Abstract
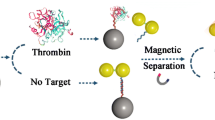
A one-step sandwich method is described for detecting proteins with magnetic nanospheres (MNs) and fluorescent nanospheres (FNs). Thrombin is selected as a model analyte to validate the method. Two DNA aptamers (Apt 29 and Apt 15 targeting two different exosites of thrombin) are chosen as recognition elements to modify MNs and FNs. The superparamagnetic MN-Apt 29 conjugate is used to separate and concentrate thrombin. The FN-Apt 15 conjugate encapsulates hundreds of fluorescent quantum dots and is used as reporter to provide a stable signal. Magnetic capture and fluorescence identification are performed simultaneously to form a sandwich complex (MN-Apt 29-thrombin-FN-Apt 15) for fluorescence determination (at excitation/emission wavelengths of 380/622 nm). The method is convenient, time saving, and gives a strong signal (compared to the two-step method where capture and identification are performed in two steps). The one-step method presented here is completed within 30 min and has a 3.5 ng·mL−1 (97 pM) detection limit. The method is reproducible, has an intra-assay variability of 1.5%, and an inter-assay variability of 4.9%. Other serum proteins (HSA, CEA, PSA, and AFP) do not interfere. The method was also applied to analyze serum samples. Almost the same fluorescence intensity was measured when analyzing 1% serum samples (compared to buffer samples).

Magnetic nanospheres with excellent superparamagnetic property and fluorescent QD-based nanospheres were prepared and used in a one-step sensitive method for detecting thrombin. The method exhibits good reproducibility, high specificity, and good selectivity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Protein biomarkers are one of most important kinds of biomarkers. Their detection is of great significance to clinical application [1]. For instance, thrombin is a central enzyme involved in platelet aggregation and blood coagulation. It plays important roles in hemostasis, inflammation, thrombosis, angiogenesis, tumor growth and metastasis. It has become an important therapeutic and biomarker for diseases associated with coagulation abnormalities [2, 3]. So far, in practical applications, chromatography/mass spectrometry-based methods and antibody-based immunoassays (e.g., enzyme-linked immunosorbent assay, ELISA) are still the most commonly used diagnostic methods for protein detection [4,5,6]. These methods have good accuracy. But they suffer from the disadvantages of time consuming, laborious and complex manipulation, high cost, or requiring sophisticated instruments and skilled technical staffs. Hence, sensitive, convenient, and reliable method for detecting protein biomarkers has always been pursued.
With the rapid progress of nanoscience and nanotechnology, numerous nanomaterials have been applied to biological detection and medical diagnosis. Quantum dots (QDs) and magnetic nanoparticles have shown great superiority and potential [7, 8]. QDs are newly developed versatile fluorescent nanoparticles. They have unique optical properties, such as good photochemical stability, high photoluminescence quantum yields, size/composition-dependent and narrow emission spectra with broad absorption, etc. QDs have become excellent report probes for improving detection sensitivity and multiplexed analysis [9, 10]. Magnetic nanoparticles exhibit superparamagnetic properties at room temperature. They behave just like a giant paramagnetic atom, and they have fast magnetic response and negligible remanence. These properties avoid magnetic nanoparticles agglomerating and allow them to be manipulated easily just by a magnet [11, 12]. Magnetic nanoparticles-based separation techniques have been used for efficient enrichment and purification in complex sample analysis. They greatly reduce interference and enhance signal sensitivity [13, 14].
Nowadays, QDs and magnetic nanoparticles have been widely applied to protein detections. However, despite their promising potential, some challenges still exist in practical operation. For instance, their optical or magnetic properties are usually affected during the modification or biofunctionalization process. They suffer from the problems of aggregation, nonspecific adsorption, or colloidal instability in complex matrices [7, 9, 15]. In our previous work, by incorporating QDs and magnetic nanoparticles with polymer nanospheres, we constructed fluorescent and magnetic nanospheres (FNs and MNs) [16, 17]. The FNs and MNs inherit the unique fluorescence and magnetic properties of QDs and magnetic nanoparticles. Moreover, they also show superiority: high stability, amplified signal, great versatility, and convenient manipulation. Accordingly, the FNs and MNs may be expected as promising tools for protein biomarker detection.
In the present work, a one-step convenient and sensitive method for detecting protein biomarkers is developed utilizing our prepared MNs and FNs. Thrombin is used as a model protein. Aptamers are used as the recognition elements. The aptamers exhibit the superiority over antibodies. They have good affinity and specificity, convenient generation, low cost, easy modification, and high resistance against denaturation [18]. In the method, MNs and FNs are respectively modified with Apt 29 (29-base DNA aptamer targeting the heparin-binding exosite of thrombin) and Apt 15 (15-base DNA aptamer targeting the fibrinogen-recognition exosite of thrombin) [19, 20]. Then, the MN-Apt 29 and FN-Apt 15 are used to specifically capture and recognize thrombin simultaneously. This method is completed within 30 min, and has a detection limit of 3.5 ng·mL−1 (97 pM). It is more time-saving and can get higher detection signal than the two-step method. (Capture and identification are performed separately in the two step method.) Besides, this method exhibits good reproducibility, high specificity, and can be applied in serum samples. Thus, this method has potential application in practice.
Experimental
Materials and reagents
Carboxyl-terminated poly(styrene/acrylamide) polymer nanospheres (Pst-AAm-COOH) were prepared by emulsifier-free polymerization method [21]. Hydrophobic oleic acid-capped magnetic nanoparticles were synthesized by high-temperature pyrolysis [17]. Hydrophobic trioctylphosphine oxide (TOPO)-capped CdSe/ZnS QDs (emission wavelength: 620 nm) were purchased from Wuhan Jiayuan QDs Co., Ltd. (http://www.qds.net.cn/). Branched poly(ethylene imine) (PEI), bovine serum albumin (BSA), tetraethyl orthosilicate (TEOS), (3-aminopropyl)triethoxysilane (APTES), polyvinylpyrrolidone (PVP), human serum albumin (HSA), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), and N-hydroxysuccinimide (NHS) were purchased from Sigma-Aldrich. (https://www.sigmaaldrich.com/china-mainland.html). Human α-thrombin was bought form Haematologic Technologies, Inc. (https://www.haemtech.com/). Streptavidin (SA) was purchased from Amresco. (http://www.amresco-inc.com/). Difco skim milk was got from Becton, Dickinson and Company. (http://www.bd.com/). Carcinoembryonic antigen (CEA), Human alpha fetoprotein (AFP), and prostate specific antigen (PSA) were purchased from Nanjing sanchen biotechnology Co., Ltd. (http://njsc2015.bioon.com.cn/). Fetal bovine serum (FBS) was bought from Gibco. (https://www.thermofisher.com/cn/zh/home/brands/gibco.html). The biotinylated 15-base DNA aptamer (Apt 15) and 29-base DNA aptamer (Apt 29) against Human α-thrombin were purchased from Invitrogen Corp. (https://www.thermofisher.com/cn/zh/home/brands/invitrogen.html). The aptamers have the following sequences, and an oligodeoxythymidine (oligo(dT)) spacer (12 T) was added to reduce the binding steric hindrance:
-
Apt 15: Biotin-5′-TTT TTT TTT TTT GGT TGG TGT GGT TGG-3′
-
Apt 29: Biotin-5′-TTT TTT TTT TTT AGT CCG TGG TAG GGC AGG TTG GGG TGA CT-3′
Apparatus
Transmission electron microscopy (TEM) images were got by an electron microscope (FEI Tecnai G2 20 TWIN). Fluorescence images were acquired by an inverted fluorescence microscope (Ti-U, Nikon, Japan) mounted with a CCD camera (Nikon digital sight DS-U3). Magnetic hysteresis loops were determined by a Lake Shore 7410 vibrating sample magnetometer. Magnetic separation was performed with a commercial magnetic scaffold (Invitrogen, 12320D, the field strength on the surface of the magnetic scaffold is 325 ± 25 mT). Fluorescence emission spectra were recorded with a fluorescence spectrometer (Hitachi, F-7000).
Construction of thrombin-targeting MNs and FNs
The MNs and FNs were respectively prepared with embedding method and assembly method according to our previous work. The detail procedures are described in the Electronic Supporting Material (S.1). The MNs and FNs were first modified with SA by carbodiimide chemistry, and then coupled with the biotinylated DNA aptamer via biotin-streptavidin conjugation (Scheme 1). Typically, 5 mg of MNs or FNs were dispersed in 1 mL of phosphate buffer (0.01 M pH 7.2) containing 50 mM EDC to react with 0.1 mg of SA. They were shaken gently at room temperature. After 4 h reaction, the MNs were separated by magnetic attraction and the FNs were separated by centrifugation. They were washed five times by phosphate buffer to remove any unreacted SA. Then, the SA conjugated MNs and FNs were respectively incubated with 10 μL of biotin-Apt 29 (100 mM) and 10 μL of biotin-Apt 15 (100 mM) for 2 h at room temperature. Afterwards, the resultants were washed with phosphate buffer to remove any unreacted aptamer. The thrombin-targeting MNs and FNs (MN-Apt 29 and FN-Apt 15) were obtained. Finally, they were blocked with 1% BSA- phosphate buffer and stored at 4 °C for use.
Thrombin detection protocols
Two kinds of strategies (one-step method and two-step method) were investigated for detecting thrombin, as illustrated in Scheme 2. In the one-step method, 0.10 mg of MN-Apt 29 and 0.90 mg of FN-Apt 15 were simultaneously added to 1 mL of sample (thrombin in buffer containing 50 mM Tris, pH 7.4, 140 mM NaCl, 1 mM MgCl2, 5 mM KCl, and 0.1% skim milk). After a gentle shaking for 30 min, the sandwich complexes were separated with a magnetic scaffold. They were washed four times by the buffer and once by the buffer without skim milk. Finally, they were resuspended in 400 μL of skim milk-free buffer for fluorescence spectrum measurements (Excitation wavelength: 380 nm; Emission wavelength: 622 nm). In the two-step method, 0.10 mg of MN-Apt 29 was first added to the sample solution for incubation (30 min) to capture thrombin. Then, the MN-Apt 29-thrombin complexes were separated with a magnetic scaffold and resuspended in the buffer. Then, 0.90 mg of FN-Apt 15 was added. After incubation for 30 min, the complexes were washed similarly with the one-step strategy and detected by a fluorescence spectrometer.
Detection of thrombin in serum samples
Thrombin (0.15 μg) was spiked into 1 mL of diluted FBS (10, 100, and 1000 times dilution of FBS with the above buffer) as synthetic samples. MN-Apt 29 and FN-Apt 15 were added to the serum samples following the above one-step detection procedure. Control experiments were performed under the same condition except no thrombin was added.
Results and discussion
Choice of materials
Protein biomarkers usually exist in complex body fluid with low concentrations. This makes their detection suffer from the problems of interference and weak signal. Hence, on the one hand, our prepared MNs with excellent superparamagnetic properties, high stability, and efficient recognition capacity are used to capture target proteins from complex matrices [17]. This is expected to remove the contaminants, and achieve purification and enrichment of target proteins. On the other hand, our prepared FNs encapsulating hundreds of QDs are used as reporters [16]. This is expected to give highly amplified signal. The characterizations of the FNs and MNs are shown in the Electronic Supporting Material (S.2) to confirm their good fluorescence and magnetic properties. Based on these considerations, we infer that the combination of MNs and FNs can improve the sensitivity of protein detection.
Selection of the favourable detection protocol
DNA aptamers against thrombin have been extensively investigated. Two specific aptamers (donated as Apt 29 and Apt 15) have been used most widely. Apt 29 contains 29 bases targeting the heparin-binding exosite of thrombin. Apt 15 contains 15 bases mainly interacting with the fibrinogen-recognition exosite of thrombin [19, 20]. In this method, Apt 29 and Apt 15 are chosen for the sandwich assay: Apt 29 modified MNs are used to capture thrombin, and Apt 15 modified FNs are used to identify thrombin. It is noteworthy that during the modification, an oligo(dT) spacer (12T) rather than a commonly used dodecyl is introduced between the nanospheres and the aptamers. That’s because a negatively charged, flexible oligo(dT) spacer can induce much more effective binding than a dodecyl spacer [22]. For thrombin detection, one-step method and two-step method were both investigated (Scheme 2). The detection results are shown in Fig. 1. It can be seen that the one-step strategy achieve higher signal-to-noise ratio (the ratio of the fluorescence intensity detected from positive sample to that from blank sample, S/N) compared with the two-step strategy. As mentioned above, Apt 29 and Apt 15 mainly recognize different sites of the thrombin. Thus, there is little competition over the binding sites between them. However, in the two-step method, MN-Apt 29 is first added to the samples and preferentially binds with thrombin. The formed MN-Apt 29-thrombin complexes may need overcoming larger steric hindrance to bind with FN-Apt 15. This probably induces less FN-Apt 15 binding and lower fluorescence signal compared with the one-step strategy where free thrombin interacting with FN-Apt 15. Apart from the enhanced signal and higher sensitivity, the one-step method also exhibits more convenient and more time-saving manipulation. Thus, one-step strategy is chosen for thrombin detection rather than two-step strategy. In addition, we investigated the effect of exchanging capture aptamer and identification aptamer on the detection signal. MN-Apt 15 and FN-Apt 29 were used for one-step detecting thrombin. Compared with the initial design (one-step method with MN-Apt 29 and FN-Apt 15), the latter shows lower S/N (Fig. 1). The MNs have larger size (350 nm) compared with the FNs (200 nm), as shown in Fig. S2 in the Electronic Supporting Material. Larger size makes the MNs have larger steric hindrance and lower flexibility in bio-recognition. As reported, Apt 29 binds to thrombin with 20 to 50-fold higher affinity than Apt 15 [19, 20]. Thus, we presume that using MN-Apt 29 for capture can compensate for the disadvantages of MNs in recognizing thrombin, thereby achieving higher detection signal than using MN-Apt 15. Based on the above results, the one-step method with MN-Apt 29 and FN-Apt 15 is selected as the favourable protocol for detecting thrombin.
Optimization of the method
The following parameters were optimized: (a) Blocking condition; (b) Incubation temperature; (c) MN-Apt 29 dosage; (d) FN-Apt 15 dosage; and (e) Incubation time. Respective data and Figures (S.3) are given in the Electronic Supporting Material. The following experimental conditions are chosen: (a) a blocking condition of 0.1% skim milk; (b) an incubation temperature of 30 °C; (c) a MN-Apt 29 dosage of 0.10 mg; (d) a FN-Apt 15 dosage of 0.90 mg; and (e) an incubation time of 30 min.
Reliability and specificity
To investigate the specificity of this method, this method was used to detect 0.5 μg·mL−1 thrombin as positive samples, several types of serum proteins (HSA, CEA, PSA, and AFP, 7.5 μg·mL−1) as negative controls, and blank buffer samples. Fluorescence intensities collected from the six samples are shown in Fig. 2a, and typical fluorescence spectra are in Fig. 2b. The positive samples exhibit strong FN fluorescence. No obvious FN peak is found in the negative samples and their fluorescence intensity is almost as low as that of the blank samples. These results suggest that those nonspecific proteins with concentrations even more than 10 times higher than that of the thrombin do not produce detectable fluorescence signal. These confirm that the aptamer-based sandwich method has good specificity and selectivity. The intra-assay variability (variation coefficient of the results from the parallel samples detected with the same batch of thrombin-targeting nanospheres) and inter-assay variability (variation coefficient of the results from the parallel samples detected with different batches of thrombin-targeting nanospheres) are calculated as 1.5 and 4.9%. These indicate this method has good reproducibility.
Quantitative analysis and detection limit
Quantitative analysis is achieved by measuring the fluorescence intensity of the sandwich complexes. The fluorescence intensities of samples at different thrombin concentrations were recorded by a fluorescence spectrometer. As shown in Fig. 3, a linear relationship is exhibited at the concentrations from 10 to 400 ng·mL−1. Furthermore, by calculating the ratio of 3 times the standard deviation of the blanks to the slope of the linear equation of the calibration curve, the limit of the detection (LOD) is calculated as 3.5 ng·mL−1 (LOD = 3SD/slope, n = 11). Based on the reported molecular weight of the thrombin (36 kDa) [23], the one-step method quantitatively detects thrombin with the concentrations ranging from 0.3 to 11.1 nM with a LOD of 97 pM.
Detection of thrombin in serum samples
Thrombin is a plasma protein, and thus it is essential that this new method can be applied in serum. To assess the potential application of this one-step sandwich method in practice, FBS was used to mimic real samples for detection. As shown in Fig. 4, the fluorescence intensities measured from the 1000-fold and 100-fold diluted serum samples (0.1% FBS and 1% FBS) are very close to those obtained with plain buffer samples. The fluorescence signals from the 10-fold diluted serum samples (10% FBS) obviously decrease. The negative 0.1 and 1% FBS samples and the buffer samples exhibit similar low background signals. The negative 10% FBS samples generate slightly higher signals. These results suggest that serum diluted 100 times and higher dose not significantly interfere with the detection signal. But lower dilutions of serum reduce the detection signal attributing to the complex matrices in the serum. Therefore, to ensure detection accuracy, serum samples should be diluted 100 times for the one-step sandwich detection. This is acceptable given the high sensitivity of this method.
Conclusions
In summary, we report a one-step sandwich method for detecting thrombin. MN-Apt 29 and FN-Apt 15 are used to capture and identify thrombin simultaneously. This method can be completed within 30 min, and has high sensitivity with picomolar detection limit. Compared with most of the recently reported methods for thrombin detection (Table 1), our method has lower or comparable detection limit, and consumes much less time. (The method based on CdTe quantum dots and Pt nanoparticles achieves much higher sensitivity, yet it takes too much time; The method with MnO2 nanosheets just needs 15 min, yet it has much lower sensitivity.) Besides, our method has good reproducibility, high specificity, and can be manipulated conveniently with low cost. It dose not require sophisticated instruments, complex pretreatment or highly trained personnel. Finally, this method is successfully applied to 100-fold diluted serum samples, indicating its potential application in practice. Furthermore, the concept of this method can be expanded to other biomarker detection and can be designed for simultaneously detecting multiple biomarkers with multicolor reporters. While it should be noted that this sandwich strategy has a major limitation. The target protein should have at least two binding sites for respectively combining MNs and FNs to form the sandwich structure. Nonetheless, this method is still expected for sensitive, reliable and rapid detection of thrombin and facilitating research on diseases associated with coagulation abnormalities.
References
Wu L, Qu X (2015) Cancer biomarker detection: Recent achievements and challenges. Chem Soc Rev 44(10):2963–2997. https://doi.org/10.1039/c4cs00370e
Coughlin SR (2000) Thrombin signalling and protease-activated receptors. Nature 407(6801):258–264. https://doi.org/10.1038/35025229
Ebrahimi S, Rahmani F, Behnam-Rassouli R, Hoseinkhani F, Parizadeh MR, Keramati MR, Khazaie M, Avan A, Hassanian SM (2017) Proinflammatory signaling functions of thrombin in cancer. J Cell Physiol 232(9):2323–2329. https://doi.org/10.1002/jcp.25753
Cai W, Tucholski T, Chen B, Alpert AJ, McIlwain S, Kohmoto T, Jin S, Ge Y (2017) Top-down proteomics of large proteins up to 223 kDa enabled by serial size exclusion chromatography strategy. Anal Chem 89(10):5467–5475. https://doi.org/10.1021/acs.analchem.7b00380
Zhao Y, Liu G, Yuan X, Gan J, Peterson JE, Shen JX (2017) Strategy for the quantitation of a protein conjugate via hybrid immunocapture-liquid chromatography with sequential HRMS and SRM-Based LC-MS/MS analyses. Anal Chem 89(9):5144–5151. https://doi.org/10.1021/acs.analchem.7b00926
Liew OW, Yandle TG, Chong JP, Ng YX, Frampton CM, Ng TP, Lam CS, Richards AM (2016) High-sensitivity sandwich ELISA for plasma NT-proUcn2: Plasma concentrations and relationship to mortality in heart failure. Clin Chem 62(6):856–865. https://doi.org/10.1373/clinchem.2015.252932
Zhang Y, Guo YM, Xianyu YL, Chen WW, Zhao YY, Jiang XY (2013) Nanomaterials for ultrasensitive protein detection. Adv Mater 25(28):3802–3819. https://doi.org/10.1002/adma.201301334
Chinen AB, Guan CM, Ferrer JR, Barnaby SN, Merkel TJ, Mirkin CA (2015) Nanoparticle probes for the detection of cancer biomarkers, cells, and tissues by fluorescence. Chem Rev 115(19):10530–10574. https://doi.org/10.1021/acs.chemrev.5b00321
Zrazhevskiy P, Sena M, Gao XH (2010) Designing multifunctional quantum dots for bioimaging, detection, and drug delivery. Chem Soc Rev 39(11):4326–4354. https://doi.org/10.1039/b915139g
Fayyadh TK, Ma FY, Qin C, Zhang XW, Li W, Zhang XE, Zhang ZP, Cui ZQ (2017) Simultaneous detection of multiple viruses in their co-infected cells using multicolour imaging with self-assembled quantum dot probes. Microchim Acta 184(8):2815–2824. https://doi.org/10.1007/s00604-017-2300-6
Lu AH, Salabas EL, Schuth F (2007) Magnetic nanoparticles: Synthesis, protection, functionalization, and application. Angew Chem Int Ed 46(8):1222–1244. https://doi.org/10.1002/anie.200602866
Wu L, Mendoza-Garcia A, Li Q, Sun S (2016) Organic phase syntheses of magnetic nanoparticles and their applications. Chem Rev 116(18):10473–10512. https://doi.org/10.1021/acs.chemrev.5b00687
Colombo M, Carregal-Romero S, Casula MF, Gutierrez L, Morales MP, Bohm IB, Heverhagen JT, Prosperi D, Parak WJ (2012) Biological applications of magnetic nanoparticles. Chem Soc Rev 41(11):4306–4334. https://doi.org/10.1039/c2cs15337h
Xiao DL, Lu T, Zeng R, Bi YP (2016) Preparation and highlighted applications of magnetic microparticles and nanoparticles: A review on recent advances. Microchim Acta 183(10):2655–2675. https://doi.org/10.1007/s00604-016-1928-y
Wen CY, Xie HY, Zhang ZL, Wu LL, Hu J, Tang M, Wu M, Pang DW (2016) Fluorescent/Magnetic micro/nano-spheres based on quantum dots and/or magnetic nanoparticles: Preparation, properties, and their applications in cancer studies. Nano 8:12406–12429. https://doi.org/10.1039/C5NR08534A
Hu J, Zhang ZL, Wen CY, Tang M, Wu LL, Liu C, Zhu L, Pang DW (2016) Sensitive and quantitative detection of C-reaction protein based on immunofluorescent nanospheres coupled with lateral flow test strip. Anal Chem 88(12):6577–6584. https://doi.org/10.1021/acs.analchem.6b01427
Wen CY, Jiang YZ, Li XY, Tang M, Wu LL, Hu J, Pang DW, Zeng JB (2017) Efficient enrichment and analyses of bacteria at ultralow concentration with quick-response magnetic nanospheres. ACS Appl Mater Interfaces 9(11):9416–9425. https://doi.org/10.1021/acsami.6b16831
Meng HM, Liu H, Kuai H, Peng R, Mo L, Zhang XB (2016) Aptamer-integrated DNA nanostructures for biosensing, bioimaging and cancer therapy. Chem Soc Rev 45(9):2583–2602. https://doi.org/10.1039/c5cs00645g
Liu JW, Cao ZH, Lu Y (2009) Functional nucleic acid sensors. Chem Rev 109(5):1948–1998. https://doi.org/10.1021/cr030183i
Tasset DM, Kubik MF, Steiner W (1997) Oligonucleotide inhibitors of human thrombin that bind distinct epitopes. J Mol Biol 272(5):688–698. https://doi.org/10.1006/jmbi.1997.1275
Wen CY, Tang M, Hu J, Wu LL, Pang DW, Zeng JB, Li XY (2016) Determination of the absolute number concentration of nanoparticles and the active affinity sites on their surfaces. Anal Chem 88(20):10134–10142. https://doi.org/10.1021/acs.analchem.6b02613
Lao Y-H, Peck K, Chen L-C (2009) Enhancement of aptamer microarray sensitivity through spacer optimization and avidity effect. Anal Chem 81(5):1747–1754. https://doi.org/10.1021/ac801285a
Davie EW, Kulman JD (2006) An Overview of the Structure and Function of Thrombin. Semin Thromb Hemost 32:3–15. https://doi.org/10.1055/s-2006-939550
Cao Y, Wang Z, Cao J, Mao X, Chen G, Zhao J (2017) A general protein aptasensing strategy based on untemplated nucleic acid elongation and the use of fluorescent copper nanoparticles: Application to the detection of thrombin and the vascular endothelial growth factor. Microchim Acta 184(10):3697–3704. https://doi.org/10.1007/s00604-017-2393-y
He J, Li G, Hu Y (2017) Aptamer-involved fluorescence amplification strategy facilitated by directional enzymatic hydrolysis for bioassays based on a metal-organic framework platform: Highly selective and sensitive determination of thrombin and oxytetracycline. Microchim Acta 184(7):2365–2373. https://doi.org/10.1007/s00604-017-2263-7
Sui N, Wang L, Xie F, Liu F, Xiao H, Liu M, Yu WW (2016) Ultrasensitive aptamer-based thrombin assay based on metal enhanced fluorescence resonance energy transfer. Microchim Acta 183(5):1563–1570. https://doi.org/10.1007/s00604-016-1774-y
Wang G-L, Hu X-L, Wu X-M, Dong Y-M, Li Z-J (2015) Fluorescent aptamer-based assay for thrombin with large signal amplification using peroxidase mimetics. Microchim Acta 183(2):765–771. https://doi.org/10.1007/s00604-015-1703-5
Ma M, Zheng X (2015) Preparation of brightly fluorescent silica nanoparticles modified with lucigenin and chitosan, and their application to an aptamer-based sandwich assay for thrombin. Microchim Acta 182(13–14):2193–2199. https://doi.org/10.1007/s00604-015-1554-0
Lin Z, Pan D, Hu T, Liu Z, Su X (2015) A near-infrared fluorescent bioassay for thrombin using aptamer-modified CuInS2 quantum dots. Microchim Acta 182(11–12):1933–1939. https://doi.org/10.1007/s00604-015-1526-4
Zhu Y, Hu XC, Shi S, Gao RR, Huang HL, Zhu YY, Lv XY, Yao TM (2016) Ultrasensitive and universal fluorescent aptasensor for the detection of biomolecules (ATP, adenosine and thrombin) based on DNA/Ag nanoclusters fluorescence light-up system. Biosens Bioelectron 79:205–212. https://doi.org/10.1016/j.bios.2015.12.015
Kuang L, Cao SP, Zhang L, Li QH, Liu ZC, Liang RP, Qiu JD (2016) A novel nanosensor composed of aptamer bio-dots and gold nanoparticles for determination of thrombin with multiple signals. Biosens Bioelectron 85:798–806. https://doi.org/10.1016/j.bios.2016.05.096
Yu J, Yang L, Liang X, Dong T, Liu H (2015) Bare magnetic nanoparticles as fluorescence quenchers for detection of thrombin. Analyst 140(12):4114–4120. https://doi.org/10.1039/c5an00519a
Wang C, Zhai W, Wang Y, Yu P, Mao L (2015) MnO2 nanosheets based fluorescent sensing platform with organic dyes as a probe with excellent analytical properties. Analyst 140(12):4021–4029. https://doi.org/10.1039/c5an00581g
Xu Z, Huang X, Dong C, Ren J (2014) Fluorescence correlation spectroscopy of gold nanoparticles, and its application to an aptamer-based homogeneous thrombin assay. Microchim Acta 181(7):723–730. https://doi.org/10.1007/s00604-013-1132-2
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 21505157), the Applied Basic Research Projects of Qingdao (No. 17-1-1-79-jch, 15-9-1-94-JCH), the Fundamental Research Funds for the Central Universities (No. 17CX02055), the Natural Scientific Foundation of Shandong (No. ZR2016BQ23), and the Open Project Foundation in Hubei Key Laboratory of Medical Information Analysis & Tumor Diagnosis and Treatment (No. PJS140011611).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare that they have no competing interests.
Electronic supplementary material
ESM 1
(DOCX 5.06 MB)
Rights and permissions
About this article
Cite this article
Wen, CY., Bi, JH., Wu, LL. et al. Aptamer-functionalized magnetic and fluorescent nanospheres for one-step sensitive detection of thrombin. Microchim Acta 185, 77 (2018). https://doi.org/10.1007/s00604-017-2621-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-017-2621-5