Abstract
An ultrasensitive enzyme-free electrochemical sandwich DNA biosensor is described for the detection of ssDNA oligonucleotides. A DNA sequence derived from the genom of Helicobacter pylori was selected as a model target DNA. The DNA assay was realized through catching target DNA on capture DNA immobilized gold electrode; then labeling the target DNA with reporter DNA (rpDNA) and initiator DNA (iDNA) co-modified gold nanoparticles (AuNPs). The high density of iDNAs serves as one of the amplification strategies. The iDNA triggers hybridization chain reaction (HCR) between two hairpins. This leads to the formation of a long dsDNA concatamer strand and represents one amplification strategy. The electrochemical probe [Ru(NH3)5L]2+, where L stands for 3-(2-phenanthren-9-ylvinyl)pyridine, intercalated into dsDNA chain. Multiple probe molecules intercalate into one dsDNA chain, serving as one amplification strategy. The electrode was subjected to differential pulse voltammetry for signal acquisition, and the oxidation peak current at −0.28 V was recorded. On each AuNP, 240 iDNA and 25 rpDNA molecules were immobilized. Successful execution of HCR at the DNA-modified AuNPs was confirmed by gel electrophoresis and hydrodynamic diameter measurements. Introduction of HCR significantly enhances the DNA detection signal intensity. The assay has two linear ranges of different slopes, one from 0.01 fM to 0.5 fM; and one from 1 fM to 100 fM. The detection limit is as low as 0.68 aM. Single mismatch DNA can be differentiated from the fully complementary DNA. Conceivably, this highly sensitive and selective assay provides a general method for detection of various kinds of DNA.

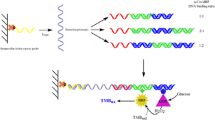
Schematic representation of the detection and the amplification principles of the electrochemical sandwich DNA assay. Purple curl: Captured DNA; Green curl: Reporter DNA; Orange curl: HCR initiator DNA; Yellow solid-circle: Gold nanoparticle; H1 and H2: Two hairpin DNA; [Ru(NH3)5L]2+: Signal probe.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As a molecule test approach, DNA detection has been of great importance in many applications, such as molecular biology, forensic science, and clinical diagnostics. The DNA levels in physiological samples are usually very low [1, 2]. The prerequisites for realizing DNA detection in physiological samples include high sensitivity and sequence-specificity. By primer-mediated enzymatic amplification, polymerase chain reaction (PCR) method has significantly advanced DNA detection sensitivities [3, 4], playing significant role in many applications. However, the PCR procedure is complex, expensive (requiring enzyme), and time-consuming. In addition, the DNA quantification range with PCR is narrow [5, 6].
The efforts have been focused on the development of DNA biosensors possessing ≤ femtomolar detection limits to meet the requirements for very low level DNA detection [2, 6,7,8,9,10]. For enhancing the DNA biosensor sensitivity, various strategies have been developed, mainly including nanomaterial-based signal amplification [8,9,10,11], and isothermal sequence specific amplification [2, 12,13,14,15,16]. Appropriate nanomaterials can offer superior specific surface area to facilitate biomolecule immobilization, and excellent signal transducing properties. Au nanoparticles (AuNPs) have become one of the most-employed nanomaterials for fabricating sensitive nucleic acid biosensors. AuNPs possess the advantages of large surface to volume ratio, excellent biocompatibility, terrific electrocatalytic activity, and excellent chemical and physical stability [6, 9, 17, 18]. In contrast, isothermal sequence amplification strategies can be divided into two categories: enzymatic, and enzyme-free. Although the enzymatic strategies, such as rolling circle amplification, can amplify specific DNA sequences with high sensitivity, they encounters some inevitable drawbacks such as high cost and instability. Hybridization chain reaction (HCR) is an enzyme free amplification strategy that is operated with the alternate assembly of two DNA hairpins in the presence of target to form a DNA concatamer [13,14,15, 19]. The HCR strategy shows high sensitivity and selectivity without participation of enzyme molecules, thereby avoiding the limitations of thermo unstable protein enzymes and the requirements of specific recognition site for nicking endonuclease [20]. It has been used very often as an amplification strategy for fabrication of various biosensors towards the analysis of nucleic acids, proteins and small molecules [19, 21,22,23,24].
However, up to now, only few DNA biosensors can reach or approach attomolar detection limits [2, 6, 17]. We have developed an enzyme-free electrochemical sandwich DNA biosensor towards a 43-mer single strand target DNA (tDNA) sequence specific in Helicobacter pylori (H. pylori) [9]. The detection limit is down to 1 fM. The DNA biosensor is simple, repetitive, sensitive, and selective. H. pylori infection is the main cause of gastroduodenal diseases including peptic ulcer disease, gastritis and gastric cancer in human [25]. In many countries, H. pylori infection is one of the most frequent and persistent bacterial infections because of its widespread prevalence and persistent living strategies in human stomach [26]. Therefore, sensitive and selective detection of H. pylori DNA sequence is of great importance for accurate diagnosis, large epidemiological studies, and point of care detection of gastroduodenal diseases. However, the detection sensitivity with the detection limit of fM is still not good enough to realize PCR-free detection of H. pylori in real physiological samples [27].
In this study, HCR was introduced to the electrochemical sandwich DNA biosensor as one of the amplification strategies to further enhance the detection sensitivity (Scheme 1). Two sequences of single strand DNA (ssDNA) were modified on AuNPs, one is the reporter DNA (rpDNA) that is complementary to one end of the 43-mer tDNA, and the other is an HCR initiator DNA (iDNA). The presence of tDNA would result in the formation of a sandwich DNA structure (iDNA-AuNP-rpDNA/tDNA/cpDNA, where cpDNA denotes for capture DNA) on the electrode. The iDNA strands were modified on the AuNPs at high density, which serves as one of the amplification strategies. They can autonomously trigger HCR between two hairpins (H1 and H2) to form many long strands of double strand DNA (dsDNA) concatamer. Thereafter, the signal indicator [Ru(NH3)5L]2+, where L denotes for 3-(2-phenanthren-9-ylvinyl)-pyridine, intercalated with the dsDNA. Multiple [Ru(NH3)5L]2+ molecules can intercalate on one dsDNA chain [28], serving one of the three amplification strategies. Finally, the electrode was subject to differential pulse voltammetry (DPV) measurement to record the electrochemical oxidation current of the intercalated signal indicator, for signal acquisition. The modification of rpDNA and iDNA on the AuNPs was characterized, and the initiation of HCR at the iDNA-AuNP-rpDNA nanoparticles was verified. The biosensor fabrication conditions were optimized. The detection sensitivity and selectivity of the optimum DNA biosensor was evaluated.
Experimental
Materials
Hydrogen tetrachloroaurate (III) hydrate, tris-(2-carboxyethyl)phosphine hydrochloride (TCEP), hexaamineruthenium (III) chloride (RuHex), N,N-dimethylformamide (DMF), and 6-mercapto-1-hexanol (MCH) were purchased from Alfa Aesar (Massachusetts, U.S.A., www.alfa.com). The [Ru(NH3)5L]2+ complex was prepared by mixing equimolecular amount of [Ru(NH3)5(H2O)]2+ and L solutions for 30 min at room temperature [29]. The [Ru(NH3)5(H2O)]2+ and L were synthesized according to the literature [30, 31] with some modifications [10] (Supplementary Material). KNO3 (99.99% metals basis grade) was obtained from Aladdin Industrial (Shanghai, China, www.aladdin-e.com). Agarose was from Shanghai Yubo Bio Tech Co., Ltd. (Shanghai, China, www.shybio.net). DNA Marker (100–1500 bp) was from TransGen Biotech (Beijing, China, www.transgen.com.cn). Ethidium bromide (EB) was purchased from Solarbio Biosience & Techology (Beijing, China, www.solarbio.com). Deionized water was obtained from a Millipore water system, and used throughout the experiment.
The compositions of the buffer solutions, including Tris−HCl buffer (10 mM, pH 7.0), phosphate buffer solution (PB, 10 mM), TE buffer (pH 8.0) are described in the Supplementary Material. The detection buffer is the 10 mM PB supplemented with 100 mM KNO3. The immobilization buffer consists of 10 mM PB, 1.0 mM EDTA (ethylene diamine tetraacetic acid), and 0.6 M NaCl.
Oligonucleotide sequences (listed in Table 1) were custom-made by Shanghai Sangon Biotechnology Co. (www.sangon.com, China). The 43-mer tDNA sequence was selected by computational restriction endonuclease analysis on the UreB gene of H. pylori [9]. Cy5 and FAM (two fluoresceins) were labeled at the end of the rpDNA and iDNA sequences, respectively for characterization of DNA amount on the AuNPs. ncDNA and mtDNA denotes for noncomplementary target and one nucleotide mismatched target, respectively.
Instruments
AuNPs were observed with transmission electron microscopy (TEM, FEI Tecnai G2 20, www.fei.com, operated at 200 kV) and atomic force microscopy (AFM, Solver NEXT, NT-MDT Spectrum Instruments, www.ntmdt-si.com). Modification of DNA on AuNPs was confirmed by using UV − vis spectroscopy (UV-2450 spectrophotometer, Shimadzu Scientific Instrument, Japan, www.ssi.shimadzu.com). DNA modification amount on AuNPs were characterized with fluorescence spectroscopy (F-4500FL spectrofluorometer, Hitachi, Japan, www.hitachi.com). HCR reaction on AuNPs was characterized by using UV − vis spectroscopy, DLS Nano Particle Size Analyzer (NanoPlus-3, Micromeritics Instrument ltd, www.micromeritics.com), and agarose gel electrophoresis (DYY-6D electrophoresis apparatus, Beijing Liuyi Instrument Factory, China, www.bjliuyi.com). The electrophoresis gels were imaged with an imaging system (G:BOX Chemi XT4, Syngene, UK, www.syngene.com). Electrochemical measurements were performed using an electrochemical analyzer (CHI660E, Shanghai Chenhua Instrument, China, www.chinstr.com) at room temperature (∼25 °C). They were with a three-electrode system: a gold working electrode, a platinum counter electrode, and an Ag|AgCl|3 M KCl reference electrode. All potentials in this paper are quoted versus this reference electrode. The working electrode and the counter electrode are disk electrodes with a diameter of 2 mm (CHI, www.chinstr.com).
Synthesis and characterization of iDNA-AuNPs-rpDNA
AuNPs with ∼13 nm in diameter were prepared by using the well-known Frens procedure [9, 32] (Supplementary Material). The AuNPs concentration was estimated based on its absorbance at 520 nm (Supplementary Material) [33]. The iDNA-AuNPs-rpDNA nanoparticles were synthesized through coupling iDNA-SH and rpDNA-SH on AuNPs with formation of Au-S bond. Before the coupling reaction, both iDNA-SH and rpDNA-SH were treated with TCEP for 1 h to reduce residual disulfide bonds. Then 36 μL various concentrations of iDNA-SH solution (15 μM, 18 μM, 20 μM, 22 μM, and 24 μM) was mixed with 8 μL 10 μM rpDNA-SH solution, followed by addition of 140 μL 50 mM PB, 10 μL 0.1 M EDTA, and 500 μL 3.6 nM AuNPs. The mixture was incubated at 30 °C for 5 h under protection from light. Then 2 M NaCl in 10 mM PB was gradually added to a final NaCl concentration of 0.6 M within 4 h. The reaction mixture was incubated for 16 h. The product was cleaned with centrifugation twice, and then kept at 4 °C. For determination of the DNA modification amount, Fam-iDNA-SH and Cy5-rpDNA-SH were used for DNA coupling, and the supernatant was subject to fluorescence analysis. The reaction system without AuNPs was used as control. The average number of iDNA and rpDNA molecules coupled on each AuNP was calculated based on DNA fluorescence−concentration calibration plots.
Confirmation of HCR with the designed iDNA, H1, and H2 sequences
To evaluate the feasibility of the designed iDNA, H1, and H2 sequences, HCR was executed from both free iDNA and the immobilized iDNA on AuNPs. Right before HCR, 0.5 μM H1 and 0.5 μM H2 solutions in the immobilization buffer, respectively, were denatured at 95 °C for 5 min, and then cooled to room temperature. At the same time, the free iDNA solution (0.5 μM) was denatured at 95 °C for 10 min, and then cooled in ice bath immediately. The free iDNA solution or the iDNA-AuNP-rpDNA nanoparticles (30 nM AuNPs) were mixed with the H1 and H2 solutions at equal volumes. The mixtures were shaken at 30 °C for 100 min for reaction. The H1/H2/iDNA-AuNP-rpDNA nanoparticles were washed with centrifugation and then re-suspended in the immobilization buffer. The H1/H2/iDNA product was characterized by agarose gel electrophoresis. The H1/H2/iDNA-AuNP-rpDNA product was characterized by UV − vis spectroscopy, agarose gel electrophoresis, and particle size analysis. In addition, to further confirm the happening of HCR on AuNPs, the H1/H2/iDNA-AuNP-rpDNA product was treated with dithiothreitol (DTT) to release DNA from the AuNPs. The DTT treated product was then subject to 2% agarose gel electrophoresis under 120 V using EB as a staining dye.
Biosensor fabrication and target DNA (tDNA) detection
The gold electrode was cleaned by being polished with 0.3 and 0.05 μm alumina powder, and then sonicated in ethanol bath and water bath consecutively. The electrode was then activated with cyclic voltammetry (CV) in 0.5 M H2SO4. Finally, it was sonicated in water and dried under nitrogen. For immobilization, the HS-cpDNA solution was treated with TCEP, and then diluted to various concentrations (0.05, 0.09, 0.1, 0.2, and 0.5 μM) with the immobilization buffer. The TCEP treated HS-cpDNA was immobilized on the activated gold electrode by applying a potential of +0.4 at the electrode for 500 s in the various dilutions of cpDNA solution. The electrode was then passivated via submergence in 1 mM MCH solution at 30 °C for 1 h. Finally, the electrode was washed, and then soaked in the immobilization buffer, ready for tDNA detection.
For detection of tDNA, 200 μL tDNA solution of various concentrations in the immobilization buffer was added onto the cpDNA/gold electrode surface. The electrode was kept at 40 °C for various periods (1 h, 2 h, 2.5 h, 3 h, and 4.5 h) under gentle shaking for DNA hybridization. Thereafter, the electrode was washed, and 200 μL of the iDNA-AuNP-rpDNA nanoparticle solution (1.8 nM based on AuNPs) was dropped onto the electrode surface. The electrode was sealed and kept at 47 °C for various periods (1 h, 2 h, 2.5 h, and 3 h), for hybridization of the labeled rpDNA with tDNA. This step resulted in iDNA-AuNP-rpDNA/tDNA/cpDNA sandwich DNA structure. The temperatures for DNA hybridization were set under consideration of the melting temperatures of the corresponding dsDNA. The later was estimated by using Mfold web server (http://unafold.rna.albany.edu). Then, after the electrode was washed, 200 μL 0.5 μM H1 solution and 200 μL 0.5 μM H2 solution in the immobilization buffer were dropped onto the electrode for HCR. The HCR was implemented at 110 rpm and 30 °C for various time periods (60 min, 80 min, 100 min, 120 min, and 180 min). The DNA structure on the electrode was then intercalated by [Ru(NH3)5L]2+ complex from the PB buffer (8 mM, pH 7.4) containing 80 mM KNO3 and 50 μM [Ru(NH3)5L]2+. The intercalation was realized via applying linear sweep voltammetry (LSV) scanning at the electrode. The LSV was run 200 cycles from −0.6 to 0.1 V at a scan rate of 0.1 V s−1. The electrode was then washed and subject to DPV detection (from −0.6 V to +0.1 V; amplitude, 0.05 V; pulse width, 0.01 s; pulse period, 0.02 s). All DPV curves were baseline-corrected using the software embedded in the CHI660E instrument. The oxidation peak current (Ip) at the potential of −0.28 V in the DPV detection was recorded, and the values were used as DNA detection signal intensities in the following quantitative comparisons, evaluations, and calibrations. For evaluation of the sensing selectivity, control DNA sequences and some surface-active serum components, including the ncDNA, the mtDNA, human serum albumin (HSA), glucose, ascorbic acid (AA), and dopamine, were also detected.
Electrochemical characterization of the biosensor fabrication process
The biosensor fabrication process was monitored and characterized by using CV and electrochemical impedance spectroscopy (EIS) techniques. The CV experiment was executed with 5 mM K3Fe(CN)6 in 1 M KCl as a probe. The EIS measurement was performed at +0.226 V in equimolar [Fe(CN)6]3−/4−(10/10 mM) supported by 0.1 M KCl with AC frequency from 0.1 Hz to 100 kHz. The effective surface area of the bare gold electrode was determined by running CVs in the 5 mM K3Fe(CN)6 solution, according to the Randles-Sevcik equation (Detailed in Supplementary Material). The surface density of the immobilized cpDNA was measured by using chronocoulometry technique with RuHex as a probe according to the Cottrell equation, as described by Steel et al. [34] (See Supplementary Material).
Results and discussion
Synthesis and characterization of iDNA-AuNP-rpDNA nanoparticles
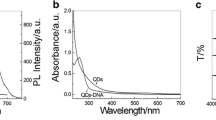
For synthesis of iDNA-AuNPs-rpDNA nanoparticles, AuNPs were firstly synthesized and characterized. Both TEM and AFM micrographs (Fig. S-1 in Supplementary Material) show that the AuNPs are monodispersed spherical particles. They have a narrow particle size distribution. The diameter of the AuNPs is 13 ± 1 nm, analyzed by using DigitalMicrograph software. The UV − vis spectrum of AuNPs (curve a in Fig. 1) exhibits an absorption peak at 520 nm. This is a surface plasmon resonance (SPR) band, characteristic for ~13 nm diameter AuNPs [35]. The UV − vis result is consistent with the TEM and the AFM images. After modification with iDNA and rpDNA (curve b in Fig. 1), only a modest shift of the SPR peak (from 520 to 523 nm) is observed, suggesting that the modified AuNPs are highly dispersed. The absorption peak at 260 nm (a characteristic absorption peak of nucleic acids) became pronounced with the DNA modification. This result suggests successful coupling of DNA on AuNPs.
The immobilization amount of rpDNA and iDNA on AnNPs was then determined. The fluorescence spectra and the fluorescence−concentration calibration plots of Cy5-rpDNA-SH and FAM-iDNA-SH are illustrated in the Supplementary Material (Fig. S-2 and Fig. S-3, respectively). The fluorescence analysis data (Table S-1, Supplementary Material) indicate that the feeding ratio of iDNA: rpDNA: AuNP = 333: 33: 1 (i.e., iDNA: rpDNA = 10:1) produces an iDNA-AuNPs-rpDNA with the composition of iDNA: rpDNA: AuNP = 240: 25: 1. This feeding ratio results in an optimum tDNA detection signal, as shown in the following Section 3.5. The immobilized iDNA: rpDNA ratio is similar with the feeding ratio, suggesting that these DNA molecules exhibit similar reaction rates with AuNPs.
Confirmation of HCR
Successful execution of HCR is prerequisite for the DNA biosensor. The gel electrophoresis image of the H1/H2/iDNA system shows many obvious bands of higher molecular weights (in the range of ~100 bp – 500 bp), in addition to a very vague band at the position of H1 and H2 (smaller than 100 bp) (Fig. 2a). This result indicates that HCR happens with the free iDNA. The immobilized iDNA on AuNPs can also successfully initiate HCR. Firstly, the UV-vis absorbance (at 260 nm) of the H1/H2/iDNA-AuNP-rpDNA HCR product (curve c in Fig. 1) is larger than that of the iDNA-AuNP-rpDNA nanoparticles, suggesting a higher DNA amount on AuNPs after HCR. Secondly, the dynamic diameter of the H1/H2/iDNA-AuNPs-rpDNA is ca. 99.4 nm (Fig. 3b), drastically larger than that of iDNA-AuNP-rpDNA nanoparticles (ca. 44.9 nm, Fig. 3a). Finally, the gel electrophoresis results further confirms successful HCR with the immobilized iDNA. As shown in Fig. 2a, the DTT treated H1/H2/iDNA-AuNPs-rpDNA HCR product shows the bands at the similar positions with the H1/H2/iDNA system. It should be noted that the iDNA-AuNPs-rpDNA shows a ‘black’ band, because the EB staining is optically quenched by the AuNPs. In contrast, in the images taken with a photo camera (Fig. 2b), the iDNA-AuNPs-rpDNA exhibits a wine-red band, coming from the plasma resonance of AuNPs. The H1/H2/iDNA-AuNPs-rpDNA HCR product shows many wine-red bands at larger molecular weight positions than iDNA-AuNPs-rpDNA, further confirming successful HCR with the immobilized iDNA. For the DTT treated H1/H2/iDNA-AuNPs-rpDNA HCR product, the blue-purple band at the sample loading position is from the aggregated AuNPs. Releasing of DNA by DTT replacement causes the aggregation of AuNPs.
Gel electrophoresis images that was (a) imaged on the G:BOX Chemi XT4 imaging system, and (b) taken with a photo camera. M: DNA molecular markers. The black band (in a) and the wine-red band (in b) of the iDNA-AuNPs-rpDNA nanoparticles are pointed in red circles. The feeding ratio of iDNA-SH to rpDNA-SH in preparation of the iDNA-AuNPs-rpDNA was 10:1. The HCR time was 100 min
Probing the biosensor fabrication process
In order to monitor and characterize the biosensor fabrication process, EIS and CV techniques were used. EIS with Fe(CN)63−/4− as probes was used to monitor the inter-facial resistance during the biosensor fabrication process. In the Nyquist plots (Fig. 4a), the semicircles at high frequency domain are mainly contributed by the charge-transfer resistance (Rct) and the double-layer capacitance (Cdl) of the electrode surface. The Rct values were determined with the assistance of ZSimpWin software (Princeton Applied Research) using two modified Randles circuits (Inset I for curve a, b, and c; Inset II for curve d, e, and f, in Fig. 4a). During the fabrication of the biosensor, the Rct value almost monotonically increased, except a slight decrease with tDNA hybridization on the MCH-cpDNA/gold electrode. Detailedly, the cpDNA/gold electrode exhibits an Rct value of 920 Ω (curve b), much bigger than that of the bare gold electrode (45 Ω, curve a). ssDNA is a non-conductive and negatively-charged molecule. Immobilization of cpDNA interferes the [Fe(CN)6]3−/4− anion probes from accessing the electrode surface. The surface density of the immobilized cpDNA was estimated through determination of the effective surface area of the electrode and the RuHex amount bound at the immobilized cpDNA (Fig. S-4 in Supplementary Material). It is ∼5.90 × 1012 molecules cm−2. After passivated with MCH, the Rct value of the electrode increases to 2990 Ω (curve c), indicating that the defects of the cpDNA layer are passivated. In contrast, capturing tDNA results in a slight decrease of the Rct value to 2790 Ω (curve d). The decrease in Rct may be the result of an Rct increase (due to the non-conductivity and negative-charge of tDNA), and an Rct decrease due to the higher electrical conductivity of dsDNA than ssDNA. Thereafter, both hybridization of the iDNA-AuNPs-rpDNA nanoparticles with the captured tDNA (curve e), and HCR (curve f) leads to the increases of the Rct values to 3290 Ω and 3860 Ω, respectively. Firstly, introduction of the nanoparticle complex sterically hinders the EIS probes from access to the electrode. Secondly, the negative charges of the introduced DNA electrostatically repels the probes from accessing the electrode surface.
(a) The EIS, and (b) the cyclic voltammograms of (a) bare gold, (b) cpDNA/gold, (c) MCH-cpDNA/gold, (d) tDNA/MCH-cpDNA/gold, (e) iDNA-AuNPs-rpDNA/tDNA/MCH-cpDNA/gold, and (f) H1/H2/iDNA-AuNPs-rpDNA/tDNA/MCH-cpDNA/gold electrodes, in (a) [Fe(CN)6]3−/4- (10/10 mM), and (b) 5 mM K3[Fe(CN)6], scan rates: 0.05 V s−1. The feeding ratio of iDNA-SH to rpDNA-SH in preparation of the iDNA-AuNPs-rpDNA was 10:1. The concentration of cpDNA and tDNA was 10−7 M and 10−13 M, respectively. The incubation time of tDNA as well as iDNA-AuNPs-rpDNA on the electrode were both 3 h. The HCR time was 2 h
The cyclic voltammograms (CVs) of the electrodes using [Fe(CN)6]3− as a probe are illustrated (Fig. 4b). The bare gold electrode displays a pair of well-defined redox peaks (curve a). The peak potential separation (ΔEp) is 62 mV, indicating a reversible electrochemical redox reaction of the probe. With the cpDNA immobilization and MCH passivation, the redox peak currents decreases from 25.8 Ampere (shorted as A) (curve a) to 23.6 A (curve b) and 13.7 A (curve c), respectively. These are accompanied by the increases of ΔEp to 73 mV and 201 mV, respectively. Both cpDNA immobilization and MCH passivation decrease the electrode electron transfer rates. In contrast, hybridization of tDNA with the immobilized cpDNA results in a very slight increase of the redox peak currents (14.1 A, curve d). For the iDNA-AuNPs-rpDNA hybridization (curve e), and the HCR (curve f) steps, the redox peak currents slightly decrease to 12.5 A and 11.2 A, respectively. The CV results are in agreement with the above EIS results.
Feasibility of signal amplification through HCR
HCR can form DNA concatamers with initiation of alternate assembly of two hairpins in the presence of iDNA. The enhancement effect of signal amplification with HCR in comparisons to that with our previous biobarcode (i.e. hpDNA) [9] was then evaluated. The DPV signals of the biosensors in response to 100 fM tDNA are shown in Fig. 5. The DPV oxidation peak at the peak potential of ca. -0.28 V is from the electrochemical oxidation of the intercalated [Ru(NH3)5L]2+ molecules. The DPV signal of the HCR-based DNA biosensor (curve c) is significantly stronger than that of the hpDNA-based DNA biosensor (curve b), and conceivably larger than that without the execution of HCR (curve a). The many long DNA concatamers (about 100–500 bp, as determined in the above gel electrophoresis measurement) formed in the HCR process can accommodate much more [Ru(NH3)5L]2+ molecules than the 15 bp hpDNA (sequence shown in Table 1), thus dramatically enhancing the DPV detection signal.
The DPV signals in response to 100 fM tDNA of the sandwich biosensors of various compositions. (a) With iDNA as a biobarcode without HCR (i.e. iDNA-AuNPs-rpDNA/tDNA/MCH-cpDNA/gold), (b) with hpDNA as a biobarcode (i.e. hpDNA-AuNPs-rpDNA/tDNA/MCH-cpDNA/gold, (c) with iDNA initiating HCR, forming long DNA concatamers as biobarcode (i.e. H1/H2/iDNA-AuNPs-rpDNA/tDNA/MCH-cpDNA/gold). The feeding ratio of iDNA-SH to rpDNA-SH in preparation of the iDNA-AuNPs-rpDNA was 10:1. The concentration of cpDNA and tDNA was 10−7 M and 10−13 M, respectively. The incubation time of tDNA as well as iDNA-AuNPs-rpDNA on the electrode were both 3 h. The HCR time was 2 h. Inset: the corresponding statistic Ip values with standard deviation as the error bar (independent biosensor preparation and detection number, n = 3). ** and *** represent that the p values are less than 0.01 and 0.001, respectively, in the t test
Biosensor optimization and analytical performances
The biosensor fabrication conditions were then optimized. The DPV detection signals in response to 100 fM tDNA reach peak values at the HS-cpDNA concentration of 9 × 10−8 M (Fig. S-5A), the iDNA: rpDNA feeding ratio of 10:1 (Fig. S-5B), and the HCR time of 100 min (Fig. S-5C), respectively. In contrast, the DPV signals attain to plateaus with the tDNA incubation time and the iDNA-AuNPs-rpDNA incubation time increased to 3 h (Fig. S-5D) and 2.5 h (Fig. S-5E), respectively.
At the optimum fabrication conditions, the analytical performances of the DNA biosensor were evaluated. The DPV curves for various concentrations of tDNA are illustrated in Fig. 6a. The semilogarithmic plot of DPV peak current (Ip) versus the logarithmic tDNA concentration (CtDNA) (insets in Fig. 6a) exhibits two linear ranges: from 0.01 fM to 0.5 fM, and from 1 fM to 100 fM. The regression equations are Ip (μA) = 0.984 + 0.053 × lg(CtDNA) (M), and Ip (μA) = 7.82 + 0.51 × lg(CtDNA) (M), with regression coefficients of 0.9538 and 0.9993, respectively. The lower detection limit (LOD) of the DNA sensor (S/N = 3) is 0.68 × 10−18 M (i.e. 0.68 aM), much lower than that of our hpDNA-based DNA biosensor (1 fM) [9]. The LOD value of the DNA biosensor is compared with those of some nanostructure-based electrochemical and electrochemiluminescence sandwich DNA sensors [6, 7, 9, 10, 17] (Table S-2). The DNA sensor reported in this paper exhibits dramatically higher sensitivity than most of the reported values. The ultrahigh detection sensitivity is attributed to the triple-amplification strategies: (a) Many iDNA molecules (about 240) are immobilized on each AuNP. (b) Each single iDNA gives rise to formation of a long DNA concatamer (about 100 bp – 500 bp long) via HCR. (c) Each dsDNA chain can be intercalated by many [Ru(NH3)5L]2+ probes. This gives a strong electrochemical signal. It is known that about four bases of dsDNA bind one probe [28]. The presentation of two linear ranges in the calibration plot (Fig. 6a) should be a result of the ‘semilogarithmic’ scale function. For reasoning, the plot of Ip versus CtDNA is illustrated (Fig. S-6). It can be seen that the Ip value increases faster at lower CtDNA. This phenomenon is very often and reasonable for chemical/biological analysis. At low substrate/analyte concentration, their conversion ratio, here the DNA hybridization efficiency, is higher than that at high substrate concentration. However, in the ‘semilogarithmic’ plot, the ‘logarithmic’ function of the CtDNA does not reflect an equidifferent scale. At low CtDNA value, the ‘CtDNA difference’ for the same value of ‘logarithmic difference’ is much smaller than that at high CtDNA value, resulting in a lower linear slope at the low lgCtDNA value in the ‘semilogarithmic’ plot.
DPV curves recorded from the DNA biosensor, (a) in response to difference concentrations of tDNA (from curve a to h, the CtDNA was 0, 0.01, 0.05, 0.50, 1, 10, 50, and 100 fM, respectively), and (b) in response to (a) blank PB, (b) 10−13 M ncDNA, (c) 10−13 M mtDNA, and (d) 10−13 M tDNA. Inset in A: the semilogarithmic plots of Ip vs. lg(CtDNA). Plot I and Plot II in the inset are the two linear ranges with standard deviation as the error bar, and dotted lines showing 95% confidence intervals. Inset in B: bar plot of the corresponding statistic Ip values. The repetitive experimental number (i.e. independent biosensor preparation and detection number, n) was 3. ** and *** represent that the p values are less than 0.01 and 0.001, respectively, in the t test
The selectivity of the HCR-based sandwich DNA sensor was evaluated by detecting the control DNA sequences (Fig. 6b) and some human serum components (Fig. S-7), including 10−13 M ncDNA, 10−13 M mtDNA, 10 mg mL−1 HSA, 4.4 mM glucose, 10.0 mM AA, and 10.0 mM dopamine. The DPV signal towards the ncDNA is very small (Ip, 69.7 nA). The intensity is similar to that towards the blank sample (63.6 nA). For the mtDNA, the detection signal is 127.8 nA, about twice the signal towards the ncDNA, but one-tenth of that towards the complementary tDNA (1.39 μA). The DPV signals towards the human serum components are similar or less than that towards the blank sample. These results indicate that the DNA sensor exhibits very good sensing selectivity to tDNA. One single mismatched DNA can be differentiated from the complementary tDNA. The signal molecule [Ru(NH3)5L]2+ binds with the H1/H2/iDNA-AuNPs-rpDNA/tDNA/MCH-cpDNA biosensor interface mainly through dsDNA intercalation binding mode [9], thus can greatly reduce background noise and interferences. The sample detection time with the DNA biosensor is about 7 h. This detection time is not very ideal, but acceptable for clinical diagnosis. Further improvement for DNA biosensor design and fabrication is ongoing in our group.
Conclusions
An ultrasensitive and selective electrochemical biosensor for detecting DNA sequences has been developed. The biosensor achieves the high sensitivity by multiplying three amplification strategies: AuNPs serving as the nano-substrate for immobilization of high amount iDNA; HCR converting the iDNA to long dsDNA concatamer; electrochemical active signal molecule [Ru(NH3)5L]2+ intercalating into dsDNA with multiple intercalators on each dsDNA chain. In addition, direct intercalative binding of the [Ru(NH3)5L]2+ signal indicator obviates complicated covalent modification procedures in biosensor fabrication, and minimizes background and interferences in detection. This ultrasensitive DNA biosensor should be able to detect H. pylori in physiological samples, and the sensor fabrication approach may provide a general platform for detection of various interest DNA in many important applications.
References
Pournaghi-Azar MH, Alipoura E, Zununi S, Froohandeh H, Hejazi MS (2008) Direct and rapid electrochemical biosensing of the human interleukin-2 DNA in unpurified polymerase chain reaction (PCR)-amplified real samples. Biosens Bioelectron 24:524–530
Huang S, Feng MM, Li JW, Liu Y, Xiao Q (2018) Voltammetric determination of attomolar levels of a sequence derived from the genom of hepatitis B virus by using molecular beacon mediated circular strand displacement and rolling circle amplification. Microchim Acta 185:206
Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N (1985) Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230:1350–1354
Makrigiorgos GM, Chakrabarti S, Zhang Y, Kaur M, Price BD (2002) A PCR-based amplification method retaining the quantitative difference between two complex genomes. Nat Biotechnol 20:936–939
Schweitzer B, Kingsmore S (2001) Combining nucleic acid amplification and detection. Curr Opin Biotechnol 12:21–27
Nam JM, Stoeva SI, Mirkin CA (2004) Bio-bar-code-based DNA detection with PCR-like sensitivity. J Am Chem Soc 126:5932–5933
Yu XX, Chai Y, Jiang J, Cui H (2012) Sensitive ECL sensor for sequence-specific DNA from Mycobacterium tuberculosis based on N-(aminobutyl)-N-ethylisoluminol functionalized gold nanoparticles labeling. J Photochem Photobiol A 241:45–51
Dong XY, Mi XN, Zhang L, Liang TM, Xu JJ, Chen HY (2012) DNAzyme-functionalized Pt nanoparticles/carbon nanotubes for amplified sandwich electrochemical DNA analysis. Biosens Bioelectron 38:337–341
Cui HF, Xu TB, Sun YL, Zhou AW, Cui YH, Liu W, Luong JHT (2015) Hairpin DNA as a biobarcode modified on gold nanoparticles for electrochemical DNA detection. Anal Chem 87:1358–1365
Ye Y, Liu Y, He S, Xu X, Cao X, Ye Y, Zheng H (2018) Ultrasensitive electrochemical DNA sensor for virulence invA gene of Salmonella using silver nanoclusters as signal probe. Sens Actuator B-Chem 272:53–59
Thiruppathiraja C, Kamatchiammal S, Adaikkappan P, Santhosh DJ, Alagar M (2011) Specific detection of Mycobacterium sp. genomic DNA using dual labeled gold nanoparticle based electrochemical biosensor. Anal Biochem 417:73–79
Hsieh KW, Patterson AS, Ferguson BS, Plaxco KW, Soh HT (2012) Rapid, sensitive, and quantitative detection of pathogenic DNA at the point of care through microfluidic electrochemical quantitative loop-mediated isothermal amplification. Angew Chem-Int Edit 51:4896–4900
Chen Y, Xu J, Su J, Xiang Y, Yuan R, Chai YQ (2012) In situ hybridization chain reaction amplification for universal and highly sensitive electrochemiluminescent detection of DNA. Anal Chem 84:7750–7755
Wang XZ, Ge L, Yu YF, Dong SS, Li F (2015) Highly sensitive electrogenerated chemiluminescence biosensor based on hybridization chain reaction and amplification of gold nanoparticles for DNA detection. Sens Actuator B-Chem 220:942–948
Chen Z, Liu Y, Xin C, Zhao J, Liu S (2018) A cascade autocatalytic strand displacement amplification and hybridization chain reaction event for label-free and ultrasensitive electrochemical nucleic acid biosensing. Biosens Bioelectron 113:1–8
Deng X, Wang C, Gao Y, Li J, Wen W, Zhang X, Wang S (2018) Applying strand displacement amplification to quantum dots-based fluorescent lateral flow assay strips for HIV-DNA detection. Biosens Bioelectron 105:211–217
Shi AQ, Wang J, Han XW, Fang X, Zhang YZ (2014) A sensitive electrochemical DNA biosensor based on gold nanomaterial and graphene amplified signal. Sens Actuator B-Chem 200:206–212
Wang J, Li HG, Li TT, Ling LS (2018) Determination of bacterial DNA based on catalytic oxidation of cysteine by G-quadruplex DNAzyme generated from asymmetric PCR: application to the colorimetric detection of Staphylococcus aureus. Microchim Acta 185:410
Li Z, Mao GB, Du MY, Tian SB, Niu LQ, Ji XH, He ZK (2019) A fluorometric turn-on aptasensor for mucin 1 based on signal amplification via a hybridization chain reaction and the interaction between a luminescent ruthenium(II) complex and CdZnTeS quantum dots. Microchim Acta 186:233
Zhou C, Zou H, Sun C, Ren D, Chen J, Li Y (2018) Signal amplification strategies for DNA-based surface plasmon resonance biosensors. Biosens Bioelectron 117:678–689
Ikbal J, Lim GS, Gao ZQ (2015) The hybridization chain reaction in the development of ultrasensitive nucleic acid assays. Trends Anal Chem 64:86–99
Dirks RM, Pierce NA (2004) Triggered amplification by hybridization chain reaction. Proc Natl Acad Sci U S A 101:15275–15278
Ge SG, Zhao JG, Wang SP, Lan FF, Yan M, Yu JH (2018) Ultrasensitive electrochemiluminescence assay of tumor cells and evaluation of H2O2 on a paper-based closed-bipolar electrode by in-situ hybridization chain reaction amplification. Biosens Bioelectron 102:411–417
Zhou Q, Lin YX, Zhang KY, Li MJ, Tang DP (2018) Reduced graphene oxide/BiFeO3 nanohybrids-based signal-on photoelectrochemical sensing system for prostate-specific antigen detection coupling with magnetic microfluidic device. Biosens Bioelectron 101:146–152
Cover TL, Blaser MJ (2009) Helicobacter pylori in health and disease. Gastroenterology 136:1863–1873
Salama NR, Hartung ML, Muller A (2013) Life in the human stomach: persistence strategies of the bacterial pathogen helicobacter pylori. Nat Rev Microbiol 11:385–399
Chen LL, Cui HF, Fan SF, Li ZY, Han SY, Ma X, Luo SW, Song XJ, Lv QY (2018) Detection of helicobacter pylori in dental plaque using a DNA biosensor for noninvasive diagnosis. RSC Adv 8:21075–21083
García T, Revenga-Parra M, Abruna H, Pariente F, Lorenzo E (2008) Single-mismatch position-sensitive detection of DNA based on a bifunctional ruthenium complex. Anal Chem 80:77–84
García T, Casero E, Revenga-Parra M, Martín-Benito J, Pariente F, Vázquez L, Lorenzo E (2008) Architectures based on the use of gold nanoparticles and ruthenium complexes as a new route to improve genosensor sensitivity. Biosens Bioelectron 24:184–190
Brown AP, Anson FC (1977) Molecular anchors for the attachment of metal complexes to graphite electrode surfaces. J Electroanal Chem 83:203–206
Kuehn CG, Taube H (1976) Ammineruthenium complexes of hydrogen sulfide and related sulfur ligands. J Am Chem Soc 98:689–702
Frens G (1973) Controlled nucleation for the regulation of the particle size in monodisperse gold suspensions. Nature (London). Phys Sci 241:20–22
Rance GA, Marsh DH, Khlobystov AN (2008) Extinction coefficient analysis of small alkanethiolate-stabilised gold nanoparticles. Chem Phys Lett 460:230–236
Steel AB, Herne TM, Tarlov MJ (1998) Electrochemical quantitation of DNA immobilized on gold. Anal Chem 70:4670–4677
Storhoff JJ, Elghanian R, Mucic RC, Mirkin CA, Letsinger RL (1998) One-pot colorimetric differentiation of polynucleotides with single base imperfections using gold nanoparticle probes. J Am Chem Soc 120:1959–1964
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (NSFC 21345007), the Plan for Scientific Innovation Talent of Henan Province to H. F. Cui (Grant number 154200510007), the Natural Science Foundation of Henan Province (Grant number 182300410314), and the Henan Open-up and Collaboration Program of Science and Technology (Grant number 132106000070).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
ESM 1
(DOCX 773 kb)
Rights and permissions
About this article
Cite this article
Lv, MM., Fan, SF., Wang, QL. et al. An enzyme-free electrochemical sandwich DNA assay based on the use of hybridization chain reaction and gold nanoparticles: application to the determination of the DNA of Helicobacter pylori. Microchim Acta 187, 73 (2020). https://doi.org/10.1007/s00604-019-3999-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-019-3999-z