Abstract
A three-dimensional structured porous graphene oxide-polyethylenimine bead (pGP) is synthesized for immobilizing gold nanoparticles and modifying glutathione molecules (denoted as pGP/AuG). The pGP/AuG has open pore structure, honeycomb-like channels, and excellent hydrophilicity. By taking advantages of the porous structure, abundant binding sites, and multivalent interactions between glycopeptides and both glutathione molecules and free amino groups, the pGP/AuG is adopted to the selective enrichment of N-linked glycopeptides with low limit of detection (2 fmol), high enrichment selectivity (1:500), binding capacity (333.3 mg/g), recovery yield (91.3 ± 2.1%), and repeatability (< 6.0% RSD) using matrix-assisted laser desorption/ionization time of flight mass spectrometry detection method. Furthermore, the practical applicability of pGP/AuG is evaluated, in which 209 N-glycosylated peptides corresponding to 128 N-glycosylated proteins are identified from 1 μL human serum in three independent analysis procedures, suggesting the great potential for application in glycoproteome fields.
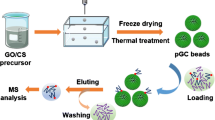
Graphical abstract

Schematic presentation of preparation for porous graphene oxide-based hydrophilic beads (pGP/AuG) with honeycomb-like microstructure. The pGP/AuG was successfully used for enriching and identifying glycopeptides from actual biological sample.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As one of the most ubiquitous and complex post-translational modifications, protein glycosylation plays indispensable roles in many cell biology processes [1]. Aberrant protein glycosylation has been proven to associate with many human diseases [2]. The comprehensive identification of glycoproteins contributes to understand their biological functions and discover clinical biomarkers. Mass spectrometry (MS) has become a most effective tool in proteomic research [3]. However, some challenges still existed in MS-based glycoproteomics analysis: the low abundance of glycopeptides/glycoproteins, the inherently inhomogeneous glycans, and the serious ion suppression of non-glycopeptides [4, 5]. As a consequence, it is highly imperative to develop the sample enrichment technologies for selective capture and preconcentration of glycopeptides/glycoproteins for glycoproteomics research.
The affinity materials are developed according to the four main modes: hydrophilic interaction liquid chromatography (HILIC), lectin affinity chromatography, boronic acid chemistry, and covalent hydrazide chemistry [6]. Among them, the HILIC method has received increasing attentions in glycoproteomics research [7]. In general, the hydrophilic functional molecules, such as amino group [8], maltose [9], polysaccharide [10], chitosan [11], and amino acids [12], were grafted on the substrates to prepare the hydrophilic materials. The multifarious substrate materials include magnetic bead [13], mesoporous silica [14], metal-organic frameworks [15], and graphene oxide [16, 17]. For some HILIC materials, the cumbersome and multistep reaction processes result in low efficiency of material functionalization. The inferior surface modification leads to low content of hydrophilic groups, low enrichment selectivity, binding capacity, and detection sensitivity. The direct immobilization of hydrophilic mercapto molecules on metal oxide/metal materials is a candidate for enriching glycopeptides [18,19,20,21].
Graphene oxide (GO) is a typical two-dimensional layered material with high specific surface area, good stability, and abundant functional groups [22]. Peptides, proteins, and even nanomaterials have been immobilized on GO [23], and these composites proven excellent performances in the selective extraction/adsorption of target biomolecules [24,25,26,27]. To further improve the hydrophilicity and specificity for glycopeptide recognition, various hydrophilic molecules were grafted on GO, and these materials achieved the selective capture of low-abundant glycopeptides [28,29,30,31]. In short, these most GO composites were two-dimensional architectures, the enrichment performances were still limited. Moreover, three-dimensional GO-based porous materials with macroporous structure have been synthesized and applied for pollutant removal with high adsorption capacity and short time [32, 33]. There is still no research about GO-based porous beads with macroporous channels being applied for glycopeptide enrichment.
In this research, the porous GO-based bead (pGP) with honeycomb-like microstructure is prepared, and it modified with zwitterionic hydrophilic molecules and being applied to the specific capture of low-abundant glycopeptides. GO served as the skeleton material for keeping the porous microstructure. Polyethylenimine played important roles both for stabilizing the three-dimensional porous architecture and further immobilizing Au nanoparticles. As an example, the L-glutathione was selected and self-assembled onto Au nanoparticles to form the pGP/AuG bead. The immobilized glutathione molecules and free amino groups of PEI were considered to provide abundant binding sites and multivalent interactions with glycopeptides. The enrichment performance of pGP/AuG beads was evaluated using both standard glycoprotein tryptic digest and actual biological specimen. These results demonstrate that the pGP/AuG beads have low limit of detection, high selectivity, recovery yield, and repeatability and are promising for glycoproteomics study.
Experiment
Chemicals and materials
Graphene oxide (GO) was provided by Suzhou Tanfeng Graphene Tech. Inc. (Suzhou, China, http://graphenechina.cnpowder.com.cn/). Tetrachloroauric (III) acid hydrate and L-glutathione were purchased from Aladdin Industrial Co., Ltd. (Shanghai, China, https://www.aladdin-e.com/). Human immunoglobulin G (IgG), branched polyethylenimine (PEI, Mn = 10,000), poly (ethylene glycol) diglycidyl ether (PEGDE, Mn = 500), chicken avidin, trypsin, dithiothreitol (DTT), iodoacetamide (IAA), urea, ammonium bicarbonate, bovine serum albumin (BSA), trifluoroacetic acid (TFA), and 2,5-dihydroxybenzoic acid (DHB) were obtained from Sigma-Aldrich (St Louis, Mo, USA, https://www.sigmaaldrich.com). Acetonitrile (ACN) was received from Merck (Darmstadt, Germany, https://www.merckgroup.com/cn-zh). PNGase F was purchased from New England Biolabs (Ipswich, MA, USA, https://www.neb.uk.com/). The healthy human serum sample was obtained from Sixth People’s Hospital of Shanghai. The utilization of human serum abided by guidelines of the Ethics Committee of the Hospital. Water was purified by a Milli-Q system (Millipore, Bedford, MA, USA).
Preparation of the pGP/AuG beads
The porous GO/PEI (pGP) beads were synthesized through an ice-templated assembly process. Twenty grams of GO aqueous slurry (10 mg/g), 2.4 g of PEI aqueous solution (250 mg/g), and 600 μL of PEGDE were mixed and magnetically stirred for 2 h. Subsequently, the GO/PEI slurry was squeezed from a syringe needle with the internal diameter of 0.25 mm and injected into liquid nitrogen solution. The droplets were rapidly frozen in 1 min and sank in the bottom of stainless steel bottle. After collection and lyophilization for 12 h in a freeze dryer, the obtained pGP beads were thermally treated for 8 h at 60 °C in an oven.
Then, 0.5 g of the pGP beads was mixed with 50 mL of tetrachloroauric (III) acid hydrate aqueous solution (2.5 mg/mL). After magnetically stirred for 2 h, the resulting product of the pGP/Au was separated, cleaned with water, and re-dispersed in 50 mL of an aqueous solution containing 2 g of glutathione. The mixture was magnetically stirred at 50 °C for 12 h. Finally, the product of the pGP/AuG was separated and cleaned with water for three times.
Material characterization
The structure and morphology of product were observed with scanning electron microscope (Hitachi TM 3000, Hitachi S-4800, Japan) and transmission electron microscope (FEI TF20, Japan). Raman spectrum was recorded with a Raman spectrometer (LabRam HR Evolution). X-ray powder diffraction pattern was tested using an X-ray diffractometer (Rigaku D/Max 2500 diffractometer, Cu Kα radiation, λ = 1.54 Å, 40 kV). X-ray photoelectron spectroscopy (XPS) spectrum was obtained on an X-ray photoelectron spectrometer (Thermo ESCALAB 250 XI, USA). Fourier transform infrared (FTIR) spectrum was recorded on a Nicolet 6700 (Thermo Fisher Scientific, USA). The pore structure, pore size distribution, and surface area were investigated on a Micromeritics mercury porosimeter (AutoPore 9510, USA). The water contact angle was tested on a DSA 100 goniometer. The porous beads were ground to powder before test.
Sample preparation, glycopeptide enrichment, mass spectrometry analysis, and database search
Human IgG was dissolved in NH4HCO3 buffer (1 mL, 50 mM, pH = 8.3) with a concentration of 1 mg/mL. After the addition of 20 μg of trypsin, the mixture was incubated at 37 °C for 17 h. BSA (5 mg) was dissolved in NH4HCO3 buffer (2.5 mL, 50 mM, pH = 8.3) containing 8 mM urea. Then, 50 μL of DTT (1 M) was added and incubated at 60 °C for 45 min. Subsequently, 15 mg of IAA was added and incubated for 1 h in darkness. Finally, the mixture was diluted to 25 mL with NH4HCO3 buffer (50 mM, pH = 8.3) and digested with 100 μg of trypsin at 37 °C for 17 h.
Human serum (1 μL) was mixed with 50 μL of NH4HCO3 buffer (50 mM, pH = 8.3); the mixture was centrifuged at 12,000 rpm for 5 min. The supernatant was carefully collected and denaturized at 100 °C for 10 min. Subsequently, 100 μL of DTT (1 M) was added and incubated at 60 °C for 1 h. Then, 7.5 mg of IAA was mixed and incubated for 1 h in darkness. Finally, 60 μg of trypsin was added and incubated at 37 °C for 17 h. The resulting tryptic digests were lyophilized and kept at − 20 °C for further use.
For the selective enrichment of glycopeptides from standard glycoprotein tryptic digests, pGP/AuG (2 mg) and human IgG tryptic digests (3 μg, with or without BSA tryptic digest) were dispersed in loading buffer (500 μL, ACN/H2O/TFA, 95:4.9:0.1, v/v/v). After 10 min of vibration incubation, the pGP/AuG beads were naturally sedimentated, separated, and washed with loading buffer for two times with 3 min each time; the captured glycopeptides were released with elution buffer (200 μL, ACN/H2O/TFA, 30:69.9:0.1, v/v/v) for 10 min. Soon after, the obtained eluate was lyophilized for deglycosylation or redissolution in water and analyzed by matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS).
For the selective enrichment of glycopeptides from human serum specimen, pGP/AuG (5 mg) was dispersed in loading buffer (2 mL, ACN/H2O/TFA, 95:4.9:0.1, v/v/v) and incubated with human serum tryptic digests for 30 min. The adsorbed impurities were washed with 1 mL of loading buffer for three times with 3 min each time. Then, the captured glycopeptides were released with elution buffer (500 μL, ACN/H2O/TFA, 30:69.9:0.1, v/v/v) for 15 min. The eluate was lyophilized, deglycosylated, and further analyzed by nano-LC-MS/MS.
The processes of deglycosylation, MALDI TOF MS experiment, nano-LC-MS/MS analysis, and database search are in Electronic supplementary material.
Results and discussion
Synthesis and characterization of pGP/AuG beads
The preparation procedure of porous GO-based zwitterionic hydrophilic bead (pGP/AuG) is displayed in Fig. 1. First, the porous graphene oxide/polyethylenimine (pGP) beads with honeycomb-like microchannel architecture were synthesized by combing the ice-templated assembly process, lyophilization, and subsequent thermal cross-linking approaches. Second, Au nanoparticles were in situ reduced and anchored on the surface of pGP in the presence of polyethylenimine (PEI). Subsequently, the selected zwitterionic hydrophilic L-glutathione molecules were self-assembled onto Au nanoparticles via Au–S bonds. It was worth noting that PEI played two important roles in the process. The three-dimensional porous structure was stabilized by forming the electrostatic interactions between GO and PEI and covalent interactions between amino groups in branched PEI and epoxy groups in intermediate poly (ethylene glycol) diglycidyl ether (PEGDE). Moreover, the PEI acted as reducing and stabilizing reagent for immobilizing Au nanoparticles. GO served as the skeleton material for keeping the porous microstructure. With the characteristics of honeycomb-like channels, open pore structure, and abundant hydrophilic molecules, the pGP/AuG beads were further used as the new affinity adsorbent for the selective enrichment of low-abundant glycopeptides from complex biological sample for mass spectrometry-based glycoproteomics study.
The morphologies of pGP and pGP/AuG beads are characterized by scanning electron microscopy (SEM) and transmission electron microscopy (TEM). As shown in Fig. S1, the pGP beads are approximate spheres with partial surface shrinkage; the average diameter is calculated to be about 1.8 mm. The enlarged images show the abundant pores with honeycomb-like channels on the surface of pGP beads (Fig. S2). The pores are formed during ice crystals growing approach when GO and PEI-mixed liquid drop is added into the liquid nitrogen solution. Several other multifunctional materials with microhoneycomb structure have been prepared by the ice crystal-templated strategy [34]. As displayed in Figs. 2a, b and S3, after the immobilization of Au nanoparticles, the porous architecture of pGP/AuG bead is similar to that of pGP bead. Compared with the smooth surface of pGP bead (Fig. 2c), abundant Au nanoparticles are anchored on the surface of pGP/AuG bead in the magnified SEM micrograph (Fig. 2d). Moreover, as presented in Fig. 2e, the edges of GO sheets are spread out in TEM image of pulverized pGP bead. By comparison, abundant Au nanoparticles are dispersed and anchored on the surface of pulverized pGP/Au bead in Fig. 2f. The average diameter size of Au nanoparticles is calculated to be 65 nm according to the TEM micrograph. These results prove the successful immobilization of Au nanoparticles.
The pore structure, porosity, and specific surface area of pGP/Au beads are investigated by mercury intrusion porosimetry (MIP). The analysis results are presented in Fig. S4; the pore volume and specific surface area of pGP/Au beads are determined to 3.91 mL/g and 203.0 m2/g, respectively. The porosity of pGP/Au beads is determined to 66.9%. In addition, the pGP/Au beads show the micropores with the average diameter (4 V/A) of 1190 nm. The high porosity, specific surface area, and open pore structure contribute to the adsorption performance of pGP/Au beads.
For demonstrating the GO component in pGP beads, the Raman spectra of GO and pGP were recorded. As shown in Fig. S5, the two typical bands at 1334 cm−1 (D-band) and 1587 cm−1 (G-band) of GO are observed. The result proves the existence of GO in pGP beads.
The X-ray diffraction (XRD) patterns of pGP and pGP/Au beads are shown in Fig. 3a. There is a wide diffraction peak located at 2θ value of 20.5° for polymers such as PEI or PEGDE. In comparison, after loading with Au nanoparticles, four new sharp diffraction peaks at 2θ values of 38.2°, 44.4°, 64.6°, and 77.6° are apparent. These peaks are assigned to the (111), (200), (220), and (311) planes of crystalline Au (JCPDS 01-1172), respectively, indicating the decoration of Au nanoparticles on the pGP beads.
X-ray photoelectron spectra (XPS) pattern is recorded to examine the surface chemical component. As displayed in Fig. 3b, two binding energies at 84.0 eV and 87.7 eV are observed in the XPS spectrum of pGP/Au bead. These peaks can be attributed to the Au 4f7/2 and Au 4f5/2 of the Au(0). In addition, the signal peak of Au element appeared in the energy-dispersive spectroscopy (EDS) (Fig. 3c). These results also demonstrate the successful attachment of Au nanoparticles on the pGP beads.
Fourier transform infrared (FTIR) spectroscopy analysis is adopted to confirm the surface modification of glutathione molecules. In Fig. 3d, compared with the FTIR spectrum of pGP/Au, the fingerprint peaks corresponding to glutathione molecules are clearly observed in the FTIR spectrum of pGP/AuG. The result reveals the successful modification of glutathione molecules on Au-immobilized pGP beads.
In the experimental stage, we observed that both GO and PEI played important roles in synthesizing GO-based porous beads with stable structure. For the GO porous beads without or low content of PEI component, a mass of fragments was observed in the aqueous solution after shaking for 24 h. When the mass ratio of PEI/GO was over 2:1, the obtained pGP beads can retain the whole structure in aqueous solution. Almost no fragment appeared in the solution, and the structures and morphologies of pGP beads retain well (Fig. S6). The stable structure can be attributed to two factors. The electrostatic interactions between carboxylic acid groups on GO and amino groups on PEI. Additionally, the intermediate PEGDE with double-epoxy groups was adopted to covalent cross-linking with branched PEI; the ring-opening polymerization reaction happened between epoxy groups and amino groups in the thermal environment. On the other hand, the PEI beads comprising PEGDE component were prepared as a control group. SEM micrographs are shown in Fig. S7. In comparison with the morphology of pGP beads, the approximately spherical architecture and porous structure of PEI beads are seriously shrank and collapsed. The pore volume and specific surface area of PEI beads are determined to be 1.12 mL/g and 48.6 m2/g, respectively. The porosity of PEI beads is determined to be 19.8%. It can be inferred that the GO nanosheets served as the skeleton material for keeping the porous microstructure of pGP beads in the material preparation and treatment processes.
The hydrophilicity is investigated by determining the water contact angle of pGP/AuG beads. The beads were pulverized in a mortar. As shown in Fig. S8, the water contact angle is measured to 0°, revealing the excellent hydrophilic feature of pGP/AuG beads. The stability was studied by incubating the beads in water for 3 days; the pGP, pGP/Au, and pGP/AuG beads retain 99.5, 99.8, and 99.7% of the original weight, respectively. These materials have good stabilities. Therefore, the unique porous structure and abundant amino groups and zwitterions molecules of pGP/AuG beads make it a promising adsorbent for glycopeptide enrichment.
Glycopeptide enrichment performance of pGP/AuG beads
Specifically, the glycopeptides with linked glycans have higher hydrophilicity than those of non-glycopeptides. Hence, a HILIC enrichment mode is adopted for pGP/AuG beads in this study. The enrichment process is simply presented in Fig. 1c. The protein tryptic digests were incubated with pGP/AuG beads; after the selective adsorption and washing procedure, the captured glycopeptides were eluted, and the eluate was analyzed by MS. The loading buffer was optimized, the result is shown in Fig. S9, and the related discussion is shown in the Electronic Supp. Material (ESM).
The MALDI-TOF mass spectra of human IgG tryptic digest without enrichment and with enrichment of several materials are presented in Fig. 4. Abundant non-glycopeptide peaks with strong signal intensities dominate the spectrum, which strongly inhibit the detection of glycopeptides. None of the glycopeptide peaks is observed in Fig. 4a. Fortunately, after enrichment by pGP/AuG beads, the non-glycopeptides almost disappeared, and 25 glycopeptide peaks with increased signal intensities and signal-to-noise (S/N) ratios are obviously identified in Fig. 4b. The detailed information of detected glycopeptides are shown in Table S1. In addition, two hydrophilic materials are adopted for glycopeptide enrichment. As shown in Fig. 4 c and d, only 20 and 21 glycopeptide peaks with clean background are detected after enrichment by pGP beads and commercial hydrophilic beads. The signal intensities and S/N ratios of identified glycopeptide peaks using pGP/AuG are higher than those of pGP and commercial hydrophilic. For example, the signal intensities and S/N ratios of a detected glycopeptide peak with m/z at 2764.2 after enrichment by pGP/AuG, pGP, and the commercial hydrophilic beads are 7968 and 1097, 2345 and 222, and 4175 and 260, respectively. The high enrichment efficiency of pGP/AuG beads can be due to the high content of hydrophilic molecules, and multivalent hydrophilic interactions between glycopeptides and both glutathione molecules and free amino groups on pGP/AuG beads. Moreover, the enrichment results of pGP/AuG, pGP/Au, pGP, and PEI comprising PEGDE are shown in Fig. S10; the corresponding discussion is given in the Electronic Supp. Material (ESM).
MALDI-TOF mass spectra of human IgG tryptic digests, a without enrichment, b with enrichment of pGP/AuG beads, c with enrichment of pGP beads, d with enrichment of commercial zwitterionic hydrophilic beads, e with enrichment of pGP/AuG beads and deglycosylated by PNGase F, f with enrichment of commercial zwitterionic hydrophilic beads and deglycosylated by PNGase F. The detected glycopeptides are labeled with “★”
To confirm the glycopeptides enriched by pGP/AuG beads and commercial hydrophilic beads that belong to N-linked glycopeptides, the obtained eluate is further deglycosylated by PNGase F. As shown in Fig. 4 e and f, it can be seen that the peaks with m/z over 2200 all disappear, which reveals that all the enriched peptides are N-linked glycopeptides. In addition, the amino acid sequences of two remaining deaminated peptides are EEQFN#STFR and EEQYN#STYR.
Limit of detection, binding capacity, enrichment selectivity, and repeatability of pGP/AuG beads
The limit of detection is investigated using different concentrations of human IgG tryptic digest. As presented in Fig. 5, nine and ten glycopeptide peaks are detected from 20 to 5 fmol/μL human IgG tryptic digest after enrichment by pGP/AuG beads (Fig. 5a, b). When the concentration is reduced to 2 fmol/μL, five glycopeptide peaks with S/N ratios over 3 can still be detected (Fig. 5c). Therefore, the limit of detection of pGP/AuG beads is as low as 2 fmol/μL. Besides, there is a linear relationship between the detected S/N ratio and the concentration of human IgG tryptic digest in a certain range from 2 to 10 fmol/μL. When the S/N ratio is set as 3, the concentration can be calculated to 0.47 fmol/μL. The result suggests that the pGP/AuG beads have good ability to detect N-linked glycopeptides with low concentration.
The enrichment selectivity of pGP/AuG beads toward low-abundant glycopeptides from a complex mixture is evaluated. The mixture of tryptic digests of human IgG and BSA is used as the testing sample. As presented in Fig. 6a, for the direct analysis of the mixture of human IgG and BSA tryptic digests at a mass ratio of 1:200, the glycopeptide peaks are hardly detected. Importantly, after enrichment with pGP/AuG beads, most of the observed peaks belonging to glycopeptide peaks, and 19 glycopeptide peaks are detected in Fig. 6b. When the mass ratio of human IgG and BSA tryptic digests increases to 1:500, 15 glycopeptides are captured and identified after enrichment by pGP/AuG beads (Fig. 6c). These analysis results prove that the pGP/AuG beads have good enrichment ability for glycopeptides even in the presence of abundant non-glycopeptides.
The binding capacity of pGP/AuG beads is determined by enriching the glycopeptides from a fixed amount of human IgG tryptic digest using different amount of pGP/AuG beads. As shown in Fig. 7, the signal intensities of six high-abundant glycopeptides increase first and reach the maximum values when the mass ratio of pGP/AuG beads to human IgG digests is 1: 3. Soon after, the signal intensities have slightly improved with further increasing the mass ratio. Therefore, the binding capacity of pGP/AuG beads is calculated to be about 333.3 mg/g.
The enrichment recovery yield is determined using the quantitative approach of stable isotopic dimethyl labeling method. As shown in Table S2, the average enrichment recovery yield of two deaminated peptides (EEQFN#STFR and EEQYN#STYR) is determined to be 91.3 ± 2.1% (n = 6).
The material recycling performance of pGP/AuG beads is studied. As shown in Fig. S11, the obtained mass spectra show the similar situations. The signal intensities of three high-abundant glycopeptide peaks have slightly declined after five consecutive uses. The relative standard deviations (RSDs) of the detection signal intensity are below 6.0%. In addition, Fig. S12 shows the SEM and TEM images of pGP/AuG beads after five repeated uses. The structure and morphology have slightly changed in comparison with the original material. These results reveal the stable structure and enrichment ability of pGP/AuG beads.
Table 1 shows the comparisons between previous nanomaterials for the selective enrichment of glycopeptides and the pGP/AuG beads. The pGP/AuG beads have the highest loading capacity among these materials. In addition, the limit of detection, specificity, and recovery yield of pGP/AuG beads are comparable with these materials. This method has some unique features. It provides a universal approach to prepare porous enrichment materials. The raw materials are not limited to inorganic and organic component, one-dimensional and two-dimensional materials. In the porous microstructure formation process, no additional pore-forming agent is needed. Therefore, the time-consuming material purification step is avoided. Additionally, in comparison with the similar material [11], the pGP/AuG beads exhibit the better enrichment performance. Several functional nanomaterials and functional molecules containing thiol groups can be immobilized on the pGP beads for preparing more hydrophilic affinity materials for enriching low-abundant glycopeptides.
Glycopeptide enrichment and identification from real biological specimen
Inspired by the outstanding performance in glycopeptide enrichment, a real complex biological sample was employed to demonstrate the practicability of pGP/AuG beads. Human serum contains several glycoproteins, which are potential biomarkers for disease diagnosis. The glycoproteomics research of human serum is significant but also challenging. In this research, 1 μL of human serum was digested with trypsin and treated with pGP/AuG beads. After washing and elution processes, the obtained eluate was deglycosylated using PNGase F and further analyzed by nano LC-MS/MS. As a result, 209 N-glycosylated peptides derived from 128 N-glycosylated proteins were identified by three independent analysis. The detailed information is shown in Table S3. The enrichment and identification performance of pGP/AuG beads is comparable with or even higher than several previously reported hydrophilic materials, such as COF-functionalized magnetic graphene composite (1 μL human serum, 85 N-glycoproteins, 232 N-glycopeptides) [15], graphene oxide/polyethylenimine-carrageenan (2 μL human serum, 56 N-glycoproteins, 56 N-glycopeptides) [26], and magHN/Au-GSH nanofiber (1 μL human serum, 104 N-glycoproteins, 246 N-glycopeptides) [35]. These analysis results demonstrate that the as-prepared pGP/AuG beads have feasible ability in capturing low-abundant glycopeptides from complex biological specimen for mass spectrometry-based glycoproteomics study.
Additionally, although the developed method shows high efficiency in the selective enrichment and identification of low-abundant glycopeptides from complex biological samples, it still has some limitations. For example, the limit of detection and specificity are unsatisfied. The average diameter and micropores of pGP/AuG beads are relatively large; more efforts are needed to prepare the micrometer or nanometer size materials with nano-sized porous microstructure. The enrichment ability of the method may be further improved through these efforts.
Conclusions
Porous graphene oxide-based hydrophilic bead with honeycomb-like microstructure is synthesized and modified with gold nanoparticle-glutathione for capturing low-abundant glycopeptides from biological samples. By combing the physicochemical characteristics of open pore structure, honeycomb-like channels, excellent hydrophilicity, abundant free amino groups, and zwitterionic molecules, the pGP/AuG beads showed high efficiency in selective enrichment of glycopeptides, including high selectivity, low limit of detection, high binding capacity, recovery yield, and repeatability. The pGP/AuG beads have been successfully applied for human serum N-glycoproteomics analysis. A limitation of the pGP/AuG beads is the millimeter-sized diameter; further efforts for preparing micrometer or nanometer size materials are needed. This study provides an alternative approach to prepare porous hydrophilic materials and their applications in glycosylated biomarker research.
References
Hart GW, Copeland RJ (2010) Glycomics hits the big time. Cell 143(5):672–676
Pinho SS, Reis CA (2015) Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer 15(9):540–555
Ruhaak LR, Xu G, Li Q, Goonatilleke E, Lebrilla CB (2018) Mass spectrometry approaches to glycomic and glycoproteomic analyses. Chem Rev 118(17):7886–7930
Palaniappan KK, Bertozzi CR (2016) Chemical glycoproteomics. Chem Rev 116(23):14277–14306
Qing GY, Yan JY, He XN, Li XL, Liang XM (2019) Recent advances in hydrophilic interaction liquid interaction chromatography materials for glycopeptide enrichment and glycan separation. Trac-Trend Anal Chem 124:115570
Zhang Y, Zhang C, Jiang H, Yang P, Lu H (2015) Fishing the PTM proteome with chemical approaches using functional solid phases. Chem Soc Rev 44(22):8260–8287
Sun N, Wu H, Chen H, Shen X, Deng C (2019) Advances in hydrophilic nanomaterials for glycoproteomics. Chem Commun 55(60):10359–10375
Wang Y, Wang J, Gao M, Zhang X (2015) An ultra hydrophilic dendrimer-modified magnetic graphene with a polydopamine coating for the selective enrichment of glycopeptides. J Mater Chem B 3(44):8711–8716
Ma W, Xu L, Li Z, Sun Y, Bai Y, Liu H (2016) Post-synthetic modification of an amino-functionalized metal-organic framework for highly efficient enrichment of N-linked glycopeptides. Nanoscale 8(21):10908–10912
Zhan Q, Zhao H, Hong Y, Pu C, Liu Y, Lan M (2019) Preparation of a hydrophilic interaction liquid chromatography material by sequential electrostatic deposition of layers of polyethylenimine and hyaluronic acid for enrichment of glycopeptides. Microchim Acta 186:600–609
Li K, Zhao B, Yu Q, Xu J, Li X, Wei D, Qian L, Liu G, Wang W (2020) Porous graphene oxide/chitosan beads with honeycomb-biomimetic microchannels as hydrophilic adsorbent for the selective capture of glycopeptides. Microchim Acta 187:324–333
Fu D, Liu Y, Shen A, Xiao Y, Yu L, Liang X (2019) Preparation of glutathione-functionalized zwitterionic silica material for efficient enrichment of sialylated N-glycopeptides. Anal Bioanal Chem 411:4131–4140
Zhang L, Yue X, Li N, Shi H, Zhang J, Zhang Z, Dang F (2019) One-step maltose-functionalization of magnetic nanoparticles based on self-assembled oligopeptides for selective enrichment of glycopeptides. Anal Chim Acta 1088:63–71
Chen H, Li Y, Wu H, Sun N, Deng C (2019) Smart hydrophilic modification of magnetic mesoporous silica with zwitterionic L-cysteine for endogenous glycopeptides recognition. ACS Sustain Chem Eng 7(2):2844–2851
Peng J, Hu Y, Zhang H, Wan L, Wang L, Liang Z, Zhang L, Wei RA (2019) High anti-interfering profiling of endogenous glycopeptides for article human plasma by the dual-hydrophilic metal-organic framework. Anal Chem 91(7):4852–4859
Wang JX, Li J, Yan G, Gao M, Zhang X (2019) Preparation of a thickness-controlled Mg-MOFs-based magnetic graphene composite as a novel hydrophilic matrix for the effective identification of the glycopeptide in the human urine. Nanoscale 11(8):3701–3709
Wang JX, Li J, Gao MX, Zhang XM (2017) Self-assembling covalent organic framework functionalized magnetic graphene hydrophilic biocomposites as an ultrasensitive matrix for N-linked glycopeptide recognition. Nanoscale 9(30):10750–10756
Ma YF, Wang LJ, Zhou YL, Zhang XX (2019) A facilely synthesized glutathione-functionalized silver nanoparticle-grafted covalent organic framework for rapid and highly efficient enrichment of N-linked Glycopeptides. Nanoscale 11(12):5526–5534
Xia C, Jiao F, Gao F, Wang H, Lv Y, Shen Y, Zhang Y, Qian X (2018) Two-dimensional MoS2-based zwitterionic hydrophilic interaction liquid chromatography material for the specific enrichment of glycopeptides. Anal Chem 90(11):6651–6659
Wang Z, Wu R, Chen H, Sun N, Deng C (2018) Synthesis of zwitterionic hydrophilic magnetic mesoporous silica materials for endogenous glycopeptide analysis in human saliva. Nanoscale 10(11):5335–5341
Jiang B, Wu Q, Deng N, Chen Y, Zhang L, Liang Z, Zhang Y (2016) Hydrophilic GO/Fe3O4/Au/PEG nanocomposites for highly selective enrichment of glycopeptides. Nanoscale 8(9):4894–4897
Dreyer DR, Park S, Bielawski CW, Ruoff RS (2010) The chemistry of graphene oxide. Chem Soc Rev 39(1):228–240
Li D, Zhang W, Yu X, Wang Z, Su Z, Wei G (2016) When biomolecules meet graphene: from molecular level interactions to material design and applications. Nanoscale 8(47):19491–19509
Hong Y, Zhao H, Pu C, Zhan Q, Sheng Q, Lan M (2018) Hydrophilic phytic acid-coated magnetic graphene for titanium (IV) immobilization as a novel hydrophilic interaction liquid chromatography-immobilized metal affinity chromatography platform for glyco- and phosphopeptide enrichment with controllable selectivity. Anal Chem 90(18):11008–11015
Su J, He X, Chen L, Zhang Y (2018) Adenosine phosphate functionalized magnetic mesoporous graphene oxide nanocomposite for highly selective enrichment of phosphopeptides. ACS Sustain Chem Eng 6(2):2188–2196
Sun Y, Lin Y, Sun W, Han R, Luo C, Wang X, Wei Q (2019) A highly selective and sensitive detection of insulin with chemiluminescence biosensor based on aptamer and oligonucleotide-AuNPs functionalized nanosilica@graphene oxide aerogel. Anal Chim Acta 1089:152–164
Cheng G, Wang ZG, Denagamage S, Zheng SY (2016) Graphene-templated synthesis of magnetic metal organic framework nanocomposites for selective enrichment of biomolecules. ACS Appl Mater Interfaces 8(16):10234–10242
Chen Y, Sheng Q, Hong Y, Lan M (2019) Hydrophilic nanocomposite functionalized by carrageenan for the specific enrichment of glycopeptides. Anal Chem 91(20):4047–4054
Jiao F, Gao F, Wang H, Deng Y, Zhang Y, Qian X, Zhang Y (2017) Ultrathin au nanowires assisted magnetic graphene-silica ZIC-HILIC composites for highly specific enrichment of N-linked glycopeptides. Anal Chim Acta 970:47–56
Wu R, Li L, Deng C (2016) Highly efficient and selective enrichment of glycopeptides using easily synthesized MagG/PDA/au/L-Cys composites. Proteomics 16:1311–1320
Feng X, Deng C, Gao M, Yan G, Zhang X (2018) Novel synthesis of glucose functionalized magnetic graphene hydrophilic nanocomposites via facile thiolation for high-efficient enrichment of glycopeptides. Talanta 179:377–385
Liu Y, Yang D, Shi Y, Song L, Yu R, Qu J, Yu ZZ (2019) Silver phosphate/graphene oxide aerogel microspheres with radially oriented microchannels for highly efficient and continuous removal of pollutants from wastewaters. ACS Sustain Chem Eng 7(26):11228–11240
Yu RM, Shi YZ, Yang DZ, Liu YX, Qu J, Yu ZZ (2017) Graphene oxide/chitosan aerogel microspheres with honeycomb cobweb and radially oriented microchannel structures for broad spectrum and rapid adsorption of water contaminants. ACS Appl Mater Interfaces 9(26):21809–21819
Fu Q, Si Y, Duan C, Yan Z, Liu L, Yu J, Ding B (2019) Highly carboxylated, cellular structured, and underwater superelastic nanofibrous aerogels for efficient protein separation. Adv Funct Mater 29:1808234
Huan W, Zhang J, Qin H, Huan F, Wang B, Wu M, Li J (2019) A magnetic nanofiber-based zwitterionic hydrophilic material for the selective capture and identification of glycopeptides. Nanoscale 11(22):10952–10960
Acknowledgments
The Authors received funding support from Zhejiang Public Welfare Technology Research Plan/Rural Agriculture (NO. LGN18B010001), the Open Project of the Key Laboratory of Modern Toxicology of Ministry of Education, Nanjing Medical University (NO. NMUAMT201807), Major Medical and Health Science and Technology Program of National Health and Health Commission-Zhejiang Province (No. WKJ-ZJ-1912), Natural Science Foundation of Zhejiang Province (Nos. Y20H180008, Y13H070008), and Natural Science Foundation of Ningbo (Nos. 2018A610234, 2017A610146).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOC 3806 kb)
Rights and permissions
About this article
Cite this article
Li, J., Huan, W., Xu, K. et al. Gold nanoparticle-glutathione-functionalized porous graphene oxide-based hydrophilic beads for the selective enrichment of N-linked glycopeptides. Microchim Acta 187, 518 (2020). https://doi.org/10.1007/s00604-020-04519-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-020-04519-w