 7.4 and 0.5 µL of 5 mM PQQ were added to the SiO2-NP (340 µL) suspension. The mixture was incubated for 1 h at room temperature with slow mixing every 10 min. Then, the solution was washed by adding 1650 μL of 25 mM HEPES buffer, pH 7.4, shaking for 1 min (mild mixing), and centrifuged for 3 min at 1000 rpm, 20 °C. A sediment (formed with PQQ-GDH/SiO2-NPs) was dispersed with 340 µL of a 25 mM HEPES buffer, pH 7.4, and then a supernatant was removed, and the residual was stored at 8 °C till usage.
Modification of the sensor’s plate with PQQ-GDH/SiO2 NPs
A PQQ-GDH/SiO2-NP suspension (20 μL) was placed on each microfiber glass paper-based spot and left for drying at room temperature for 20 min using hairdryer (low speed, cold air). The sensor’s plates modified with PQQ-GDH/SiO2-NPs were stored at 8 °C until usage.
Procedure of sensor’s analysis
For visualization of the PQQ-GDH activity, 20 μL of reagent solution based on a 25 mM HEPES buffer, pH 7.4, with 1 mM PMS and glucose (in different concentrations or analyzed real samples with different dilutions), was simultaneously dropped on each circle of the sensor’s plate using multichannel pipette. The enzymatic reaction of glucose consumption with the simultaneous PMS reduction was performed for 10 min in dark at room temperature. Then the sensor’s plate was washed twice (firstly by dipping and then by mild shaking for 2 min in 25 mM HEPES buffer, pH 7.4). It was then placed into the UV Fluorescence Analysis Cabinet with a UV lamp operating at a 365-nm wavelength and power of 150 µW (Fig. S1A in the SI). A smartphone camera was then used in manual mode (ISO 400; speed 1/15) in order to take photos of the occurring fluorescence. These were changed to grayscale photo variant and processed with standard program software ImageJ to estimate fluorescence intensity of each sensing spot (Fig. S3 in the SI). The numerical data was then transferred to either Excel or OriginLab, where corresponding graphs were created. The final signal processing steps could be performed with a smartphone or with a regular computer, as convenient.
All experiments were replicated at least 4 times (using 2 sensor plates with two of the same spots roads).
Preparation and analysis of serum samples
Taking into account that the normal level of glucose in blood is about 5 mM when the linear frames of the developed sensor are 0.025–0.7 mM glucose, it is necessary to dilute the tested serum samples. The samples were diluted with a solution of 25 mM HEPES buffer, pH 7.4, 1 mM PMS (final concentrations), in 10, 16, and 50 times. For example, to prepare × 10 serum dilution, 20 μL of serum was added to 180 μL of 28 mM HEPES buffer, pH 7.4, with 1.1 mM PMS and mixed. Then, 20 μL of each diluted serum sample (× 10; × 16; × 50) was simultaneously dropped on each circle of the sensor’s plate using a multichannel pipette. For the first spot of the sensor plates, used as blank, instead of a diluted serum, samples were dropped a “blind probe” (20 μL of 25 mM HEPES buffer, pH 7.4, 1 mM PMS). The time of enzymatic reaction, procedure of assay, and signal processing was the same as described above (see the Procedure of sensor’s analysis section).
In the case of serum sample analysis using fluorimetric Glucose Assay Kit (Cell Biolabs, Inc.), the sample dilution and analysis were performed in accordance with the producer protocol. The results were obtained using fluoromiter (λex/em = 530/585 nm).
Results and discussion
The operation of the studied optical biosensor with fluorescence detection is based on an ability of pyrroloquinoline quinone-dependent glucose dehydrogenase (PQQ-GDH) to transfer electrons from glucose through a PQQ coenzyme to low-molecular weight electron acceptors. This enzyme ability is widely used for spectrophotometric analysis of the PQQ-GDH activity itself or for glucose analysis. The reaction proceeds with electron transfer from enzymatically reduced PQQ cofactor (PQQH2) to dichlorophenol indophenol (DCPIP) as a final electron acceptor with assistance of phenazine methosulfate (PMS) operating as a mediator [18]. The initial oxidized form of DCPIP is intensively blue with a maximum absorption at 600 nm, while the reduced DCPIP is colorless. However, because of low absorptivity of DCPIP, this method suffers from low sensitivity with a limit of detection (LOD) above 1 mM glucose (Fig. S4 in the SI). It is known that fluorimetry is significantly more sensitive in comparison with spectrophotometry based on the optical absorbance changes; however, it requires usage of highly fluorescent dyes. We propose a new optical analytical approach based on fluorescence detection of a highly fluorescent dye — the reduced form of PMS produced as the result of the PQQ-GDH-catalyzed reaction in presence of glucose (Fig. 1). Under UV 340 nm excitation, aqueous solutions of the cationic PMSox show blue fluorescence (λem 465 nm), whereas its reduced form (PMSred) shows intensively green fluorescence (λex/em = 365/526 nm) [28]. Moreover, due to its hydrophobic characteristics, enzymatically generated PMSred is concentrated around the immobilized enzyme, avoiding its flowing out from the analyzed spot and supporting a stable level of the substrate-dependent fluorescence intensity. Notably, the DCPIP electron acceptor used in the optical absorbance measurements (Fig. S4 in the SI) is not needed when the fluorescent signal from the PMSred is measured.
Schematics of the biocatalytic process resulting in the fluorescent output proportional to the analyte (glucose) concentration. Abbreviations: Glc, glucose; GlcA, gluconic acid; PQQ and PQQH2, oxidized and reduced forms of pyrroloquinoline quinone, respectively; PMSox and PMSred, phenazine methosulfate oxidized and reduced forms, respectively; λex and λem, wavelength maximum of the excitation and emission of PMSred fluorescence, respectively
Successful immobilization of enzymes is a critical and challenging part of biosensor development and maintenance of their structural stability [29]. Recently, we reported on construction of nonconductive interfaces for investigation of switchable biocatalytic reactions controlled by interfacial pH changes produced by orthogonal biocatalytic processes [30]. In the present study, we mostly reproduced the successful immobilization procedure used earlier but with some important variations [30]. Silica oxide nanoparticles (SiO2-NPs) were first silanized with (3-aminopropyl)triethoxysilane (APTES) providing amino groups for the next modification steps. Then, the amino groups of the silane layer were cross-linked with glutaric dialdehyde resulting in stabilization of the SiO2-NPs in the form of a multilayer. Since not all aldehyde groups were used for the SiO2-NPs cross-linking, the rest of them was reacted with amino groups of lysine residues in the PQQ-GDH enzyme backbone resulting in the enzyme immobilization around the nanoparticles. Notably, the reaction of amino groups with the aldehyde groups resulted in the formation of Schiff-base bonds which provided stable cross-linking due to multi-point binding [29]. The immobilization procedure allowed the formation of only a single layer of the enzyme around the SiO2-NPs that was important for easy diffusional access of the enzyme substrate/co-substrate (glucose and PMSox, respectively). On the other hand, to ensure maximum load of the enzyme, microporous glass microfiber paper was used as a support for the enzyme-functionalized SiO2-NPs.
The resulting glass microfiber support modified with the SiO2-NPs functionalized with PQQ-GDH was analyzed using field emission scanning electron microscopy (SEM) and confocal microscopy (Fig. 2). The SEM micrograph demonstrates the formation of a multilayer PQQ-GDH/SiO2-NP structure penetrating deep into microfiber glass paper support (Fig. 2A). To estimate how many layers are forming the biorecognition sensor film, confocal microscopy of the PQQ-GDH/SiO2-NP-modified microfiber glass paper support was performed (Fig. 2B). For this, the PQQ-GDH/SiO2-NP-modified microfiber glass support was cut in a 0.3 mm slice, with the next visualization of the enzymatic activity in its cross-section according to the reaction shown schematically in Fig. 1. It can be seen in Fig. 2B that the enzymatically reduced PMS coloring the glass paper support in typical blue color goes ca. 125 µm inside the support. Taking into account that the average diameter of the SiO2-NP corresponds to 200 nm, the estimated biorecognition layer of the sensor consisted of ca. 625 single monolayers of the PQQ-GDH/SiO2-NPs. This multilayer PQQ-GDH/SiO2-NP formation supports an increasing local fluorescence intensity of the hydrophobic PMSred, significantly improving sensitivity of the biosensor. Moreover, the deep penetration of the PQQ-GDH/SiO2-NPs into the microfiber glass paper prevents their washing out, as well as capillary forces inside of the support keep the surrounding wet, providing high stability of the biocatalytic film.
A The SEM micrograph of the glass fiber support loaded with enzyme-modified SiO2-NPs; the nanoparticles multilayer and glass fibers are visible. B The image obtained with a confocal microscope visualizing enzymatic activity using reduced PMS (PMSred) as a fluorescent dye. Conditions: 25 mM HEPES buffer, pH 7.4; 20 mM glucose; 1 mM PMS; λex = 410 nm
The steps of the biosensor response analysis are shown in Fig. 3A. Notably, the optical output signals in the form of fluorescence were detected with a photo camera of a standard smartphone with further conversion of the obtained images to the gray format and quantification using a simple app (see the technical details in the Procedure of sensor’s analysis section). The concentration of PMS was optimized experimentally as 1 mM (Fig. S5 in the SI). Using duplicate biosensing spots on the sensor’s plate with the same concentration of glucose applied allowed the estimation of analysis accuracy (statistical error) (Fig. 3A). The fluorescent signals produced upon application of different glucose concentrations were normalized versus the background fluorescence produced in the absence of glucose (Δ fluorescence intensity). Then, the increase of the fluorescence produced by the biocatalytic reaction was considered as the biosensor output represented as relative fluorescence units (RFU) (Fig. 3B and C). The sensing spot with no applied glucose, producing the background fluorescence, was used in all sensing strips to exclude variability of the relative output signals because of minor differences in the strip preparation. In other words, it was used for the internal calibration of the optical output signals on each biosensing strip.
A Photos of the biosensing system obtained at different steps of its preparation: (a) as prepared, prior to application of glucose; (b) after applying glucose at different concentrations on each sensing spot; (c) after rinsing the sensing spots with 25 mM HEPES buffer, pH 7.4; (d) after converting the previous photo to the gray format. Glc and the numbers correspond to the used glucose concentration. B The fluorescence output (the increase from the background level) as the function of the glucose concentration, derived from the images shown in A. C The relative fluorescence output (%) in the presence of glucose 0.5 mM taken as 100% compared to the output toward of several possible interferants (all applied at 0.5 mM concentration). The error bars of the figure indicate accuracy of the mean value
In order to check any possible difference between the fluorescence produced in a soluble state (in a control experiment) and the immobilized state of the biosensor system, both states were analyzed, and similar fluorescence intensity was found (see Fig. S6 in the SI).
The optical glucose biosensor demonstrated the LOD (3δ) about 25 µM, with the output linearity in the glucose concentration range from 50 to 0.75 mM (Fig. S7 in the SI). The KM apparent value (KMapp) was found as 1.2 mM glucose with maximal Δ fluorescence intensity at the substrate (glucose) saturation near 220 RFU (Fig. 3A).
The glucose biosensor demonstrated good selectivity to a number of the most common compounds appearing in human serum (Fig. 3C). It was shown that only ascorbate and citrate in 0.5 mM concentration affected on biosensor output (about 10% and 4%, respectively). However, it should be noted that the average concentration of both compounds in a human serum is in significantly lower concentration than was taken for the analysis (about 60 µM for ascorbate [31] and up to 200 µM for citrate [32]) when the normal glucose level is up to 11 mM [33]. Therefore, the interference with ascorbate and citrate can be negligible, at least under physiologically relevant conditions. The biosensor was tested on human serum samples, and the obtained results are highly correlated (r = 0.977) with the reference fluorometric approach using a commercial analytical kit (Glucose Assay Kit from Cell Biolabs, Inc.) as well as with the glucose content indicated by producer (Sigma-Aldrich) (Fig. S8 in the SI).
The main sensor characteristics were compared with the recently developed analogues smartphone-adapted sensor systems (Table S1 in the SI). In general, the LOD and linearity frames of the developed biosensor are quite similar to most of them. The only biosensor based on combination of glucose oxidase (GOx), silver nanoparticles, and Zr-based MOF is characterized with significantly lower LOD value when compared with that described in this work (0.5 mM and 25 mM glucose, respectively) [18]. On the other hand, the sensor suffers from a short linearity (0.2 vs 1.0 mM glucose) that makes it less applicable in contrast to developed by us, for analysis of the samples with high glucose content (e.g., blood) [18]. Another GOx-based sensor, which linear frames are very broad (0–6 mM glucose), that theoretically could be used for the detection of all human samples (tears, saliva, sweat, urine, blood) even without their dilution, requires to use fluorescent microscope that exclude possibility of point-of-care analysis [22]. So, taking into account that the proposed sensor system is sufficiently sensitive for analysis of glucose content in all the human liquids and does not require any complicated or expensive equipment or specific skills, it would be interesting for the final consumer for accurate point-of-care glucose analysis.
Conclusions
The developed optical biosensor is characterized with a 40-fold lower LOD and one order of magnitude lower KMapp value in comparison with classical spectrophotometric analysis based on PQQ-GDH/PMS/DCPIP system with the optical absorbance measurements. However, it is tenfold less sensitive than the commercial fluorometric kit (Glucose Assay Kit from Cell Biolabs, Inc.) based on glucose oxidase (Fig. S8A in the SI). Nevertheless, in the point of view that the developed optical sensor does not require any expensive devices like fluorometers, the analysis is fast and easy for everyone without specific skills; it could be very prospective for routine point-of-care glucose analysis for diabetics. Importantly, the main signal transducing/reading part of the system is a standard smartphone with a photo camera and simple app for the image processing.
It should be noted that despite the fact that all components of the studied system are not novel, particularly the use of a smartphone as a signal detecting device that is broadly known, the combination of the enzyme immobilization method, the use of PMS as a fluorophore, and the smartphone signal detector resulted in the high quality and easy use biosensor, justifying novelty of the developed technology. However, despite the fact that the glucose analysis with the new procedure was successful, the real motivation of the study was in establishing the analytical platform using the PQQ-GDH enzyme (wild type in the present study). Based on the obtained results, the next step in the project will include the use of the artificial chimeric enzymes switchable from inactive to active states upon formation of a complex with different analytes, which include peptides (e.g., M13 peptide, skeletal muscle myosin light chain kinase peptide), proteins (such as α-amylase and human serum albumin), and different drugs (such as methotrexate, tacrolimus, rapamycin, and cyclosporin) [23, 24, 25, 26]. Importantly, the chimeric enzymes are produced by genetic engineering using the same PQQ-GDH enzyme as the biocatalytic unit linked to a biorecognition part. The present analytical system for the optical (fluorescent) analysis of glucose represented only the development of the platform producing signals with the use of the PQQ-GDH enzyme. We are particularly interested in the application of a smartphone as a signal-reading device for the point-of-care and end-user systems. The more sophisticated systems with the artificial chimeric enzymes are presently in the study, and the results will be published elsewhere.
References
Venn RF (ed) (2008) Principles and practice of bioanalysis. CRC Press, Boca Raton
Scheller FW, Hintsche R, Pfeiffer D, Schubert F, Riedel K, Kindervater R (1991) Biosensors: Fundamentals, applications and trends. Sens Actuat B 4:197–206
Hnasko R (2015) ELISA - Methods and protocols. Springer Science, New York
Huang X, Xu D, Chen J, Liu J, Li Y, Song J, Ma X, Guo J (2018) Smartphone-based analytical biosensors. Analyst 143:5339–5351
Kanchi S, Sabela MI, Mdluli PS, Inamuddin, Bisetty K (2017) Smartphone based bioanalytical and diagnosis applications: a review. Biosens Bioelectron 102:136–149
McCracken KE, Yoon J-Y (2016) Recent approaches for optical smartphone sensing in resource-limited settings: a brief review. Anal Methods 8:6591–6601
Rateni G, Dario P, Cavallo F (2017) Smartphone-based food diagnostic technologies: a review. Sensors 17:art. No. 1453
Roda A, Michelini E, Zangheri M, Di Fusco M, Calabria D, Simoni P (2016) Smartphone-based biosensors: a critical review and perspectives. Trends Anal Chem 79:317–325
Xu X, Akay A, Wei H, Wang S, Pingguan-Murphy B, Erlandsson B-E, Li X, Lee W, Hu J, Wang L (2015) Advances in smartphone-based point-of-care diagnostics. Proc IEEE 103:236–247
Zhang D, Liu Q (2016) Biosensors and bioelectronics on smartphone for portable biochemical detection. Biosens Bioelectron 75:273–284
Jia Y, Sun H, Li X, Sun DK, Hu T, Xiang N, Ni ZH (2018) Paper-based graphene oxide biosensor coupled with smartphone for the quantification of glucose in oral fluid. Biomed Microdevices 20:art. No. 89
Soni A, Jha SK (2017) Smartphone based non-invasive salivary glucose biosensor. Anal Chim Acta 996:54–63
Elsherif M, Hassan MU, Yetisen AK, Butt H (2019) Hydrogel optical fibers for continuous glucose monitoring. Biosens Bioelectron 137:5–32
Lin YR, Hung CC, Chiu HY, Chang BH, Li BR, Cheng SJ, Yang JW, Lin SF, Chen GY (2018) Noninvasive glucose monitoring with a contact lens and smartphone. Sensors (Basel) 18:3208
Choi J, Bandodkar AJ, Reeder JT, Ray TR, Turnquist A, Kim SB, Nyberg N, Hourlier-Fargette A, Model JB, Aranyosi AJ, Xu S, Ghaffari R, Rogers JA (2019) Soft, skin-integrated multifunctional microfluidic systems for accurate colorimetric analysis of sweat biomarkers and temperature. ACS Sensors 4:379–388
Erenas MM, Carrillo-Aguilera B, Cantrell K, Gonzalez-Chocano S, Vargas-Sansalvador IM, Orbe-Payá I, Capitan-Vallvey LF (2019) Real time monitoring of glucose in whole blood by smartphone. Biosens Bioelectron 136:47–52
Vaquer A, Barón E, de la Rica R (2021) Detection of low glucose levels in sweat with colorimetric wearable biosensors. Analyst 146:3273–3279
Guo L, Chen S, Yu YL, Wang JH (2021) A Smartphone optical device for point-of-care testing of glucose and cholesterol using Ag NPs/UiO-66-NH2-based ratiometric fluorescent probe. Anal Chem 93:16240–16247
Alizadeh N, Salimi A, Hallaj R (2019) A strategy for visual optical determination of glucose based on a smartphone device using fluorescent boron-doped carbon nanoparticles as a light-up probe. Mikrochim Acta 187:14
Sun K, Yang Y, Zhou H, Yin S, Qin W, Yu J, Chiu DT, Yuan Z, Zhang X, Wu C (2018) Ultrabright polymer-dot transducer enabled wireless glucose monitoring via a smartphone. ACS Nano 12:5176–5184
Deng M, Song G, Zhong K, Wang Z, Xia X, Tian Y (2022) Wearable fluorescent contact lenses for monitoring glucose via a smartphone. Sens Actuators B Chem 352:131067
Song C, Yang Y, Tu X, Chen Z, Gong J, Lin C (2021) A smartphone-based fluorescence microscope with hydraulically driven optofluidic lens for quantification of glucose. IEEE Sens J 21:1229–1235
Guo Z, Smutok O, Johnston WA, Ayva CE, Walden P, McWhinney B, Ungerer JPJ, Melman A, Katz E, Alexandrov K (2021) Circular permutated PQQ-glucose dehydrogenase as an ultrasensitive electrochemical biosensor. Angew Chem Int Ed 61:e202109005
Bollella P, Edwardraja S, Guo Z, Whitfield J, Melman A, Alexandrov K, Katz E (2021) Connecting artificial proteolytic and electrochemical signaling systems with caged messenger peptides. ACS Sensors 6:3596–3603
Guo Z, Smutok O, Johnston WA, Walden P, Ungerer JPJ, Peat TS, Newman J, Parker J, Nebl T, Melman A, Suderman RJ, Katz E, Alexandrov K, K, (2021) Design of a methotrexate-controlled chemical dimerization system and its use in bio-electronic devices. Nature Commun 12:article No. 7137
Guo Z, Johnston WA, Whitfield J, Walden P, Cui Z, Wijker E, SelvakumarEdwardraja S, Lantadilla IR, Ely F, Vickers C, Ungerer JPJ, Alexandrov K (2019) Generalizable protein biosensors based on synthetic switch modules. J Am Chem Soc 141(20):8128–8135
Oubrie A, Rozeboom HJ, Kalk KH, Olsthoorn AJ, Duine JA, Dijkstra BW (1999) Structure and mechanism of soluble quinoprotein glucose dehydrogenase. EMBO J 18:5187–5194
Stockert JC, Carou MC, Casas AG, GarcíaVior MC, EzquerraRiega SD, Blanco MM, Espada J, Blázquez-Castro A, Horobin RW, Lombardo DM (2020) Fluorescent redox-dependent labeling of lipid droplets in cultured cells by reduced phenazine methosulfate. Heliyon 6:art. No. e04182
Willner I, Katz E (2000) Integration of layered redox-proteins and conductive supports for bioelectronic applications. Angew Chem Int Ed 39:1180–1218
Wells PK, Smutok O, Melman A, Katz E (2021) Switchable biocatalytic reactions controlled by interfacial pH changes produced by orthogonal biocatalytic processes. ACS Appl Mater Interfaces 13:33830–33839
Padayatty SJ, Levine M (2016) Vitamin C physiology: the known and the unknown and Goldilocks. Oral Dis 22:463–493
Costello LC, Franklin RB (2016) Plasma citrate homeostasis: how it is regulated; and its physiological and clinical implications. An important, but neglected, relationship in medicine. HSOA J Hum Endocrinol 1:art. No. 005
Woerle HJ (2004) Glucose physiology, normal. In: Huhtaniemi I (ed) Encyclopedia of Endocrine Diseases. Academic Press, Cambridge, pp 263–270
Funding
This work was supported in part by the Human Frontier Science Program, project grant RGP0002/2018 to OS and EK, and by the US Department of Defense award W81XWH2010708 to EK. This paper is dedicated to the memory of Prof. Artem Melman (deceased in 2021) who was one of the leading scientists working on this project, as well as on many other research projects.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
This paper is dedicated to the memory of Artem Melman.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wells, P.K., Melman, A., Katz, E. et al. Fluorescent sensor based on pyrroloquinoline quinone (PQQ)-glucose dehydrogenase for glucose detection with smartphone-adapted signal analysis. Microchim Acta 189, 371 (2022). https://doi.org/10.1007/s00604-022-05466-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-022-05466-4